Abstract
Although adeno-associated virus (AAV)-mediated gene therapy has been hindered by the small viral packaging capacity of the vector, trans-splicing AAV vectors are able to package twice the size of the vector genome. Unfortunately, the efficiency of current trans-splicing vectors is very low. Here we show that rational design of the gene splitting site has a profound influence on trans-splicing vector-mediated gene expression. Using mRNA accumulation as a guide, we generated a set of efficient trans-splicing vectors and achieved widespread expression of the 6-kb ΔH2-R19 mini-dystrophin gene in skeletal muscle of mdx mice, a model for Duchenne muscular dystrophy. The dystrophic phenotype was ameliorated in both adult and aged mice. This demonstrates the use of trans-splicing vectors to efficiently express a large therapeutic structural protein. This strategy should be applicable to other large therapeutic genes or large transcription regulatory elements.
AAV is an extremely promising gene therapy vehicle1-3. However, being one of the smallest DNA viruses, it cannot carry a large therapeutic expression cassette. A 5-kb genome is considered the upper limit for a single AAV virion4. Based on the inherent capacity of AAV genome concatamerization5-7, the trans-splicing dual vector approach was developed 5 years ago to deliver up to a 10-kb genome8-11. Using this approach, a large gene is split into two parts. The 5′ gene fragment and a tail-tagged splicing donor sequence are packaged into a donor virus. The 3′ gene fragment and a preceding splicing acceptor sequence are packaged into an acceptor virus. Protein expression is achieved through co-infection, viral genome recombination and splicing.
Encouraging preliminary results were achieved for certain secreted proteins such as erythropoietin and Factor VIII8,12. Unfortunately, transduction efficiency has been too low to reach the therapeutic level for a structural protein11. Moderate improvement was achieved by using novel AAV serotypes or engineering the AAV inverted terminal repeat (ITR) sequence13,14. Nonetheless, the challenge remains to translate the exciting potential of trans-splicing vectors to a real disease model. Recently, our lab and others have systematically investigated the potential barriers14,15. Our results suggest that the accumulation of mRNA, rather than the simultaneous uptake of both donor and acceptor viruses, is the predominant rate-limiting step15.
A unique feature of trans-splicing vectors is transcription and splicing across the double-D (dD)-ITR junction in the reconstituted viral genome6. We hypothesized that mRNA production in the reconstituted genome is influenced by the DNA sequences flanking the irregular dD-ITR structure. Rational selection of the gene splitting site and optimization of the intron and exon splicing signals may improve overall transduction efficiency of trans-splicing vectors. In this study, we tested this hypothesis with the 6-kb ΔH2-R19 mini-dystrophin (ΔH2) gene.
The ΔH2 minigene was developed from a truncated, but highly functional, patient-derived gene (Δexon17-48)16,17. The patient with the exon 17-48 deletion displayed an extremely mild phenotype and was ambulant beyond age 60 (ref. 16). The ΔH2 minigene is nearly identical to the full-length dystrophin cDNA except for a perfectly phased deletion in the rod domain (from hinge 2 to spectrin-like repeat 19). Transgenic expression of the ΔH2 minigene completely prevented dystrophy in mdx muscle and resulted in muscle strength indistinguishable from that of the wild-type animal17. These studies suggest that the ΔH2 minigene may represent a promising alternative to the full-length cDNA, to rescue dystrophic pathology. Because of the large size of the cDNA, the ΔH2 minigene represents a great system to explore limitations and improvements in trans-splicing AAV vectors.
To determine the optimal gene splitting site, we first screened the splicing signals from a series of endogenous exon/intron/exon junctions in the ΔH2 minigene by RNase protection assay (RPA). Considering the size constraint of a single AAV virion, any exon boundary between exons 51 and 65 would be appropriate to divide the ΔH2 minigene. We initially selected three sites that have the highest consensus splicing values and the most favorable thermodynamic parameters for U1snRNA binding18. These exon/intron/exon junctions are 56/56/57, 60/60/61 and 63/63/64. We also picked the 53/53/54 junction, which has an average U1snRNA binding value of the entire dystrophin gene18.
