The structure of Rv0802c was determined in an unliganded form to 2.0 Å resolution utilizing single-wavelength anomalous dispersion from a samarium soak that resulted in a single bound Sm3+:citrate2 complex.
Keywords: Rv0802c, Mycobacterium tuberculosis, GCN5-related N-acetyltransferases
Abstract
Gene rv0802c from Mycobacterium tuberculosis encodes a 218-amino-acid protein and is annotated as a hypothetical protein with homology to GCN5-related N-acetyltransferases. The structure of Rv0802c was determined in an unliganded form to 2.0 Å resolution utilizing single-wavelength anomalous dispersion from a samarium soak that resulted in a single bound Sm3+:citrate2 complex. The structure confirms that Rv0802c exhibits the GCN5-related N-acetyltransferase fold and revealed a tetramer composed of a dimer of dimers with approximate 222 symmetry. In addition, a bound acetate ion indicated that Rv0802c may utilize a unique acyl donor for the family. The subsequent determination of the structure of Rv0802c in complex with succinyl-CoA to 2.3 Å resolution suggests that Rv0802c is the first known GCN5-related N-acetyltransferase family member to utilize succinyl-CoA as a substrate.
1. Introduction
Members of the GCN5-related N-acetyltransferase (GNAT) family typically catalyze the transfer of an acetyl group from acetyl-coenzyme A (AcCoA) to a wide range of amino-containing substrates such as Nα protein termini, the N∊ amine of lysine side chains and various small molecules (Dyda et al., 2000 ▶; Vetting, de Carvalho, Yu et al., 2005 ▶). GNATs are therefore involved in a number of important processes such as small-molecule biosynthesis, antibiotic resistance and gene regulation. The GNAT fold exhibits a remarkably flexible nature in its diversity of primary sequences: GNATs with different acceptor substrates typically display less than 30% sequence identity. This makes the annotation (what are the substrates?) and three-dimensional modeling (what residues contact the substrates?) of GNATs problematic. The human pathogen Mycobacterium tuberculosis has 21 proteins which have GNAT sequence signatures [see the acyl-CoA N-acyltransferases (Nat) superfamily at http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY]. Only six of these proteins have been partially characterized and/or annotated based on similar sequences: Rv3420c and Rv0995 (ribosomal N α-acetyltransferase proteins RimI and RimJ; Yoshikawa et al., 1987 ▶), Rv1347c (mycobactin biosynthesis; Card et al., 2005 ▶; Frankel & Blanchard, 2008 ▶), Rv0819 (mycothiol biosynthesis; Koledin et al., 2002 ▶; Vetting et al., 2003 ▶), Rv2747 (arginine biosynthesis; Errey & Blanchard, 2005 ▶) and Rv0262c (antibiotic modification/unknown cellular function; Vetting et al., 2002 ▶).
The protein Rv0802c is one of the M. tuberculosis proteins with the GNAT sequence signature but with no known structure or function. Orthologs of Rv0802c are only found in a small subset of actinomycetes. For example, Rv0802c orthologs are found in M. marinum but not in M. smegmatis or M. leprae (Fig. 1 ▶). Rv0802c has previously been cloned but not extensively characterized (Kovacs, Csanadi, Kiss et al., 2005 ▶). The structures of Rv0802c presented here suggest that Rv0802c may function as a succinyl-CoA transferase rather than an acetyl-CoA transferase.
Figure 1.
Multiple sequence alignment of Rv0802c orthologs. Putative orthologs of Rv0802c (gi|15607942) from M. marinum (gi|183984857), Nocardioides sp. JS614 (gi|119717482), Arthrobacter sp. FB24 (gi|116669736), Kineococcus radiotolerans (gi|152968286) and Streptomyces sp. TP-A0584 (gi|78042205) were aligned using the program ClustalW (Thompson et al., 1994 ▶). Residues which are identical or similar in 75% of the sequences are shaded black and grey, respectively. Secondary-structural elements are indicated above the alignment and colored according to the ribbon diagrams in Fig. 3 ▶. Residues that are important in coordinating the succinyl moiety of SucCoA are indicated by red circles below the alignment. Residues which border a putative binding pocket for the second substrate are indicated by blue circles below the alignment. Values given in parentheses are the percentage identity to the M. tuberculosis Rv0802c sequence. The Streptomyces sp. TP-A0584 sequence is also termed GodH and is an acetyltransferase involved in goadsporin biosynthesis.
2. Materials and methods
2.1. Cloning, expression and purification of Rv0802c
The M. tuberculosis gene rv0802c was amplified by PCR using the primers 5′-TTTTTTCATATGTCTCGTCACTGGCCGTTGT-3′ and 5′-TTTTTACTCGAGATAACGAGGCGGTTCCAA-3′ from M. tuberculosis genomic DNA, which incorporate NdeI and XhoI restriction-endonuclease sites (bold). The amplified DNA product was digested with NdeI and XhoI and the purified insert was ligated into purified plasmid pET-23a (Novagen) previously linearized with the same restriction enzymes, yielding an expression plasmid for M. tuberculosis Rv0802c with a C-terminal His6 tag. The plasmid pET-23a::rv0802c was then isolated and sequenced.
