Crystals of the nucleocapsid protein of human respiratory syncytial virus have been obtained in two forms, the better of which diffracted to 3.6 Å resolution and contained ten subunits in the asymmetric unit.
Keywords: HRSVN, human respiratory syncytial virus, nucleocapsid proteins
Abstract
Human respiratory syncytial virus (HRSV) has a nonsegmented negative-stranded RNA genome which is encapsidated by the HRSV nucleocapsid protein (HRSVN) that is essential for viral replication. HRSV is a common cause of respiratory infection in infants, yet no effective antiviral drugs to combat it are available. Recent data from an experimental anti-HRSV compound, RSV-604, indicate that HRSVN could be the target site for drug action. Here, the expression, purification and preliminary data collection of decameric HRSVN as well as monomeric N-terminally truncated HRSVN mutants are reported. Two different crystal forms of full-length selenomethionine-labelled HRSVN were obtained that diffracted to 3.6 and ∼5 Å resolution and belonged to space group P212121, with unit-cell parameters a = 133.6, b = 149.9, c = 255.1 Å, and space group P21, with unit-cell parameters a = 175.1, b = 162.6, c = 242.8 Å, β = 90.1°, respectively. For unlabelled HRSVN, only crystals belonging to space group P21 were obtained that diffracted to 3.6 Å. A self-rotation function using data from the orthorhombic crystal form confirmed the presence of tenfold noncrystallographic symmetry, which is in agreement with a reported electron-microscopic reconstruction of HRSVN. Monomeric HRSVN generated by N-terminal truncation was designed to assist in structure determination by reducing the size of the asymmetric unit. Whilst such HRSVN mutants were monomeric in solution and crystallized in a different space group, the size of the asymmetric unit was not reduced.
1. Introduction
Human respiratory syncytial virus (HRSV) is the major cause of lower tract infections in the paediatric population, with the most common infections being bronchiolitis and pneumonia. Most children are infected with the virus by 2 y of age. Moreover, HRSV is one of the main public health problems facing infants in the United States (Leader & Kohlhase, 2002 ▶). The World Health Organization has recognized HRSV to be responsible for 160 000 deaths per year worldwide. Unfortunately, to date there is little effective treatment; neither vaccines nor useful antiviral drugs are available. The only significant treatment available is prophylaxis with a humanized monoclonal antibody (Simoes & Groothuis, 2002 ▶). HRSV is a negative-sense single-stranded RNA virus of the family Paramyxoviridae. The 15.2 kbp HRSV genome encodes 11 proteins and is encapsidated by the HRSV nucleocapsid protein (HRSVN), which protects the viral genetic information from ribonucleases. The HRSVN–RNA complex is the functional template for the replication and transcription of the viral genome. Recent studies have focused attention on antiviral compounds such as RSV604 (Chapman et al., 2007 ▶). Escalating doses of RSV604 in tissue culture give rise to escape mutants where changes are located in the HRSVN, suggesting that the latter may be a potential target for anti-HRSV drugs.
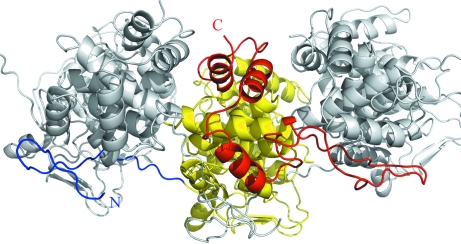
The structures of nucleocapsid proteins from vesicular stomatitis virus (VSV) and rabies virus (RB) have been determined in complex with RNA by X-ray crystallography (Albertini et al., 2006 ▶; Green et al., 2006 ▶). They form decameric and undecameric rings, respectively. Although the percentage amino-acid sequence identity is not significant, the two proteins are structurally very similar. Each protomer has two domains and binds nine nucleotides of single-stranded RNA. In the case of HRSV, the nucleocapsid also forms ring structures but only seems to bind six nucleotides per protomer (Tran et al., 2007 ▶). A 24 Å resolution structure of HRSVN obtained by electron microscopy revealed that the protein is morphologically similar to the rhabdoviruses VSV and RB (Maclellan et al., 2007 ▶). Nevertheless, a higher resolution structure of HRSVN is needed in order to compare the different nucleocapsid proteins at the atomic level and ultimately to help in the design of antiviral molecules. In this study, we present the cloning, purification and crystallization of HRSVN. Also, in order to attempt to simplify the crystallographic structure determination, we report the effect of truncating the N-terminus of HRSVN in reducing the oligomeric state to a monomer. These truncations were designed on the basis of the VSV and RB nucleocapsid crystal structures, in which the N-terminus extends from one subunit to the adjacent protomer (Fig. 1 ▶).
