Abstract
In Saccharomyces cerevisiae, mRNA encoding the cell-fate determinant Ash1p is localized to the distal tip of daughter cells. Five SHE genes are required for proper Ash1 mRNA localization, one of which encodes the myosin Myo4p. We show that three of the five She proteins, She2p, She3p, and Myo4p, colocalize with Ash1 mRNA in vivo and coimmunoprecipitate with Ash1 mRNA from cell extracts. We also find that She3p binds to Myo4p in the absence of RNA and She2p is required for binding She3p-Myo4p to Ash1 mRNA. These results suggest that She3p acts as an adapter protein that docks the myosin motor onto an Ash1–She2p ribonucleoprotein complex.
The asymmetric distribution of proteins is a vital process in cellular function and cell-fate determination. One means of generating protein asymmetry is to localize the mRNA encoding the protein to a distinct site within the cell. The localization process is specified by sequences within the mRNA, which are generally found in the 3′ untranslated regions (UTRs), and mediated by cytoskeletal filaments that are required for transport and subsequent anchoring of the mRNA at its final destination (1, 2). The transport, anchoring, and translation regulation of localized transcripts are governed by proteins that form large ribonucleoprotein complexes with the mRNAs (1, 3).
In Saccharomyces cerevisiae, Ash1p, a repressor of mating-type switching, is asymmetrically distributed to the daughter cell nucleus through the localization of Ash1 mRNA to the distal tip of the daughter cell (4, 5). Because of the ability to use both biochemical and genetic techniques in yeast, Ash1 provides an excellent model system for understanding the molecular basis of mRNA localization. Previous work identified five genes (the SHE genes) that are required for proper localization of Ash1 mRNA (4, 5). Two of these genes encoded previously identified cytoskeletal-associated proteins. SHE5(BNI1) encodes a protein involved in regulating the actin cytoskeleton (6), whereas SHE1(MYO4) encodes a myosin belonging to the myosin V family (7). The latter result suggested that Ash1 mRNA is localized to the bud tip by actomyosin-based transport. This hypothesis was strongly supported by the important observations that green fluorescent protein (GFP)-tagged Ash1 mRNA moves from the mother to the daughter cells in a linear manner and at rapid rates and that this motion does not occur in myo4Δ cells (8). Furthermore, a direct association of Myo4p with Ash1 mRNA was revealed by immunoprecipitation experiments (9).
Whereas the involvement of actin and myosin in Ash1 localization has been established, the role of the other She proteins and the mechanism of docking Myo4p to Ash1 mRNA have been less clear. Both SHE2 and SHE3 were found to be required for Myo4p to interact with Ash1 mRNA, raising the possibility that She2p and/or She3p serve as adapters that bind Myo4p to Ash1 mRNA (9). However, in another study, She2p was reported not to colocalize with Ash1 mRNA (8). Moreover, a direct interaction between Myo4p and other She proteins had not been demonstrated in prior work. To resolve these questions and further investigate the role of the She proteins, we have combined genetic analysis, in vivo colocalization, and in vitro biochemical analysis of each She protein. Our results establish that She2p, She3p, and Myo4p associate with Ash1 mRNA and demonstrate a direct interaction between Myo4p and She3p that is independent of RNA. We also show that She2p enables the She3p–Myo4p complex to bind to Ash1 mRNA.
Materials and Methods
Plasmids and Strains.
The U1Ap-GFP plasmid was constructed from AS144 which contains pHIS-GFP-lacI-NLS (a gift from Aaron Straight, Harvard University). A PCR product encoding amino acids 1–102 of U1A was generated from plasmid BT617 (a gift from Michael Rosbash, Brandeis University) and was cloned into AS144 between pHIS and GFP. The lacI sequence was replaced with a PCR product encoding glutathione S-transferase. The pHIS sequence was replaced with a PCR product containing the glycerol phosphate dehydrogenase promoter. The final construct (pGPD-U1A-GFP-GST-NLS) was subcloned into a CEN-ARS vector. U1Atag-ASH1 was constructed by subcloning pGal-ASH1 from AS174 (a gift from Anita Sil, University of California, San Francisco) into a 2-μ vector. Four repeats of the U1A binding sequence from the U1A premRNA 3′ UTR (10) were inserted into a unique BamHI site immediately downstream of the start codon. U1Atag-ADH2 was constructed by replacing ASH1 in U1Atag-ASH1 with ADH2 (including 500 nucleotides of the 3′ UTR). All strains were derived from w303. SHE-MYC strains were prepared by the method of Longtine et al. (11).