The plasmid coding for the Rv0802c gene product was transformed into Escherichia coli Rosetta2 (DE3) (Novagen) cells. Transformed cells were grown overnight in 50 ml LB broth containing 100 µg ml−1 ampicillin and 35 µg ml−1 chloramphenicol. 1 l cultures were then inoculated to an A 600 of ∼0.05. The cells were grown at 310 K to an A 600 of 0.5. Protein expression was subsequently induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and the cells were left to grow for a further 20 h at 293 K. Expression of the protein was confirmed by SDS–PAGE. All protein-purification steps were carried out at 277 K. The cell pellet (10 g) was resuspended in 50 ml 20 mM triethanolamine (TEA), 100 mM ammonium sulfate, 15 mM imidazole pH 7.8 containing one tablet of Complete protease-inhibitor cocktail (Roche). The cells were disrupted on ice by sonication. The suspension was then centrifuged (10 000g for 30 min) to remove cell debris. The supernatant was then filtered using a 0.2 µm syringe filter and applied onto a pre-equilibrated (buffer A; 20 mM TEA, 100 mM ammonium sulfate, 15 mM imidazole pH 7.8) Pharmacia 10 ml Ni–NTA column. The column was washed at 1 ml min−1 with 20 column volumes of the same buffer and then eluted with a linear gradient of imidazole (15–500 mM over 25 column volumes) in buffer A. Protein was detected with an on-line detector monitoring A 280 and column fractions were collected and analyzed by SDS–PAGE. Fractions containing the ∼25 kDa protein were pooled. The protein sample was extensively dialyzed against 20 mM TEA, 100 mM ammonium sulfate, 1 mM EDTA pH 7.8 and concentrated using a YM10 Amicon ultrafiltration membrane to a final concentration of 12 mg ml−1 and was either used immediately for crystallization experiments or stored as a 50%(v/v) glycerol stock at 253 K. Preparations of Rv0802c at elevated concentrations (>4 mg ml−1) tended to form microcrystals upon extended storage at 277 K. This was partially mitigated by increasing the pH and or the addition of substrates or products. Later preparations of Rv0802c therefore utilized 20 mM Bicine, 100 mM ammonium sulfate, 2 mM EDTA, 1 mM DTT pH 8.75 as the final dialysis buffer.
2.2. Oligomer analysis
Dynamic light-scattering experiments were performed on a DynaPro-MS/X instrument (Protein Solutions). The data were measured at 293 K with 4 and 0.4 mg ml−1 Rv0802c in buffer B (20 mM Bicine, 100 mM ammonum sulfate, 2 mM EDTA, 1 mM DTT pH 8.75).
Sedimentation-equilibrium experiments were performed using the interference optics of a Beckman XL-I analytical ultracentrifuge with six-channel centerpieces in a Ti-60 rotor at 298 K. Three concentrations each of sample (∼3.8, ∼1.5 and ∼0.5 mg ml−1) in buffer B and 2 mM succinyl-CoA (SucCoA) were equilibrated for 24 h at 9000 rev min−1. Five interference scans were globally analyzed using HeteroAnalysis v.1.1.33 (J. L. Cole & J. W. Lary, Analytical Ultracentrifugation Facility, Biotechnology Services Center, University of Connecticut, USA) to determine the weight-average molecular weight. The resolved molecular weight and the 95% joint confidence intervals are reported. The partial specific volume of 0.7317 ml g−1 (from the amino-acid composition) and the density of 1.00678 g ml−1 (from the buffer composition) were calculated using SEDNTERP v.1.06 (D. Hayes, T. Laue & J. Philo, University of New Hampshire, USA).
2.3. Crystallization and data collection
All crystallization trials were carried out by vapour diffusion under silicon oil (Fisher Scientific) in 96-well round-bottom assay plates (Costar 3795) at 291 K. Two crystal forms (unliganded and SucCoA) were obtained using various commercially available crystallization screens. Data were collected in-house at 110 K using a Rigaku/MSC R-AXIS IV++ detector mounted on an RU-H3R rotating-anode X-ray generator equipped with Osmic Blue confocal focusing mirrors and a 0.3 mm collimator running at 50 kV and 100 mA. Data were processed using DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶). Data-collection statistics are given in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution bin.