Figure 1.
Intersubunit contacts for VSV nucleocapsid protein protomers (Green et al., 2006 ▶; PDB code 2gic). The N-terminus is coloured blue and the C-terminal region red.
2. Materials and methods
2.1. Protein purification
2.1.1. Purification of HRSVN
The HRSVN gene was cloned in the pET3d expression vector (Novagen) via the restriction sites NcoI and BamHI to create the plasmid pMUT113. The latter plasmid, which codes for an N-terminally His6-tagged HRSVN, was transformed into Rosetta(DE3)pLysS for protein expression. A 60 ml Luria Broth starter culture supplemented with 50 µg ml−1 carbenicillin and 34 µg ml−1 chloramphenicol was grown overnight with shaking at 310 K. The culture was then diluted 1/100 into 6 l medium and grown at 310 K until the optical density reached 0.6 at 600 nm. At this point, IPTG was added to a final concentration of 1 mM and induction was carried out at 303 K overnight. The cells were harvested by centrifugation and the pellets were resuspended in buffer A [50 mM sodium phosphate pH 7.4, 500 mM NaCl, 0.2%(v/v) Tween 20]. The cells were disrupted by sonication and the supernatant was then clarified by centrifugation and applied onto a 5 ml nickel-chelating column (HiTrap Chelating Column; GE Healthcare) pre-equilibrated with buffer A. HRSVN was eluted with buffer A containing 500 mM imidazole using a linear gradient. The fractions containing HRSVN were applied onto a Superdex S200 gel-filtration column (GE Healthcare) pre-equilibrated with 50 mM Tris pH 7.4 and 200 mM NaCl. Purified HRSV was concentrated to 18 mg ml−1. Selenomethionine-labelled HRSVN was expressed using the methionine-auxotroph strain B834 and the protein was purified using the same protocols as used for the native protein. The presence or absence of dithiothreitol (DTT) did not seem to affect protein crystallization and thus it was not used during protein purification. The extent of selenomethionine (SeMet) incorporation into HRSVN was assessed by mass spectrometry.
2.1.2. Purification of N-terminal truncation mutants
The cDNA coding for the N-terminal truncation mutants plus an N-terminal His6 tag was obtained by standard PCR techniques using the pMUT113 plasmid (described above) as a template. The PCR products were cloned into the pET3d expression vector using NcoI and BamHI restriction sites. Protein expression was carried out in Rosetta(DE3)pLysS cells grown in Luria Broth supplemented with 50 µg ml−1 carbenicillin and 34 µg ml−1 chloramphenicol. The transformed cells were grown with shaking at 310 K to an optical density of 0.6 at 600 nm before induction with 0.25 mM IPTG and growth overnight at 301 K. The cells were harvested by centrifugation and resuspended in buffer B (50 mM Tris pH 8.0, 500 mM NaCl). The cells were lysed by sonication and the protein was purified from the soluble fraction after centrifugation by affinity chromatography using a Ni2+-chelating column. The protein was eluted with buffer B containing 500 mM imidazole using a linear gradient. Fractions shown by SDS–PAGE to contain the protein of interest were pooled, exchanged into buffer C (10 mM Tris pH 8.0, 10 mM NaCl) and applied onto a heparin-affinity column. The protein was eluted with buffer C containing 1 M NaCl using a linear gradient. Finally, fractions containing the RSVN truncation mutant were pooled and applied onto a Superdex S75 gel-filtration column pre-equilibrated with 50 mM Tris pH 7.4, 250 mM NaCl. Proteins were concentrated to 10 mg ml−1 and stored at 193 K. Preparation of SeMet-labelled protein was carried out using B834 cells using the same protocol with two exceptions: 1 mM DTT was added to all buffer solutions and purified protein was exchanged into 50 mM Tris pH 7.4, 250 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine for crystallization experiments.