Induction and Imaging of U1Atag-ASH1 RNA Particles.
Cells containing U1Ap-GFP and either U1Atag-ASH1 or U1Atag-ADH2 were grown overnight at 30°C in synthetic media containing 2% raffinose. Overnight cultures were adjusted to OD600 ≈0.5 in synthetic media containing 2% raffinose and incubated for 2 h at 30°C. Galactose was added to 0.2% and the cultures were incubated for 1–1.5 h at 30°C. Cells were examined under a Zeiss Axioplan microscope by using a X63/NA 1.4 lens. Images were captured with a cooled charged-coupled device and digital images were displayed by using Adobe Photoshop. For colocalization experiments in SHE-MYC cells, samples were fixed after induction in 3.7% formaldehyde for 1 h. Cells were washed and spheroplasted in 100 mM phosphate buffer (pH 7.0)/1.2 M sorbitol containing 30 mM β-mercaptoethanol/40 μg/ml zymolyase 100T (ICN) for 15 min at 37°C. Cells were washed and spread on polylysine-coated, multiwell test slides. Cells were incubated in 9E10 (Babco, Richmond, CA) at a 1:1,000 dilution in PBS/1% BSA for 1 h. After washing, cells were incubated with rhodamine-conjugated goat anti-mouse IgG (Boehringer Mannheim) in PBS/1% BSA for 1 h. Cells were washed and mounted in PBS/90% glycerol/1 mg/ml p-phenylenediamine/0.1 μg/ml 4′,6-diamidino-2-phenylindole.
Immunoprecipitation of Ash1 RNA and She-Myc Proteins.
SHE-MYC cells containing U1Ap-GFP and either U1Atag-ASH1 or U1Atag-ADH2 were grown overnight in synthetic media containing 2% raffinose. Cultures were adjusted to OD600 ≈0.5 in 200 ml of rich media containing 2% raffinose and incubated 2.5 h at 30°C. Galactose was added to 0.2%, the cultures were incubated 1.5 h at 30°C, and the cells were harvested by centrifugation. SHE-MYC cells were grown overnight in rich media containing 2% dextrose and adjusted to OD600 ≈0.1 in 200 ml of rich media containing 2% dextrose. Cultures were incubated for 5 h at 30°C, and the cells were harvested by centrifugation. All cells were washed twice in 25 mM Hepes-KOH (pH 7.5)/150 mM KCl/2 mM MgCl2. Cells were resuspended in the above buffer containing 20 mM vanadyl ribonucleoside complexes (Sigma)/200 units/ml RNasin (Life Technologies, Grand Island, NY)/0.1% Nonidet P-40/1 mM DTT/0.2 mg/ml heparin/1 mM PMSF/10 μg/ml each of aprotinin, leupeptin, and pepstatin. Glass beads were added and the samples were vortexed six times for 30 s. The supernatants were removed and centrifuged for 10 min at 3,000 × gmax. To immunoprecipitate U1A-ASH1 RNA or U1A-ADH2 RNA, extracts were incubated with affinity-purified anti-GFP antibodies coupled to protein A agarose for 1 h at 4°C. Beads were washed four times in 25 mM Hepes-KOH (pH 7.5)/150 mM KCl/2 mM MgCl2, and were eluted in 50 mM Tris⋅HCl (pH 8.0)/100 mM NaCl/10 mM EDTA/1% SDS for 10 min at 65°C. A portion of each eluate was extracted with phenol/chloroform and ethanol precipitated to isolate the RNA or mixed with sample buffer for Western analysis. To immunoprecipitate She-myc proteins, extracts were incubated with anti-myc antibodies (9E10, Babco) coupled to protein G Sepharose for 1 h at 4°C. Beads were washed and eluted as above. Before immunoprecipitation, some samples were incubated with 200 μg/ml RNase A for 30 min on ice. RNA immunoprecipitates were analyzed by Northern analysis with radiolabeled probes generated against Ash1 RNA and Adh2 RNA by using the Strip-EZ kit (Ambion, Austin, TX). Western blots were performed by using anti-myc antibodies (9E10, Babco) or antihemagglutinin (HA) antibodies (HA.11, Babco).