We synthesized a series of mini-cassettes to mold the reconstituted viral genomes. Each cassette contained the flanking exon sequences at the putative splitting site, the endogenous intron splicing signals and the dD-ITR junction (Fig. 1a). We then examined the splicing profile and mRNA accumulation by RPA (Supplementary Methods online)15. The 60/60/61 junction had the best calculated splicing value (Table 1). Interestingly, the 63/63/64 junction, also a well conserved splicing site, showed the highest splicing efficiency in the RPA (Fig. 1b, Table 1). The ratio of spliced to unspliced RNA in p63/63/64 was more than twofold higher than that in p60/60/61. Surprisingly, the 56/56/57 junction was the poorest site. In contrast to the distinctive bands of unspliced and spliced RNA transcripts, our RPA revealed a smear of low molecular weight products from p56/56/57 (Fig. 1b). In addition to quantifying splicing efficiency, we also measured the steady state level of spliced RNA. The 60/60/61 junction yielded the highest level of mRNA, followed by the 63/63/64 junction and the 53/53/54 junction. The 56/56/57 junction produced little detectable spliced transcript (Fig. 1b and Table 1).
Figure 1.

Endogenous splicing signal screening. The RPA was used to evaluate splicing and mRNA accumulation of the selected dystrophin exon/intron/exon junctions in the presence of the dD-ITR structure. (a) Schematic outline of the constructs used in the RPA (not drawn to scale). The endogenous intron splicing sequences were retrieved from genomic DNA by PCR. The dD-ITR was engineered between the 5′ and the 3′ splicing signals inside the intron. The location of antisense RPA probes is marked. (b) A representative photomicrograph from six independent RPA experiments. The endogenous human β-actin RNA (the control for RPA and loading) was detected by an independent probe added at the same time. *, the expected location of the RNA transcripts from p56/56/57.
Table 1.
The predicted splicing parameters and RPA results
| Exon/intron/exon combination |
||||
|---|---|---|---|---|
| 53/53/54 | 56/56/57 | 60/60/61 | 63/63/64 | |
| CVa | 0.90, 0.60 | 0.84, 0.97 | 0.94, 0.98 | 0.95, 0.90 |
| ΔDGo37b | −7.7 | −10.4 | −11.7 | −10.6 |
| Total RNA | 68.7 ± 27.8 | 2.4 ± 1.2c | 342.2 ± 24.3d | 159.9 ± 9.4c |
| Unspliced RNA | 20.0 ± 9.1 | 1.6 ± 1.2 | 87.1 ± 11.2d | 20.4 ± 9.1 |
| Spliced RNA | 48.8 ± 20.6 | 0.8 ± 0.2 | 255.1 ± 20.6d | 139.5 ± 6.4d |
| S/U ratioe | 2.5 ± 0.3 | 1.0 ± 0.6 | 3.0 ± 0.4 | 7.1 ± 0.9d |
| % of splicing | 71.1 | 31.3c | 74.5 | 87.2c |
Consensus splicing values (CV) at the 5′ and the 3′ ends of each indicated intron (from ref. 18). A higher value indicates a better match.
Free energy exchange during U1snRNA annealing (Kcal/mole). The more negative the value, the more favorable the annealing.
Values are statistically different from each other in the same category (P < 0.014).
Values are statistically different from other exon/intron/exon combinations in the same category (P < 0.016).
The ratio of spliced (S) to unspliced (U) RNA transcript.
The RPA results suggest that the 60/60/61 and the 63/63/64 junctions may represent excellent candidate sites to split the minigene. To test whether the unique sequence organization in these locations could overcome the mRNA accumulation barrier, we engineered two pairs of trans-splicing vectors based on the endogenous splicing signals from introns 60 and 63, respectively (Fig. 2a). The minigene was split precisely between exons 60 and 61, or between exons 63 and 64, as determined by RPA (Fig. 1). Donor and acceptor viruses were packaged in AAV serotype-6 capsid and delivered either individually or together to the tibialis anterior (TA) and the extensor digitorum longus (EDL) muscles of 2-month-old mdx mice (Supplementary Note and Supplementary Fig. 1 online). Mini-dystrophin was not detected in muscles infected with a single vector alone (Figs. 2b and 3b, and Supplementary Fig. 2 online). At 1 month after infection, low levels of the mini-dystrophin protein were found in AV.Donor.63 and AV.Acceptor.63 co-infected muscles (Fig. 2). The 60/60/61 junction yielded the highest level of mRNA in the RPA (Fig. 1). Consistent with this result, trans-splicing vectors developed using this junction (AV.Donor.60 and AV.Acceptor.60) yielded significantly higher mini-dystrophin expression (Fig. 2). At 3 months after infection, ∼80% (ranging from 46% to 96%) of myofibers were transduced in the injected TA muscles (Fig. 3a, Supplementary Fig. 2 online). Importantly, this high level transduction was observed in the absence of a cellular immune response (Supplementary Fig. 3 online).