2.2. Crystallization and data collection
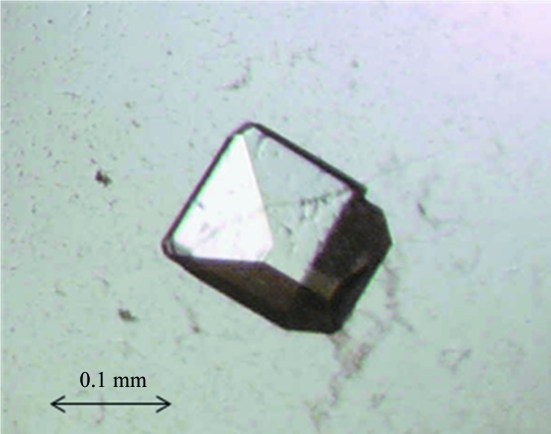
Crystallization trials were initially performed by the sitting-drop vapour-diffusion method at 292 K using several Hampton Research, Wizard and Emerald Biostructures kits. A Cartesian Technologies pipetting robot was used to set up 100 + 100 nl drops in Greiner 96-well plates which were placed in a TAP storage vault (Walter et al., 2005 ▶). HRSVN protein crystallized in Hampton Research Natrix condition No. 43 [10%(v/v) MPD, 0.05 M ammonium acetate, 0.05 M Tris pH 7.5, 0.01 M MgCl2]. Additive screens were used to identify benzamidine [1.3%(v/v) final concentration], which acted as an aid to crystal formation by improving the crystal diffraction quality. SeMet HRSVN crystals grew in Hampton Research Natrix condition No. 36 [10%(w/v) PEG 400, 0.1 M KCl, 0.05 M HEPES pH 7.0, 0.01 M CaCl2; Fig. 2 ▶). Crystals were prepared for cryoprotection by transferring them to reservoir solution containing 20%(v/v) glycerol.
Figure 2.
HRSVN crystal grown in Hampton Research Natrix condition No. 43 with benzamidine [1.3%(v/v) final concentration].
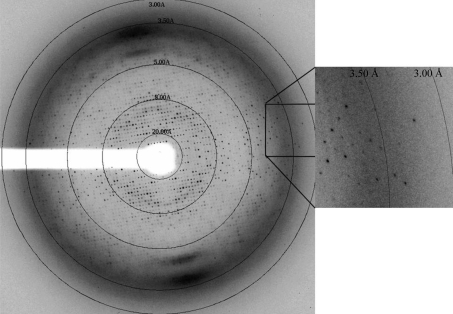
Most of the HRSVN crystals either did not diffract or only diffracted to low resolution. However, following extensive screening efforts crystals of HRSVN that diffracted to 3.6 Å resolution could occasionally be identified (Fig. 3 ▶). X-ray diffraction data for HRSVN were collected at 100 K on beamline ID29, whereas SeMet HRSVN diffraction data were collected on beamline BM14 at ESRF, Grenoble, France (Table 1 ▶). Images were indexed and integrated with DENZO and reflections were merged using SCALEPACK (Otwinowski & Minor, 1996 ▶).
Figure 3.
Snapshots of a diffraction pattern of a SeMet HRSVN crystal (P212121).
Table 1. Data-collection statistics for native and SeMet-labelled HRSVN crystals.
Values in parentheses are for the highest resolution shell.
| SeMet HRSVN | SeMet HRSVN | ||||
|---|---|---|---|---|---|
| Peak | Remote | Peak | Remote | Native HRSVN | |
| Beamline | ESRF BM14 | ESRF BM14 | ESRF ID29 | ||
| Wavelength (Å) | 0.9785 | 0.9050 | 0.9785 | 0.8855 | 0.9797 |
| Resolution range (Å) | 30.0–3.6 (3.73–3.60) | 50.0–4.4 (4.51–4.41) | 50.0–4.9 (5.07–4.90) | 55.0–3.6 (3.73–3.60) | |
| Space group | P212121 | P21 | P21 | ||
| Unit-cell parameters (Å, °) | a = 133.6, b = 149.9, c = 255.1 | a = 175.1, b = 162.6, c = 242.8, β = 90.1 | a = 175.2, b = 163.5, c = 241.5, β = 90.1 | ||
| Content of the ASU | 10 monomers | 20 monomers | 20 monomers | ||
| No. of observed reflections | 592190 | 179416 | 822080 | 240013 | 349067 |
| No. of unique reflections | 53027 | 48358 | 85301 | 62670 | 145892 |
| Redundancy | 11.2 (10.9) | 3.7 (3.5) | 9.6 (7.3) | 3.8 (3.7) | 2.9 (2.8) |
| Rmerge† (%) | 14.5 (85.6) | 11.7 (82.9) | 14.0 (44.2) | 14.7 (67.3) | 13.1 (96.6) |
| Completeness (%) | 81.7 (66.2) | 80.8 (67.7) | 99.1 (95.1) | 99.3 (99.8) | 91.7 (91.9) |
| I/σ(I) | 14.1 (2.5) | 8.0 (1.4) | 19.6 (3.4) | 9.7 (2.2) | 7.6 (1.8) |
R
merge = 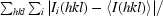
 , where Ii(hkl) is the ith observed intensity of reflection hkl and 〈I(hkl)〉 is the mean intensity for all observations i of reflection hkl.