| Unliganded | SucCoA | |
|---|---|---|
| Space group | P212121 | I222 |
| Resolution (Å) | 86–2.0 (2.07–2.0) | 104–2.3 (2.38–2.3) |
| Completeness (%) | 95.4 (79.4) | 95.9 (92.8) |
| Redundancy | 5.5 (3.2) | 3.5 (3.0) |
| 〈I/σ(I)〉 | 22.3 (4.8) | 15.3 (4.8) |
| Rmerge | 0.048 (0.178) | 0.055 (0.204) |
| Wilson B factor (Å2) | 21.7 | 26.5 |
| Model and refinement data | ||
| Resolution (Å) | 86–2.0 (2.05–2.0) | 104–2.3 (2.36–2.3) |
| Unique reflections | 66073 (3925) | 51029 (3573) |
| Rcryst (%) | 16.4 (18.2) | 16.9 (19.8) |
| Rfree (%) (5% of data) | 20.5 (24.9) | 22.2 (28.8) |
| Contents of the model | ||
| Residues (1–218 + tag) | A2–A36, A56–A212, B2–B36, B56–B211, C2–C35, C57–C211, D2–D36, D56–D211 | A2–A211, B2–B211, C2–C211, D2–D211 |
| Waters | 581 | 413 |
| Other | 1 Sm3+, 2 citrate, 5 2-ethoxyethanol, 4 sulfate, 4 acetate | 4 SucCoA |
| Total atoms | 6823 | 7416 |
| Average B factor (Å2) | ||
| Protein | 18.5 | 19.3 |
| SucCoA | — | 27.7 |
| Waters | 31.7 | 23.8 |
| R.m.s.d. | ||
| Bond lengths (Å) | 0.014 | 0.015 |
| Angles (°) | 1.55 | 1.68 |
| MOLPROBITY statistics | ||
| Ramachandran most favored/outliers (%) | 98.8/0.0 | 97.7/0.0 |
| Rotamer outliers (%) | 0.62 | 1.44 |
| Clashscore† | 4.71 (98th percentile) | 4.76 (99th percentile) |
| Overall score† | 1.24 (99th percentile) | 1.43 (99th percentile) |
Scores are ranked according to structures of similar resolution as formulated in MOLPROBITY.
Unliganded Rv0802c crystals were obtained by mixing equal volumes of Rv0802c [7 mg ml−1 in 10 mM TEA, 100 mM ammonium sulfate, 3%(v/v) glycerol, 1 mM EDTA, 2 mM DTT pH 7.8] with precipitant (150 mM sodium citrate pH 5.25) under oil. The crystals grew as rectangular rods with maximum dimensions of 0.3 × 0.15 × 0.15 mm over 1–2 weeks. The crystals were incubated (5–30 min) in 150 mM sodium citrate, 25%(v/v) 2-ethoxyethanol pH 5.25 prior to vitrification in liquid nitrogen. The crystals belonged to space group P212121, with approximate unit-cell parameters a = 70.7, b = 111.5, c = 135.2 Å. There is a tetramer in the asymmetric unit, resulting in a solvent content of 56%.
Crystals of the Rv0802c–SucCoA complex were obtained by mixing equal volumes of Rv0802c (8 mg ml−1 in 20 mM TEA, 100 mM ammonium sulfate, 1 mM EDTA, 1 mM DTT, 2 mM CoA, 4 mM SucCoA pH 8.5) with precipitant (1 M LiCl, 100 mM HEPES pH 7.0). The crystals grew as thin rectangular rods with maximum dimensions of 0.4 × 0.08 × 0.08 mm over 1–2 weeks. The crystals were incubated (∼5 min) in 1 M LiCl, 100 mM HEPES, 20 mM SucCoA, 20%(v/v) glycerol pH 7.0 prior to vitrification in liquid nitrogen. The crystals belonged to space group I222, with approximate unit-cell parameters a = 113.2, b = 135.2, c = 164.1 Å. There is a tetramer in the asymmetric unit, resulting in a solvent content of 54%.
2.4. Structure determination and refinement
A single crystal of the P212121 crystal form was first soaked in 150 mM sodium citrate, 25%(v/v) 2-ethoxyethanol pH 5.25 for 15 min prior to gradual addition of and finally transfer (over 4 min) to a solution containing 150 mM sodium citrate, 25%(v/v) 2-ethoxyethanol, 75 mM samarium(III) acetate pH 5.25. The structure was solved by single-wavelength anomalous dispersion (SAD); the heavy-atom sites were located and the initial phases were calculated using the program SOLVE (Terwilliger, 2002 ▶). The initial model was built using solvent-flattened phases calculated by the program RESOLVE (Terwilliger, 2002 ▶) and the automated fitting program MAID (Levitt, 2001 ▶). Subsequent rounds of manual rebuilding and refinement against the data were performed using the programs Coot (Emsley & Cowtan, 2004 ▶) and REFMAC (Murshudov et al., 1997 ▶), respectively. The structure of the Rv0802c–SucCoA complex was solved by molecular replacement utilizing the program AMoRe (Navaza, 1994 ▶), with a single subunit of unliganded Rv0802c as a search model.