Results
Association of She Proteins with Ash1 mRNA in Vivo.
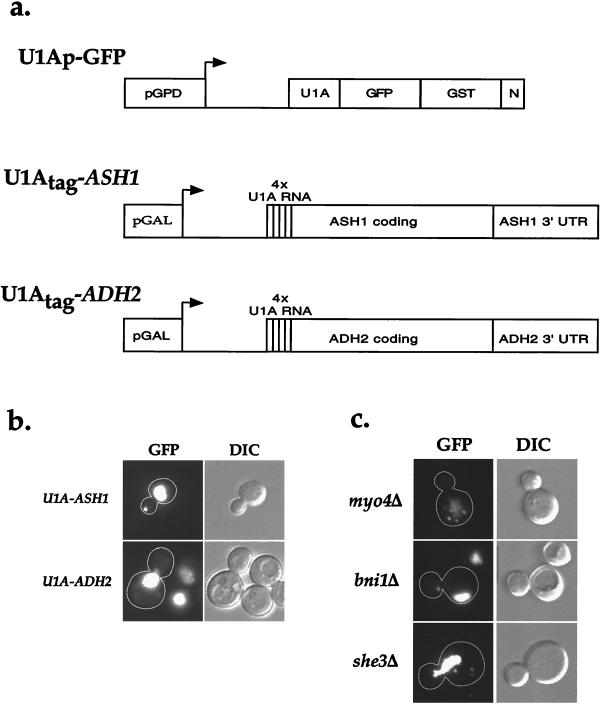
We have developed a system to visualize Ash1 mRNA in living and fixed cells and immunoprecipitate Ash1 mRNA from cell extracts (Fig. 1a). This system consists of a plasmid expressing Ash1 mRNA with a specific RNA aptamer and a second plasmid that expresses GFP fused to a protein that binds the RNA aptamer. For the RNA aptamer tag, we added four repeats of a RNA-binding sequence for the splicing protein U1A immediately downstream of the start codon in ASH1; the ASH1 promoter was replaced by the inducible galactose promoter to control expression levels (U1Atag-ASH1). A control plasmid (U1Atag-ADH2) was constructed that contained four U1A aptamer repeats inserted into ADH2. The second plasmid (U1Ap-GFP) contained the RNA-binding domain of U1A fused to the S65T mutant of the GFP; glutathione S-transferase was added for possible use in affinity purification, and a nuclear localization sequence was included at the C terminus to lower the amount of cytoplasmic fluorescence from the fusion protein. Expression of U1Ap-GFP was controlled by the constitutive glycerol phosphate dehydrogenase promoter. Two other groups have independently taken similar approaches to visualize Ash1 mRNA in vivo (8, 12). However, both groups used only the 3′ UTR of ASH1 and fused the RNA-binding protein MS2 to GFP, which bound Ash1 mRNA containing multiple copies of the MS2 RNA-binding sequence. Because the coding and 3′ UTR of ASH1 both play a role in Ash1 mRNA localization (4, 13, 14), we used the entire coding region and ≈560 nucleotides of the 3′ UTR of ASH1 for our experiments.
Figure 1.