Figure 2.

ΔH2 mini-dystrophin expression from two independent sets of trans-splicing AAV vectors. The EDL and the TA muscles of 2-month-old mdx mice were co-infected with 1 × 1010 vector genomes (vg) (for the EDL muscle) and 2 × 1010 vg (for the TA muscle) particles of each pair of trans-splicing viruses. Single vector infection (donor or acceptor alone) was also included in the cohort study (5 × 109 vg particles for the EDL muscle and 1 × 1010 vg particles for the TA muscle). Mini-dystrophin expression was evaluated 1 month later. (a) Schematic diagram of ΔH2 mini-dystrophin trans-splicing vectors (not drawn to scale). The donor virus carries the cytomegalovirus (CMV) promoter, the 5′ portion of the ΔH2 minigene and the endogenous splicing donor signal (from intron 60 and 63, respectively). The acceptor virus carries the endogenous splicing acceptor signal from the same intron (60 and 63, respectively), the remaining minigene and the polyA sequence. The viruses are accordingly named after the respective intron. (b) Western blot analysis of mini-dystrophin expression in muscle extracts with monoclonal N-terminal (specific for human dystrophin) and C-terminal antibodies, respectively. Each lane is from one infected TA muscle. Different samples were used for the N-terminal and C-terminal immunoblots. (c) Quantification of ΔH2 minigene expression by immunofluorescence staining with a human dystrophin–specific antibody. Left panels are the representative photomicrographs from the TA muscles infected with the indicated set of trans-splicing vectors. Scale bar, 200 μm. Right panel represents mini-dystrophin-positive myofibers per muscle section. n = 3–7 samples for each group. *, the difference between two sets of trans-splicing vectors was statistically significant.
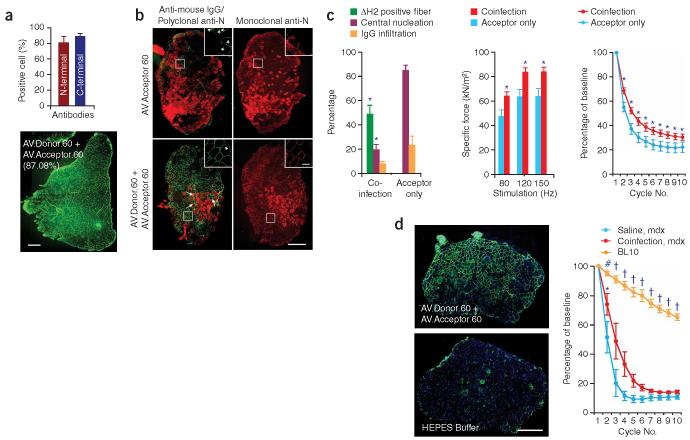
Figure 3.