In all structure refinements the initial rounds utilized strict NCS restraints, which were relaxed and finally removed in the final rounds of refinement. Waters were added to the model at positions where densities were above 3.5σ and 1.0σ in the F o − F c and 2F o − F c maps, respectively, and which could make acceptable hydrogen-bonding interactions. Translation–libration–screw (TLS) refinement was used in the last steps of refinement (Winn et al., 2001 ▶). A total of four TLS groups were used per chain and the groups were determined by submission of the PDB file to the TLSMD server (Painter & Merritt, 2006a ▶,b ▶). Starting PDB files for ligands were downloaded from the HIC-UP server (Kleywegt & Jones, 1998 ▶) and submitted to PRODRG2 (Schüttelkopf & van Aalten, 2004 ▶) for calculation of stereochemical restraints for use in REFMAC (Murshudov et al., 1997 ▶). Models were submitted to the MOLPROBITY server (Davis et al., 2004 ▶) to assess structural quality. Accessible surface-area calculations were performed with a 1.4 Å radius probe and were calculated using AREAMOL (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
3.1. Structure determination: Sm3+:citrate2 complex
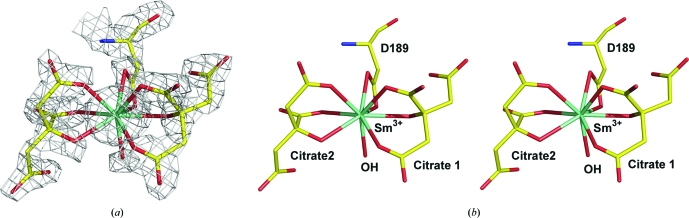
Crystals of unliganded Rv0802c were obtained in 150 mM sodium citrate pH 5.25 and typically diffracted to 3.5 Å resolution. Incubation of these crystals in cryoprotectant [150 mM sodium citrate, 25%(v/v) 2-ethoxyethanol pH 5.25] for >15 min decreased the c axis by 3–5% and increased the diffraction resolution to 2.8 Å. Typically, lanthanides (Sm3+, Gd3+, Yb3+) are not utilized as heavy-atom derivatives in the presence of citrate as the citrate ion effectively chelates the heavy atom and competes with protein acid side chains for derivatization. However, it was noted that this crystal form developed cracks when soaked in high concentrations of samarium(III) acetate (75 mM) which subsequently healed over time. The resultant samarium-soaked crystals exhibited a further shrinkage of the c axis by 1–2% and an increase in diffraction to >2.0 Å. Anomalous difference Patterson maps confirmed heavy-atom binding with a maximum peak height of 28.5σ indicative of a quality derivative. Because of the non-isomorphism of the crystals, initial phasing took place using single-wavelength anomalous dispersion (SAD) with solvent flattening. Data from a single crystal were collected on a home source and the structure was completed in a straightforward manner by automated and manual fitting. A single Sm3+ ion was found in the crystal structure which is coordinated by two citrates, one aspartate (Asp189) and a water molecule (see Fig. 2 ▶). The Sm3+ is nine-coordinate, with each citrate making three interactions and the aspartate making two interactions. The Sm3+:citrate2 complex bridges two noncrystallographic symmetry-related subunits. The region has a high density of arginine side chains (Arg164, Arg170, Arg184 and Arg192) which form numerous hydrogen bonds and electrostatic interactions with the citrates. There was no indication either in anomalous difference Fourier maps or the F o − F c difference maps that any free Sm3+ had bound to the protein. Since the Sm3+:citrate2 derivative diffracted to much higher resolution, this data set was used as the final ‘unliganded’ complex. Examination of previous data sets indicated that the binding of Sm3+:citrate2 did not affect the overall structure (data not shown).
Figure 2.
Derivatization of Rv0802c. (a) Final 2F o − F c electron density for the Sm3+:citrate2 complex contoured at 2.5σ. (b) Stereoview of the coordination geometry of the Sm3+ ion.