Visualization of U1A-ASH1 RNA in wild-type and sheΔ cells. (a) A diagram of the constructs used to visualize and immunoprecipitate Ash1 mRNA. The RNA-binding construct (U1Ap-GFP) consisted of the amino terminus of U1A fused to GFP(S65T) for visualization, glutathione S-transferase for possible use in affinity purification, and a nuclear localization sequence (N) to lower the amount of cytoplasmic fluorescence. This construct was expressed from the constitutive glycerol phosphate dehydrogenase (GPD) promoter. To provide a binding site for U1Ap-GFP, four repeats of the U1A 3′ UTR were introduced just downstream of the start codon of either the entire coding region of ASH1 and ≈500 bp of its 3′ UTR (U1Atag-ASH1) or the entire coding region of ADH2 and ≈500 bp of its 3′ UTR (U1Atag-ADH2). Both of these constructs were expressed from the inducible galactose promoter. (b) Cells expressing U1Ap-GFP and U1Atag-ASH1 contain a single large particle at the distal tip of the bud, whereas cells expressing U1Ap-GFP and U1Atag-ADH2 display an increase in cytoplasmic fluorescence but do not contain any particles. (c) sheΔ cells contain mislocalized U1A-ASH1 RNA particles. myo4Δ and she3Δ cells contain multiple particles that are distributed throughout the mother cell, whereas bni1Δ cells contain a single particle that is localized to the neck. she2Δ and she4Δ cells displayed a similar GFP fluorescence pattern as myo4Δ and she3Δ cells (not shown).
Cells containing the U1Ap-GFP plasmid and either the U1Atag-ASH1 or the control U1Atag-ADH2 plasmid were induced with galactose and then either directly visualized or fixed in paraformaldehyde 1–1.5 h later (no differences were observed between living and fixed samples). Without U1Atag-ASH1 induction, U1Ap-GFP was confined to the nucleus (data not shown). Likewise, cells expressing ASH1 or ADH2 without the U1A aptamer contained only nuclear U1Ap-GFP (data not shown). In contrast, cells expressing U1Atag-ASH1 also displayed a single large GFP particle localized to the distal tip of daughter cells. This localized particle was specific for ASH1 because cells expressing U1Atag-ADH2 displayed only uniform, cytoplasmic fluorescence (Fig. 1b). In cells expressing either ASH1 or ADH2 without the U1A sequence, the GFP signal was confined to the nucleus (data not shown). Thus, the U1A-GFP tagging method can be used to follow the localization of Ash1 mRNA to the distal tip.
To demonstrate that the U1Atag-ASH1 RNA particle is being localized by a similar mechanism as wild-type Ash1 mRNA, we analyzed the localization of the U1Atag-ASH1 RNA particle in cells deleted of the SHE genes. In myo4Δ and she3Δ cells, there were multiple GFP particles, none of which were localized (Fig. 1c). A similar staining pattern was also observed in she2Δ and she4Δ cells (data not shown). In bni1Δ cells, there was a single GFP particle, but it was mislocalized to the neck region (Fig. 1c), as was observed for endogenous Ash1 mRNA in bni1Δ cells (4, 5). Thus, similar to the results reported by Bertrand et al. (8), all five genes that are required for proper localization of endogenous Ash1 mRNA are also required to localize the U1Atag-ASH1 RNA particle.
Whereas the U1A-tagged RNA can be used to study RNA transport to the bud tip, it displayed somewhat different behavior than wild-type Ash1 mRNA. First, endogenous Ash1 mRNA forms multiple particles that localize to the bud tip (4) rather than the single particle observed with U1Atag-ASH1 RNA. Bertrand et al. (8) and Beach et al. (12), by using the MS2 system to visualize Ash1 mRNA, also observed a single particle. The ability of Ash1 mRNA to aggregate in vivo combined with its subsequent concentration at the distal tip may lead to the aberrant formation of a single particle. We, as well as Beach et al. (12), also observed that the U1Atag-ASH1 RNA particle was usually localized at the neck in late anaphase (binucleated cells). In contrast, endogenous Ash1 mRNA localizes to the distal tip in late anaphase (4, 5). The altered Ash1 mRNAs may not be properly anchored at the distal tip and can move when the actin cytoskeleton reorients toward the neck for cytokinesis.
Association of She Proteins with U1Atag-ASH1 mRNA.