, where Ii(hkl) is the ith observed intensity of reflection hkl and 〈I(hkl)〉 is the mean intensity for all observations i of reflection hkl.
3. Results and discussion
Typically, 10 mg pure HRSVN was obtained from 1 l of bacterial culture. The results from gel filtration and dynamic light scattering were in agreement with the ability of HRSVN to form decamers and/or undecamers. HRSVN stays tightly associated with Escherichia coli RNA as previously reported (Murphy et al., 2003 ▶). Isolation of RNA from the complex with HRSVN, followed by sequencing of DNA after reverse transcription, showed many species including fragments of various E. coli genes as well as HRSVN and components of the expression plasmid (unpublished data). Such heterogeneity might contribute to crystal disorder and thus attempts to remove RNA were made. Although different methods were used to try to remove RNA bound to the nucleocapsid protein, including RNAse digestion and use of high salt, none succeeded. Crystal growth was not very reproducible and could take anything between a week and a few months; moreover, the crystal morphology was very variable.
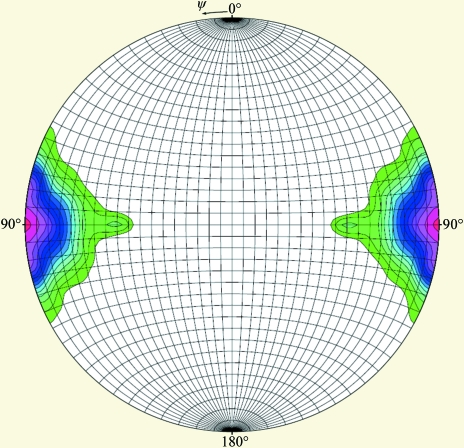
Crystals belonging to space group P21 could be obtained with either SeMet-labelled or unlabelled HRSVN, with approximate unit-cell parameters a = 175, b = 163, c = 242 Å, β = 90.1°. Crystals belonging to space group P212121 were only observed with SeMet-labelled protein and had approximate unit-cell parameters a = 133, b = 150, c = 255 Å (Table 1 ▶). The Matthews coefficients predicted ten monomers in the asymmetric unit of the P212121 form and 20 in the P21 crystals. Moreover, in the latter crystal form a pseudo-translation could be detected which would make this P21 form more difficult to solve. Further studies were mainly focused on the P212121 data sets. Tenfold symmetry rather than 11-fold symmetry was confirmed by the self-rotation function from CNS (Brünger et al., 1998 ▶; Fig. 4 ▶), indicating that the HRSVN rings were composed of decamers.
Figure 4.
Self-rotation functions, κ = 36°, with an integration radius of 60 Å and using data in the resolution range 30–3.6 Å (P212121 data set). The tenfold axis [I/σ(I) = 0.45] is aligned with the y axis. The presence of a second peak results from the crystallographic twofold symmetry along the x and z axes.
Despite the low amino-acid sequence identity of HRSVN to the VSV nucleocapsid protein, the two nucleocapsid structures are thought to be structurally similar (Maclellan et al., 2007 ▶). Decameric and monomeric VSV nucleocapsid proteins were used as models for molecular replacement for the P212121 data sets (Table 1 ▶). Unfortunately, despite the use of numerous programs such as CNS and the molecular-replacement software available in CCP4 (Collaborative Computational Project, Number 4, 1994 ▶), no convincing solution could be found even though the model tenfold axis seemed to be properly aligned with the tenfold axis determined by the self-rotation function. Therefore, work was orientated towards selenium phasing methods.
Each RSVN monomer contains 14 methionines; thus, production of SeMet-labelled protein was carried out for attempts at MAD phasing. Although SeMet was fully incorporated in the HRSVN and fluorescence scans indicated its presence, only a weak anomalous signal to 6 Å could be extracted from the data sets. Nevertheless, solving the structure by MAD would have been particularly challenging owing to the limited resolution and the large number of Se atoms in the asymmetric unit (140 for the orthorhombic crystal form).