Widespread expression of mini-dystrophin in AV.Donor.60 and AV.Acceptor.60 co-infected mdx muscle attenuates dystrophic pathology. (a) Mini-dystrophin expression in adult mdx TA muscle. 2 × 1010 vg particles of each virus were co-delivered to the TA muscles of 2-month-old male mdx mice. Expression was determined 3 months later by immunostaining. Bar graph represents the transduction efficiency quantified according to immunostaining with monoclonal antibodies. n = 6 for N-terminal antibody and n = 5 for C-terminal antibody. A representative montage composite photomicrograph from an entire TA muscle is shown in the bottom panel. The dystrophin-positive myofibers were revealed by a polyclonal N-terminal antibody. Number in parenthesis indicates the percentage of dystrophin-positive myofibers in the section. Scale bar, 500 μm. (b) Mini-dystrophin expression in adult EDL muscle prevents sarcolemma leakage. Equal amounts (5 × 109 vg particles) of each virus were co-injected into the left EDL muscles of 2-month-old male mdx mice. 5 × 109 vg particles of AV.Acceptor.60 was injected to the contralateral EDL muscles. Mini-dystrophin expression and sarcolemma integrity were examined 3 months later. Photomicrographs are from the representative EDL muscles. Sarcolemma leakage in injured myofibers is revealed by the uptake of immunoglobulin from circulation (anti-mouse lgG staining). Dystrophin expression is detected either by a polyclonal anti-N-terminal antibody (for both mini-dystrophin and revertant dystrophin) or a monoclonal anti-N-terminal antibody (only detects human dystrophin–derived mini-dystrophin). The overlay image of anti-mouse lgG staining (red) and polyclonal antibody immunostaining (green) illustrates the protection of the sarcolemma by mini-dystrophin. Monoclonal antibody immunostaining also reveals membrane leakage in injured myofibers (intense cytosolic staining, different from peripheral membrane staining of dystrophin). Inserts are high power photomicrographs of boxed areas in the corresponding panels. Arrow, immunoglobulin infiltration is prevented in transduced myofibers whereas surrounding cells were not protected. *, revertant myofibers. Scale bar, 400 μm for low power photomicrographs and 50 μm for high power inserts. (c) Quantitative evaluation of muscle pathology and muscle force in the EDL muscles described in panel b. Left panel, quantification of transduction efficiency (n = 15 for co-infection, n = 7 for acceptor only), central nucleation (n = 5 for co-infection, n = 7 for acceptor only), and lgG infiltration (n = 5 for co-infection, n = 4 for acceptor only); middle panel, specific muscle force (n = 7 pairs); right panel, resistance to eccentric contraction-induced injury (n = 7 pairs). *, the difference between co-infected muscle and single vector infected muscle was statistically significant. (d) Efficient mini-dystrophin expression in aged mdx EDL muscle moderately improved muscle function. The left EDL muscles of 1-year-old mdx mice were co-infected with 1.5 × 1010 vg particles of AV.Donor.60 and AV.Acceptor.60 (7.5 × 109 vg particles each). The right EDL muscles were mock-infected with HEPES-buffered saline. Transgene expression, muscle pathology and muscle physiology were examined 6 months later. Left panel, representative photomicrographs of the EDL muscles either co-infected with both AV.Donor.60 and AV.Acceptor.60, or mock infected with HEPES saline. Immunostaining was performed with monoclonal anti-N-terminal antibody. Scale bar, 400 μm. Right panel, resistance to eccentric contraction-induced injury (n = 3 for each group). *, the difference between co-infected muscle and saline-injected muscle was statistically significant according to paired t test. Cross, the results in BL10 muscle were significantly different from that in mdx muscle (both AAV infected and mock treated) according to one-way ANOVA and Bonferroni post hoc test. Pound, the result in BL10 was only significantly better than that of saline-injected mdx muscle according to one-way ANOVA and Bonferroni post hoc test.
To determine the functional competence of mini-dystrophin produced by trans-splicing vectors, we first evaluated whether it would assemble the dystrophin-associated glycoprotein complex (DGC). The components in all three DGC subcomplexes (the dystroglycan, the sarcoglycan and the cytoplasmic) were restored to the sarcolemma (Supplementary Fig. 4 online). Consistent with a previous study in the Δ17-48 minigene transgenic mdx mice19, neuronal nitric oxide synthase expression was not restored (Supplementary Fig. 4 online).
A more stringent test is to determine whether mini-dystrophin produced by trans-splicing vectors could ameliorate dystrophic pathology and improve muscle strength. We first examined the therapeutic effect in the EDL muscle of 2-month-old mdx mice. On average, co-infection led to mini-dystrophin expression in 49% of myofibers (ranging from 12% to 98%) (Fig. 3b,c). The percentage of centrally nucleated myofibers, a hallmark of muscle degeneration and regeneration, was reduced from 85% in AV.Acceptor.60 infected muscle to 20% in AV.Donor.60 and AV.Acceptor.60 co-infected muscle (Fig. 3c). Trans-splicing AAV-treated muscle also showed a trend toward increased sarcolemma integrity and mini-dystrophin-positive fibers were completely protected from membrane damage (Fig. 3b,c). In a physiology assay, we observed a significant improvement in the specific force of trans-splicing AAV–treated muscle P < 0.018 (Fig. 3c, Supplementary Note and Supplementary Fig. 1 online). Furthermore, these muscles became more resistant to eccentric contraction-induced injury (Fig. 3c).