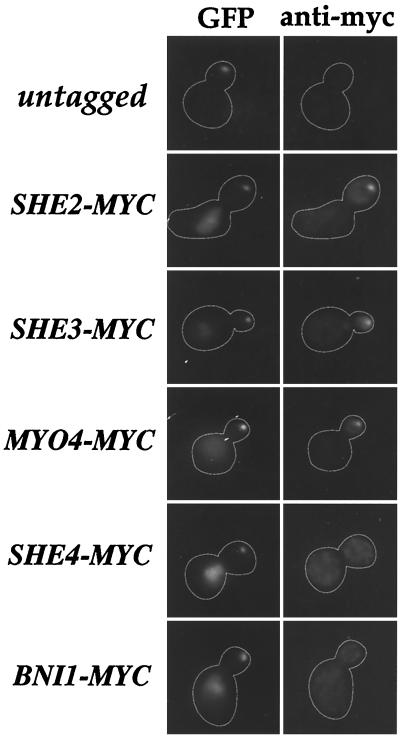
To begin to assess which of the five She proteins plays a direct role in Ash1 mRNA localization, we analyzed whether myc-tagged versions of the She proteins colocalized with the U1Atag-ASH1 RNA particle in vivo. SHE-MYC strains displayed normal localization of endogenous Ash1 mRNA, indicating that the myc-tagged She proteins were functional (data not shown). We found that She2p, Myo4p, and She3p colocalized with the GFP signal from the U1Atag-ASH1 RNA particle, whereas She4p and Bni1p did not (Fig. 2). These results suggest that She2p, Myo4p, and She3p interact with Ash1 mRNA in vivo. Bertrand et al. (8) also demonstrated colocalization of Myo4p and She3p with MS2-Ash1 mRNA particles, but reported that She2p did not colocalize.
Figure 2.
She2p, She3p, and Myo4p colocalize with the U1A-ASH1 RNA particle. Thirteen repeats of the c-myc peptide sequence were inserted at the C terminus of SHE2, SHE3, MYO4, SHE4, and BNI1 in wild-type cells (w303). All samples express both U1Ap-GFP and U1Atag-ASH1. In untagged cells, the GFP fluorescence from the U1A-ASH1 RNA particle is visible at the distal tip of the bud but no staining is detected with the anti-myc antibody. In SHE2-MYC, SHE3-MYC, and MYO4-MYC cells, the GFP fluorescence from the U1A-ASH1 RNA particle colocalizes with the anti-myc immunofluorescence. In SHE4-MYC and BNI1-MYC cells, the U1A-ASH1 RNA GFP particle is visible at the distal tip of the bud but does not colocalize with the anti-myc fluorescence.
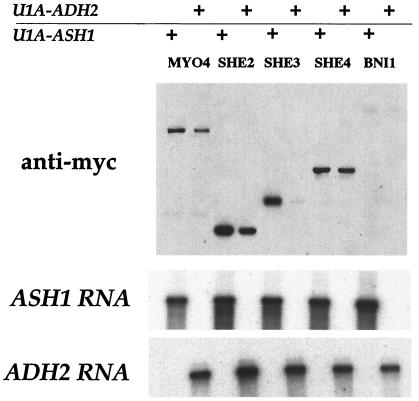
We next investigated whether U1Atag-ASH1 RNA can be used as a system for biochemical identification of Ash1 mRNA-associated proteins. To accomplish this, U1Atag-ASH1 RNA complexed with U1Ap-GFP was immunoprecipitated from whole-cell extracts with affinity-purified anti-GFP antibodies coupled to protein A agarose; the immunoprecipitate was then assayed for ASH1 mRNA by Northern analysis, and the presence of each of the myc-tagged She proteins was analyzed by immunoblotting. As a negative control, U1Atag-ADH2 RNA complexed with U1Ap-GFP was immunoprecipitated. She3p was clearly associated with U1Atag-ASH1 RNA but not U1Atag -ADH2 RNA, indicating that it is a bona fide member of an Ash1 ribonucleoprotein complex (Fig. 3). She2p and Myo4p were found in immunoprecipitates of both U1Atag-ASH1 and U1Atag-ADH2 RNAs, although there was reproducibly greater amounts of both proteins associated with U1Atag-ASH1 RNA (Fig. 3). In contrast, She4p associated equally with U1Atag-ASH1 and U1Atag-ADH2 RNAs, and Bni1p was not detected in the immunoprecipitate of either RNA. The reason for the high nonspecific background association of some She proteins with the control U1Atag-ADH2 mRNA was not clear, and efforts to significantly reduce this nonspecific binding were not successful. However, these results, taken together with the in vivo colocalization data, suggest that She2p, She3p, and Myo4p specifically associate with Ash1 mRNA, whereas Bni1p and She4p do not.