3.2. Monomer fold
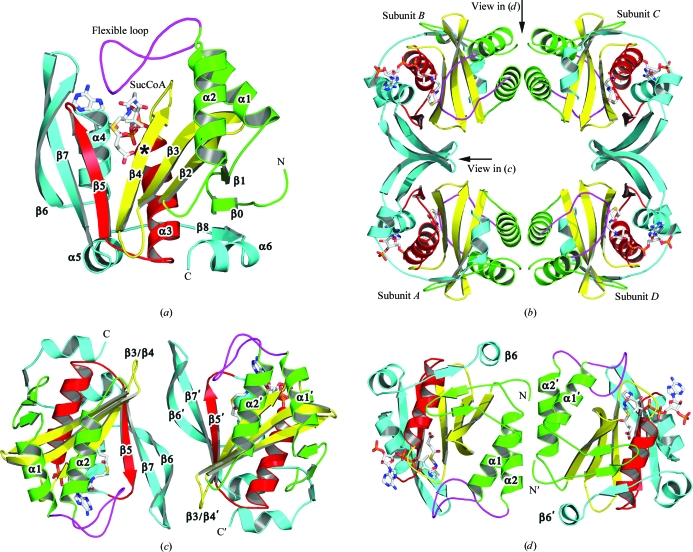
The unliganded structure confirms that Rv0802c is a member of the GCN5-related N-acetyltransferase (GNAT) family as suggested by its primary sequence. The monomer is a mixed αβ-fold with a central β-sheet partially covered by α-helices α1, α2 and α6 on one face and α3, α4 and α5 on the other (Fig. 3 ▶ a). The strands of the β-sheet are mostly antiparallel (β4 and β5 are parallel) with order β8, β0–β5, β7, β6. The β-sheet has a V-shape owing to the splaying of β4 from β5, a structural characteristic of GNAT-family members that is utilized to coordinate the pantothenate moiety of CoA (Dyda et al., 2000 ▶; Vetting, de Carvalho, Yu et al., 2005 ▶). A Rv0802c monomer was used to perform a structural alignment search utilizing the Secondary Structure Matching (SSM) server (Krissinel & Henrick, 2004 ▶). The results were culled and sorted based on the Q score, which represents the quality function of Cα alignment and takes both the alignment length and the r.m.s.d. into account. A total of 14 PDB entries were returned (>60% secondary-structure matched), with most being annotated as putative GCN5-related N-acetyltransferases. The highest Q score was to the ribosomal L7/L12 N α-acetyltransferase RimL (Vetting, de Carvalho, Roderick et al., 2005 ▶) from Salmonella typhimurium (PDB code 1s7l; Q = 0.51, r.m.s.d. = 1.81 Å, 161 Cα atoms aligned). The largest macroscopic differences between the two structures are the conformation of the α1–α2 loop and a C-terminal extension of Rv0802c. The α1–α2 loop is involved in substrate recognition (see below) and exhibits high structural variability in GNAT-family members. The C-terminal extension of Rv0802c (α5, β8 and α6) traverses 20 Å across the surface of the structure to add another strand to the N-terminal end of the β-sheet and does not appear to directly affect typical substrate-binding regions. The results of the SSM search (r.m.s.d. scores > 1.7 Å, sequence identity < 21%) suggest that the Rv0802c structure presented here is the first of this subfamily of GNAT proteins.
Figure 3.
Structure of Rv0802c. Ribbon diagrams for (a) an Rv0802c monomer, (b) a tetramer, (c) the A–B (C–D) dimer interface and (d) the A–D (B–C) dimer interface. The coordinates of the Rv0802c complex with SucCoA (sticks, colored by atom type) were used to make the diagram to illustrate the relative orientation of the four active sites and the flexible loop (α1–α2) which is not visible in the unliganded (acetate) structure. The presumed location of the active site is marked by an asterisk in (a).
3.3. Oligomeric state
Analysis of Rv0802c by dynamic light scattering yielded a monodisperse peak with a hydrodynamic radius of 46 Å, which is consistent with an oligomer of 120 kDa and with Rv0802c being a tetramer or pentamer (the molecular weight of Rv0802c is 24 983 Da). Sedimentation-equilibrium analysis of substrate-bound Rv0802c yielded a molecular weight of 107 849 Da (105 035–110 655 Da 95% joint confidence intervals), which is consistent with the protein being tetrameric in solution. A tetramer consisting of a dimer of dimers with approximate 222 symmetry was found in both crystal forms. The tetramer has a donut-like appearance, with an outer radius of 40–45 Å, an inner radius of 12 Å and a height of 35 Å (Fig. 3 ▶ b). The inner radius is punctuated by increases in its diameter to >20 Å near the location of the four active sites. Each subunit makes contacts with two other subunits but not the third. The A–B (or C–D) dimer interface is similar to that observed in other GNAT dimers such as RimL (Vetting, de Carvalho, Roderick et al., 2005 ▶), AAC(6′)-Ii (Burk et al., 2003 ▶) and tabtoxin resistance protein (He et al., 2003 ▶) (Fig. 3 ▶ c). At this interface β6 interacts with β6′ of the twofold-related monomer to form a single continuous β-sheet (the strand order at the interface is β5, β7, β6|β6′, β7′, β5′). Additional contacts are made between the β3–β4 loop and the β6–β7 loop. A total of 1505 Å2 of solvent-accessible surface area is buried upon formation of this dimer interface. In contrast, the A–D (or B–C) dimer interfaces only involve structural elements from the N-terminus (Fig. 3 ▶ d). At this interface the subunits are joined such that α2 and α2′ are offset and their helical rungs (and therefore their projected side chains) interdigitate each other. The other component of this interface is the interaction of α1 with both α1′ and the twofold-related N-terminus (Ser2′–Phe8′). A total of 2076 Å2 of solvent-accessible surface area is buried upon formation of this dimer interface, including a number of hydrophobic residues (Trp5, Trp67, Phe60, Leu63, Leu70, Pro59 and Ala71), suggesting a stable interface. Many of the residues which are buried in this interface are not conserved, suggesting that orthologs may have different oligomeric states. GNAT proteins are typically monomers or dimers, with only the yeast histone acetyltransferase yHPA2 forming a tetramer in solution (Angus-Hill et al., 1999 ▶). The oligomeric state of Rv0802c could play an important role in substrate specificity as the active-site grooves of the individual subunits are adjoined at the A–B (or C–D) interface and project towards the internal radius of the tetramer.