Old mdx muscle displays more severe muscle pathology. To further evaluate our trans-splicing vectors, we infected the EDL muscles of 1-year-old mdx mice. Surprisingly, efficient transduction was achieved at 6 months after infection (Fig. 3d). On average, the transduction efficiency reached 40% (ranging from 18% to 73%) (Supplementary Fig. 5b online). Despite a quite efficient gene transfer, we only observed moderate improvement in the eccentric contraction assay (Fig. 3d). Other pathology parameters such as central nucleation, fibrosis and macrophage infiltration were not altered (Supplementary Fig. 5 online). This finding suggests that early intervention may be critical for Duchenne muscular distropy (DMD) gene therapy. Nevertheless, our results in both adult and aged mdx muscles clearly demonstrated the therapeutic potential of trans-splicing AAV vectors for DMD gene therapy.
Until the recent development of dual vector technologies, AAV has been excluded as a candidate vehicle for large therapeutic genes. Trans-splicing vectors, though theoretically promising, have so far failed to express a large structure gene at the therapeutic level. We have recently found that the RNA processing represents a crucial barrier to efficient transduction in trans-splicing vectors15. Boosing the level of accumulated mRNA may represent a promising avenue to improve trans-splicing vector-mediated gene expression. To test this hypothesis, we examined splicing efficiency and mRNA production from a series of exon-intron-exon junctions in the 6-kb mini-dystrophin gene. Consistent with the well-documented observation that RNA transcription and splicing are influenced by DNA sequences20,21, insertion of the dD-ITR junction to different introns resulted in different effects on splicing and mRNA accumulation. Intron 63 was the most efficient splicing site whereas intron 60 yielded the highest level of mRNA. The molecular mechanisms underlying this observation remain to be defined, but it may relate to transcription blocking and/or RNA stability15,22. We also compared protein production between trans-splicing vectors that were split at either intron 60 or intron 63. Both western blot analysis and immunostaining demonstrated a higher expression in the intron 60 splitting vectors. This result strongly suggests that trans-splicing vectors should be constructed using the site that yields the highest mRNA.
In summary, we have developed a strategy to rationally design trans-splicing AAV vectors. This finding may have a broad impact on developing trans-splicing vectors for other large therapeutic genes (such as the cystic fibrosis transmembrane conductance regulator gene and the photoreceptor-specific ATP binding cassette transport gene for the treatment of cystic fibrosis and Stargardt disease, respectively) as well as the use of large complicated transcription regulatory elements in the AAV vector.
METHODS
Evaluation of the endogenous splicing signals
The exon-intron junctions flanking introns 53, 56, 60 and 63 of the human dystrophin gene were amplified by PCR from HEK293 cell genomic DNA (Supplementary Methods online). The double D-ITR junction was derived from a circular AAV genome isolated from infected muscle15. The mini-constructs for the RPA were assembled on pcDNA3.1(+) (Invitrogen) (Fig. 1). A total of four plasmids were generated including (in the order of exon/intron/exon) p53/53/54, p56/56/57, p60/60/61 and p63/63/64. The templates for RPA probes were PCR amplified from each respective plasmid and cloned in pGEM3Z (Promega) (Supplementary Methods online). RPA probes (for the dystrophin gene and the endogenous human β-actin gene) were generated by in vitro transcription as described before15. RPA reaction and RNA signal quantification were carried out according to a published protocol15. Constructs generated by PCR amplification were validated by sequencing.
AAV vector and animal procedure
Two sets of trans-splicing vectors were generated based on the endogenous splicing signals from introns 60 and 63, respectively. Each set included a donor and an acceptor. These vectors were generated by first introducing the corresponding intron splicing sequences in the ΔH2 minigene. The intermediate constructs were then inserted into previously described multiple cloning AAV cis plasmids (Supplementary Methods online). AAV-6 packaging plasmids (pMTrep2 and pCMVcap6) were gifts from A. Dusty Miller23. Pseudotyped recombinant AAV stocks were generated by quadruple plasmid transfection with the cis plasmid, pMTrep2, pCMVcap6 and pHelper (Stratagene) at a ratio of 1:1:3:3 in HEK293 cells. Crude lysate was purified through three rounds of CsCl isopycnic ultracentrifugation and titrated by slot blot as we reported before24. All animal experiments were approved by the Animal Care and Use Committee at the University of Missouri and were in accordance with National Institutes of Health guidelines. The mdx mouse strain was originally purchased from the Jackson Laboratory. The experimental mice were obtained from breeding colonies established locally (Supplementary Note and Supplementary Fig. 1 online). Both experimental and breeding mice were housed in a specific-pathogen-free animal facility. AAV was delivered to the EDL and/or TA muscles of male mdx mice using previously described protocols24,25. Both slow- and fast-type myofibers were transduced by AAV-6. However, there seemed to be a trend toward preferential transduction of fast type myofiber (Supplementary Fig. 6 online).