We reasoned that solution of the structure of the decameric HRSVN protein would be facilitated by first determining the structure of a monomeric unit. Inspection of the structures of the related VSV and RB nucleocapsid proteins suggested a strategy for the generation of a monomeric species. In the VSV and RB structures (PDB codes 2gic and 2gtt, respectively), the extended N-terminus and a region towards the C-terminus of one subunit interact with adjacent subunits (Albertini et al., 2006 ▶; Green et al., 2006 ▶; Fig. 1 ▶). Removing these residues would be expected to weaken the interaction between subunits, perhaps favouring the formation of a monomeric species. It has been shown for bovine RSVN that deletion of two or more amino acids from the N-terminus inhibited encapsidation in a minigenome system, whereas deletion of up to 27 amino acids from the C-terminus resulted in only a modest reduction in encapsidation (Khattar et al., 2000 ▶), suggesting that N-terminal truncation was more likely to give the desired result. A number of N-terminal truncation mutants were made, designated ΔNX, where X is the number of residues deleted. ΔN3, ΔN6, ΔN9, ΔN12, ΔN15 and ΔN18 were cloned. Small-scale expression was carried out, indicating that a proportion of the protein was soluble in each case. Gel filtration of Ni2+-affinity purified protein showed that ΔN3 and ΔN6 formed multimeric species, whilst for the other proteins a peak was seen with a retention time consistent with the presence of monomeric protein. The amount of soluble protein present decreased markedly as more residues were deleted, with particularly low expression of ΔN15 and ΔN18. These results suggested that ΔN9 and ΔN12 were the most suitable for crystallization trials. Larger scale expression of both unlabelled and SeMet-labelled ΔN9 and ΔN12 was carried out. Gel filtration and dynamic light scattering were consistent with the presence of monomeric protein which could be concentrated to ∼10 mg ml−1. Crystallization trials (Walter et al., 2005 ▶) were carried out and small crystals of selenomethione-labelled ΔN9 and ΔN12 formed in the presence of 2.3–2.4 M ammonium phosphate pH 8.4–8.6 after 3–14 d. Further trials with either 2 mM UMP or U6 RNA as additives yielded crystals under the same conditions. As for full-length HRSVN, growth was not very reproducible, with the majority of crystals yielding either no diffraction or only diffracting to very low resolution. A 20 × 50 × 100 µm crystal of ΔN9 HRSVN crystallized in the presence of U6 RNA showed diffraction to 14 Å resolution. The data could be indexed in space group C2, with unit-cell parameters a = 424.5, b = 160.8, c = 157.4 Å, β = 111°. Matthews cell-content analysis was consistent with the presence of 20 monomers in the asymmetric unit with a solvent content of 57%. These results suggested that the protein may be present as a multimeric species in the crystal.
Current work is focused on screening heavy atoms for phasing the HRSVN crystals of space group P212121. This method has proven to be successful for the VSV and RB nucleocapsid proteins.
References
- Albertini, A. A., Wernimont, A. K., Muziol, T., Ravelli, R. B., Clapier, C. R., Schoehn, G., Weissenhorn, W. & Ruigrok, R. W. (2006). Science, 313, 360–363. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Chapman, J. et al. (2007). Antimicrob. Agents Chemother.51, 3346–3353. [DOI] [PMC free article] [PubMed]
- Green, T. J., Zhang, X., Wertz, G. W. & Luo, M. (2006). Science, 313, 357–360. [DOI] [PubMed]
- Khattar, S. K., Yunus, A. S., Collins, P. L. & Samal, S. K. (2000). Virology, 270, 215–228. [DOI] [PubMed]
- Leader, S. & Kohlhase, K. (2002). Pediatr. Infect. Dis. J.21, 629–632. [DOI] [PubMed]
- Maclellan, K., Loney, C., Yeo, R. P. & Bhella, D. (2007). J. Virol.81, 9519–9524. [DOI] [PMC free article] [PubMed]
- Murphy, L. B., Loney, C., Murray, J., Bhella, D., Ashton, P. & Yeo, R. P. (2003). Virology, 307, 143–153. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1996). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Simoes, E. A. & Groothuis, J. R. (2002). Respir. Med.96, Suppl. B, S15–S24. [DOI] [PubMed]
- Tran, T. L., Castagne, N., Bhella, D., Varela, P. F., Bernard, J., Chilmonczyk, S., Berkenkamp, S., Benhamo, V., Grznarova, K., Grosclaude, J., Nespoulos, C., Rey, F. A. & Eleouet, J. F. (2007). J. Gen. Virol.88, 196–206. [DOI] [PubMed]
- Walter, T. S. et al. (2005). Acta Cryst. D61, 651–657. [DOI] [PMC free article] [PubMed]