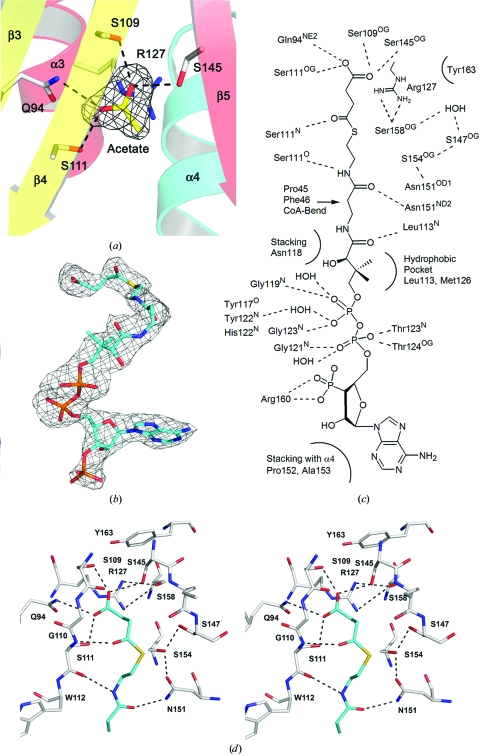
3.4. Acetate/SucCoA-binding site
A planer trilobed piece of density was observed in the unliganded Rv0802c structure (see Fig. 4 ▶ a). Based on hydrogen bonding and the nature of the surrounding side chains, the density was fitted as an acetate molecule, presumably bound and retained from the cell lysate during the one-column Ni–His6 purification. The carboxylate forms hydrogen bonds to the side chains of Gln94, Ser109, Ser111 and Ser145 and is electrostatically compensated by the guanidium group of Arg127. The interaction with Arg127 is not through hydrogen bonding (salt link), but through an atypical short-range electrostatic stacking interaction of a carboxylate and a guanidium group, with the guanidinium group held in place by hydrogen bonds to the side chain of Ser158. Interestingly, the acetate-binding site is adjacent to the splay between β4 and β5, suggesting it may be part of the substrate-binding site. Typically, the S-acetylpantetheine moiety of AcCoA forms a pseudo-antiparallel β-sheet interaction with β4, with the acetyl moiety lying in nearly the same plane as the β-sheet and in the crux of the splay between β4 and β5 (Vetting, de Carvalho, Yu et al., 2005 ▶). The acceptor amine invariantly approaches from the ‘front face’ of the β-sheet (the same side as α1–α2) for in-line nucleophilic attack on the Re face of the acetyl group of AcCoA. However, in this case the acetate-binding site is located on the ‘back face’ of the β-sheet (the same side as α3–α4). The structure of unliganded Rv0802c was superimposed on the structure of the GNAT protein TTHA1209 from Thermus thermophilus (T. Kaminishi, C. Takemoto, T. Uchikubo-Kamo, T. Terada and S. Yokoyama, unpublished work; PDB code 2cy2), the closest structural homolog returned by SSM that had been solved with an AcCoA cofactor. The carbonyl and methyl C atoms of the acetyl group of AcCoA are ∼2.2 and 1.2 Å from the methyl group of the acetate bound to Rv0802c. The location and close proximity of the acetate-binding site to the cofactor suggests that it is part of the CoA-binding site, with succinyl-CoA (SucCoA) being the most likely choice for a substrate.
Figure 4.
The SucCoA-binding site. (a) F o − F c OMIT map contoured at 4σ for the acetate ion in the unliganded structure. Protein side chains which interact with the acetate are shown as sticks. (b) F o − F c OMIT map contoured at 2.5σ for SucCoA. (c) ChemDraw illustration of the interactions of Rv0802c with SucCoA. (d) Stereoview of the SucCoA-binding site of Rv0802c in the region of the succinyl moiety. OMIT maps were calculated after ligands had been removed, the coordinates had been randomized to produce an r.m.s.d. of 0.25 Å and the randomized ligand-omitted structure had undergone positional refinement.