Protein expression and morphology
Mini-dystrophin expression was evaluated by indirect immuofluorescence staining and western blot analysis26,27. Antibodies used for these studies include Dys-3 (monoclonal, Novocastra, Newcastle, UK; 1:10 for immunostaining and 1:20 for western blot analysis, specific for human dystrophin N-terminal domain), Dys-2 (monoclonal, Novocastra, 1:30 for immunostaining and 1:100 for western blot analysis, specific for dystrophin C-terminal domain), Mandys 8 (monoclonal, Sigma; 1:100 for immunostaining, specific for exon 32) and an affinity purified polyclonal anti-dystrophin N-terminal antibody (1:600 for immunostaining only)17,25. Muscle microsome preparation was used for western blot analysis. Equal loading was confirmed by Ponseu S staining of the membrane and anti-β-catenin immunoblotting of stripped membranes (Supplementary Methods online)28. The DGC components were evaluated by immunostaining with monoclonal antibodies against β-dystroglycan (Novocastra, 1:50), β-sarcoglycan (Novocastra, 1:50), dystrobrevin (BD Bioscience; 1:200), and syntrophin (Abcam; 1:200). Transduction efficiency was determined by dividing the total number of Dys-3 positive cells (immunostaining) with the total number of myofibers (hematoxylin and eosin (H&E) staining of adjacent section). At least two montage composites of the entire cross sections in the mid-belly were quantified for each muscle sample. The percentage of centrally nucleated myofibers was determined as described before25. Standard histochemical procedures were used for HE, Masson trichrome and nonspecific esterase staining.
Muscle physiology
The EDL muscle was carefully isolated and mounted on a dual mode servomotor transducer (Aurora Scientific) in Ringer's solution. Muscle stimulation, data acquisition and data analysis were conducted with DMC/DMA software (Aurora) (Supplementary Methods online). The absolute muscle force was measured at 80, 120 and 150 Hz. The specific force was calculated by dividing the value of absolute force with muscle cross-sectional area (Supplementary Methods online)25. The eccentric contraction assay was performed according to a published protocol25.
Statistical analysis
Data are presented as the mean ± s.e.m. and analyzed with SPSS software. In studies involving two groups, comparisons were made using two-tailed t-tests (paired or unpaired depending on the situation). In studies involving multiple groups, comparisons were made using one-way ANOVA followed by Bonferroni post hoc test. Differences were considered significant when P < 0.05.
ACKNOWLEDGMENTS
We thank A. Dusty Miller for the AAV-6 packaging plasmids. We thank Scott Q. Harper and Zhuping Xu for helpful discussion and technical assistance. This work was supported by grants from the National Institutes of Health (AR-49419, D.D.) and the Muscular Dystrophy Association (D.D. and J.S.C.).