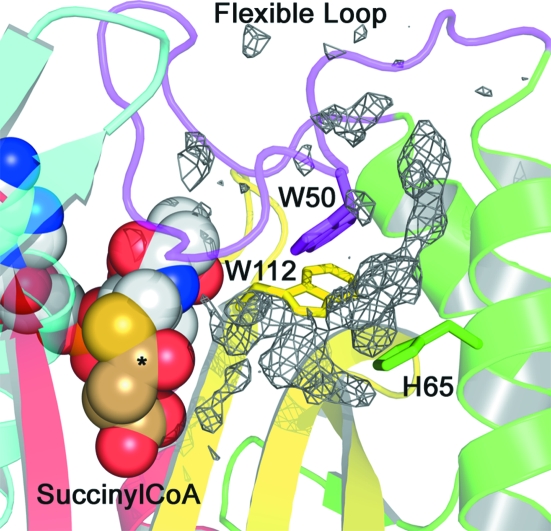
Based on these results, attempts were made to crystallize Rv0802c in complex with SucCoA. A different crystal form was obtained with the Rv0802c–SucCoA complex and the structure was determined to 2.3 Å resolution. There was excellent density for the cofactor in all four subunits of the tetramer, with the B factors for the entire cofactor and the succinyl moiety similar to those of the surrounding protein residues and suggesting high occupancy (Fig. 4 ▶ b). The carboxylate of SucCoA is bound in a pocket formed by α3, α4, β4 and β5 and superimposes absolutely upon and is coordinated identically to the acetate ion of the unliganded structure (Figs. 4 ▶ c and 4 ▶ d). The carbonyl C atom of the succinyl moiety is hydrogen bonded to a backbone amide of β4 (Ser111) in a similar manner as previously observed for numerous GNAT structures determined with AcCoA (Vetting, de Carvalho, Yu et al., 2005 ▶). Examination of the unliganded and SucCoA complexes suggests that the possible ‘acyl’ ligands for Rv0802c are limited by the distance between the acyl–β4 hydrogen bond and the tightly constrained carboxylate-binding site, with two C atoms being the minimal linker, i.e. SucCoA. The CoA portion of SucCoA is coordinated in a similar fashion to other GNAT proteins determined to date, with the major determinants of binding being the coordination of the pyrophosphate moiety by the main-chain backbone atoms of the ‘P-loop’ (β4–α3 loop) and the pseudo-antiparallel β-sheet interaction of the S-acetylpantetheine moiety with β4. The cofactor takes a C-shape, with the convex apolar surface of the C making contacts with the α1–α2 loop (residues 36–56, flexible loop) through van der Waals contacts with the side chains of Pro45 and Phe46. The loop is further stabilized by polar interactions with β6–β7 and by a perpendicular CH–π contact between Trp50 and Trp112 (on β4). The α1–α2 loop was disordered in the unliganded structure, suggesting that cofactor binding is important in stabilizing the conformation of this loop. The closing of the loop onto the cofactor results in the formation of a second pocket, which may be involved in the coordination of the amine-containing substrate. Residual electron density in F o − F c maps was found in this pocket; however, the density was of insufficient quality to determine its molecular source (Fig. 5 ▶). The difference density is bordered by Trp50, His65 and Trp112, where Trp50 and Trp112 are strictly conserved amongst Rv0802c orthologs and His65 is either His, Tyr, Phe or Trp. However, other residues which border the transition from this pocket to the SucCoA are generally not conserved.
Figure 5.
Secondary binding groove. Residual electron density (final F o − F c contoured at 2.5σ) in a secondary binding groove adjacent to the SucCoA-binding site. Conserved binding residues are shown as sticks. SucCoA is shown as CPK spheres, with the succinyl moiety shown in brown C atoms and the acyl carbon marked with an asterisk.
3.5. Possible roles
The genomic environment of Rv0802c yields little suggestion of its molecular function. Rv0802c borders several hypothetical genes (Rv0799, Rv0801, Rv0804 and Rv0805), a probable aminopeptidase (pepC-Rv0800) and several genes from the purine-biosynthetic pathway (purL-Rv0803, purF-Rv0808 and purM-Rv0809). One putative target is CFP29 (Culture Filtrate Protein 29) encoded by Rv0798c, which showed heterogeneity on two-dimensional electrophoresis that was suggestive of covalent modification; however, the authors did not determine the nature of the modification (Rosenkrands et al., 1998 ▶). The M. bovis ortholog of Rv0802c (Mb0825c) was found to copurify with the mRNA-processing enzyme RNase E, but has not been conclusively determined to be part of the mycobacterial ‘degradosome’ (Kovacs, Csanadi, Megyeri et al., 2005 ▶). The closest ortholog of Rv0802c with an annotated function is GodH from Streptomyces sp. TP-A0584 (40.9% sequence identity over 222 amino acids), which is involved in the N α-acetylation of goadsporin, a polypeptide antibiotic with thiazole and oxazole rings (Onaka et al., 2005 ▶). However, M. tuberculosis does not appear to have a goadsporin-like biosynthetic pathway. Interestingly, GodH contains three alterations to residues which coordinate the carboxylate of SucCoA in Rv0802c. Ser109 and Ser111 which coordinate the carboxylate are changed to an Asp and a Cys, respectively, while Ser158 which holds Arg127 (electrostatic stacking with carboxylate) is mutated to an alanine (Fig. 1 ▶). Therefore, in GodH a new interaction could form between Asp109 and a repositioned Arg127, filling the carboxylate-binding pocket and resulting in GodH being an acetyltransferase rather than a succinyltransferase.