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Duan D, Yue Y, Engelhardt JF. In: Lung Biology in Health and Disease, Gene Therapy in Lung Disease. Albelda SM, editor. Marcel Dekker Inc.; New York, NY: 2002. pp. 51–92. [Google Scholar]
- 2.Kay MA, et al. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat. Genet. 2000;24:257–261. doi: 10.1038/73464. [DOI] [PubMed] [Google Scholar]
- 3.Carter BJ. Adeno-associated virus vectors in clinical trials. Hum. Gene Ther. 2005;16:541–550. doi: 10.1089/hum.2005.16.541. [DOI] [PubMed] [Google Scholar]
- 4.Dong JY, Fan PD, Frizzell RA. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther. 1996;7:2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- 5.Duan D, et al. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long term episomal persistence in muscle. J. Virol. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan D, Yan Z, Yue Y, Engelhardt JF. Structural analysis of adeno-associated virus transduction intermediates. Virology. 1999;261:8–14. doi: 10.1006/viro.1999.9821. [DOI] [PubMed] [Google Scholar]
- 7.Yue Y, Duan D. Double strand interaction is the predominant pathway for intermolecular recombination of adeno-associated viral genomes. Virology. 2003;313:1–7. doi: 10.1016/s0042-6822(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 8.Yan Z, Zhang Y, Duan D, Engelhardt JF. Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. USA. 2000;97:6716–6721. doi: 10.1073/pnas.97.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, Li J, Xiao X. Overcoming adeno-associated virus vector size limitation through viral DNA heterodimerization. Nat. Med. 2000;6:599–602. doi: 10.1038/75087. [DOI] [PubMed] [Google Scholar]
- 10.Nakai H, Storm TA, Kay MA. Increasing the size of rAAV-mediated expression cassettes in vivo by intermolecular joining of two complementary vectors. Nat. Biotechnol. 2000;18:527–532. doi: 10.1038/75390. see comments. [DOI] [PubMed] [Google Scholar]
- 11.Duan D, Yue Y, Engelhardt JF. Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol. Ther. 2001;4:383–391. doi: 10.1006/mthe.2001.0456. [DOI] [PubMed] [Google Scholar]
- 12.Chao H, Sun L, Bruce A, Xiao X, Walsh CE. Expression of human factor VIII by splicing between dimerized AAV vectors. Mol. Ther. 2002;5:716–722. doi: 10.1006/mthe.2002.0607. [DOI] [PubMed] [Google Scholar]
- 13.Reich SJ, et al. Efficient trans-splicing in the retina expands the utility of adeno-associated virus as a vector for gene therapy. Hum. Gene Ther. 2003;14:37–44. doi: 10.1089/10430340360464697. [DOI] [PubMed] [Google Scholar]
- 14.Yan Z, Zak R, Zhang Y, Engelhardt JF. Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes. J. Virol. 2005;79:364–379. doi: 10.1128/JVI.79.1.364-379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Z, et al. Trans-splicing adeno-associated viral vector-mediated gene therapy is limited by the accumulation of spliced mRNA but not by dual vector coinfection efficiency. Hum. Gene Ther. 2004;15:896–905. doi: 10.1089/hum.2004.15.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.England SB, et al. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990;343:180–182. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- 17.Harper SQ, et al. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat. Med. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- 18.Sironi M, et al. Analysis of splicing parameters in the dystrophin gene: relevance for physiological and pathogenetic splicing mechanisms. Hum. Genet. 2001;109:73–84. doi: 10.1007/s004390100547. [DOI] [PubMed] [Google Scholar]
- 19.Chao DS, et al. Selective loss of sarcolemmal nitric oxide synthase in Becker muscular dystrophy. J. Exp. Med. 1996;184:609–618. doi: 10.1084/jem.184.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 21.Ibrahim el C, Schaal TD, Hertel KJ, Reed R, Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc. Natl. Acad. Sci. USA. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Halbert CL, Allen JM, Miller AD. Adeno-associated virus type 6 (aav6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of aav2 vectors. J. Virol. 2001;75:6615–6624. doi: 10.1128/JVI.75.14.6615-6624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan D, Yue Y, Yan Z, Engelhardt JF. A new dual-vector approach to enhance recombinant adeno-associated virus-mediated gene expression through intermolecular cis activation. Nat. Med. 2000;6:595–598. doi: 10.1038/75080. [DOI] [PubMed] [Google Scholar]
- 25.Liu M, et al. Adeno-associated virus-mediated micro-dystrophin expression protects young mdx muscle from contraction-induced injury. Mol. Ther. 2005;11:245–256. doi: 10.1016/j.ymthe.2004.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue Y, et al. Microdystrophin gene therapy of cardiomyopathy restores dystrophin-glycoprotein complex and improves sarcolemma integrity in the mdx mouse heart. Circulation. 2003;108:1626–1632. doi: 10.1161/01.CIR.0000089371.11664.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue Y, Skimming JW, Liu M, Strawn T, Duan D. Full-length dystrophin expression in half of the heart cells ameliorates beta-isoproterenol-induced cardiomyopathy in mdx mice. Hum. Mol. Genet. 2004;13:1669–1675. doi: 10.1093/hmg/ddh174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]