4. Conclusion
The determination of the ‘unliganded’ structure of Rv0802c, a conserved hypothetical protein from M. tuberculosis, confirmed that Rv0802c is a member of the GNAT family of proteins. In addition, the discovery of an acetate ion in the active site guided future experiments, resulting in the structural determination of the Rv0802c–SucCoA complex. This complex suggests that Rv0802c is instead a succinyltransferase, the first example in the GNAT family. While the structure of Rv0802c presented here has not furnished us with the molecular target of its activity, it has suggested an alternative acyl donor which should facilitate target identification, as there should be far fewer succinylated targets compared with acetylated targets.
Supplementary Material
PDB reference: Rv0802c, 2vzy, r2vzysf
PDB reference: succinyl-CoA complex, 2vzz, r2vzzsf
Acknowledgments
We thank the Albert Einstein College of Medicine for their support of the analytical ultracentrifuge facility and Dr Michael Brenowitz for conducting the experiments. This work was supported by NIH grants AI33696 (to JSB) and T32 AI07501 (to MWV).
References
- Angus-Hill, M. L., Dutnall, R. N., Tafrov, S. T., Sternglanz, R. & Ramakrishnan, V. (1999). J. Mol. Biol.294, 1311–1325. [DOI] [PubMed]
- Burk, D. L., Ghuman, N., Wybenga-Groot, L. E. & Berghuis, A. M. (2003). Protein Sci.12, 426–437. [DOI] [PMC free article] [PubMed]
- Card, G. L., Peterson, N. A., Smith, C. A., Rupp, B., Schick, B. M. & Baker, E. N. (2005). J. Biol. Chem.280, 13978–13986. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Davis, I. W., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2004). Nucleic Acids Res.32, W615–W619. [DOI] [PMC free article] [PubMed]
- Dyda, F., Klein, D. C. & Hickman, A. B. (2000). Annu. Rev. Biophys. Biomol. Struct.29, 81–103. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Errey, J. C. & Blanchard, J. S. (2005). J. Biol. Chem.187, 3039–3044. [DOI] [PMC free article] [PubMed]
- Frankel, B. A. & Blanchard, J. S. (2008). Arch. Biochem. Biophys.477, 259–266. [DOI] [PMC free article] [PubMed]
- He, H., Ding, Y., Bartlam, M., Sun, F., Le, Y., Qin, X., Tang, H., Zhang, R., Joachimiak, A., Liu, J., Zhao, N. & Rao, Z. (2003). J. Mol. Biol.325, 1019–1030. [DOI] [PubMed]
- Kleywegt, G. J. & Jones, T. A. (1998). Acta Cryst. D54, 1119–1131. [DOI] [PubMed]
- Koledin, T., Newton, G. L. & Fahey, R. C. (2002). Arch. Microbiol.178, 331–337. [DOI] [PubMed]
- Kovacs, L., Csanadi, A., Kiss, E. & Miczak, A. (2005). Acta Microbiol. Immunol. Hung.52, 363–371. [DOI] [PubMed]
- Kovacs, L., Csanadi, A., Megyeri, K., Kaberdin, V. R. & Miczak, A. (2005). Microbiol. Immunol.49, 1003–1007. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Levitt, D. G. (2001). Acta Cryst. D57, 1013–1019. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Onaka, H., Nakaho, M., Hayashi, K., Igarashi, Y. & Furumai, T. (2005). Microbiology, 151, 3923–3933. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006a). Acta Cryst. D62, 439–450. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006b). J. Appl. Cryst.39, 109–111.
- Rosenkrands, I., Rasmussen, P. B., Carnio, M., Jacobsen, S., Theisen, M. & Andersen, P. (1998). Infect. Immun.66, 2728–2735. [DOI] [PMC free article] [PubMed]
- Schüttelkopf, A. W. & van Aalten, D. M. F. (2004). Acta Cryst. D60, 1355–1363. [DOI] [PubMed]
- Terwilliger, T. C. (2002). Acta Cryst. D58, 1937–1940. [DOI] [PubMed]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res.22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Vetting, M. W., de Carvalho, L. P., Roderick, S. L. & Blanchard, J. S. (2005). J. Biol. Chem.280, 22108–22114. [DOI] [PubMed]
- Vetting, M. W., de Carvalho, L. P. S., Yu, M., Hegde, S. S., Magnet, S., Roderick, S. L. & Blanchard, J. S. (2005). Arch. Biochem. Biophys.433, 212–226. [DOI] [PubMed]
- Vetting, M. W., Hegde, S. S., Javid-Majd, F., Blanchard, J. S. & Roderick, S. L. (2002). Nature Struct. Biol.9, 653–658. [DOI] [PubMed]
- Vetting, M. W., Roderick, S. L., Yu, M. & Blanchard, J. S. (2003). Protein Sci.12, 1954–1959. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed]
- Yoshikawa, A., Isono, S., Sheback, A. & Isono, K. (1987). Mol. Gen. Genet.209, 481–488. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Rv0802c, 2vzy, r2vzysf
PDB reference: succinyl-CoA complex, 2vzz, r2vzzsf