Figure 3.
Immunoprecipitation of U1A-ASH1 RNA reveals an association between Ash1 mRNA and Myo4p, She2p, and She3p. Extracts were prepared from SHE-MYC cells expressing either U1Ap-GFP and U1Atag-ASH1 or U1Ap-GFP and U1Atag-ADH2. Extracts were incubated with anti-GFP antibodies coupled to protein A agarose. Immunoprecipitates were analyzed for the She proteins by Western blot and for U1A-ASH1 RNA and U1A-ADH2 RNA by Northern analysis. The Western blot was quantitated with National Institutes of Health image. The -fold increase of each She protein in U1A-ASH1 RNA immunoprecipitates compared with U1A-ADH2 RNA immunoprecipitates is Myo4p, 1.5; She2p, 1.5; She3p, 3.3; She4p, 1.1; and Bni1p, 1.1.
Immunoprecipitation of She Proteins.
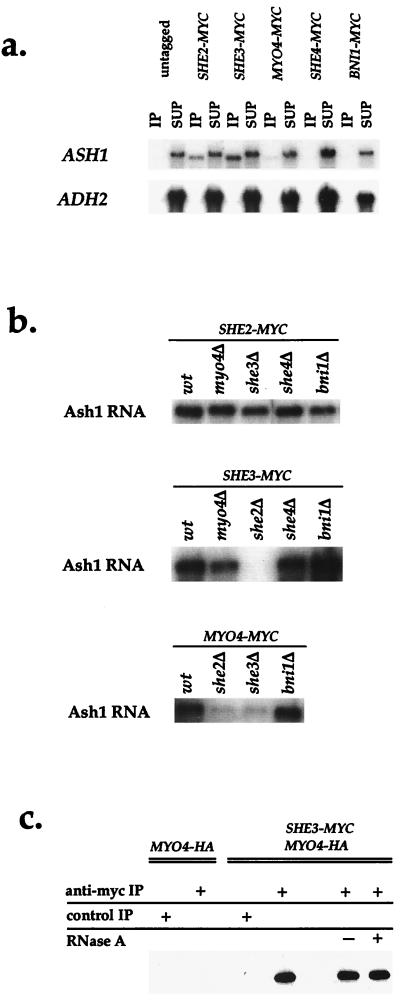
Because of the high nonspecific background in some of the RNA immunoprecipitations, we immunoprecipitated She-myc proteins and analyzed them for endogenous Ash1 or Adh2 mRNA. To detect the mRNA, we used Northern analysis, because the signal is more linear with RNA concentration than reverse transcription–PCR. As shown in Fig. 4a, both She2p and She3p immunoprecipitates contained Ash1 but not Adh2 mRNA. A small, but specific, Ash1 mRNA signal was also detected in Myo4p immunoprecipitates, and this was particularly apparent if a larger portion of the immunoprecipitate was analyzed (Fig. 4b and data not shown). One possible reason for the lower amount of Ash1 mRNA in Myo4p immunoprecipitates is that the binding of the anti-myc antibody to the C-terminal tag may interfere with Ash1 mRNA association. In contrast, neither She4p nor Bni1p coimmunoprecipitated with Ash1 mRNA. These results confirm that only She2p, She3p, and Myo4p associate with Ash1 mRNA.
Figure 4.
Analysis of the interactions between the She proteins and Ash1 mRNA. (a) She2p, She3p, and Myo4p associate with Ash1 mRNA. She-myc proteins were immunoprecipitated and analyzed for Ash1 mRNA and Adh2 mRNA by Northern blot. (b) Dependence on the other SHE genes for the interaction between She2p, She3p, Myo4p, and Ash1 mRNA. SHE2, SHE3, and MYO4 were c-myc tagged in sheΔ cells, and the proteins were immunoprecipitated and analyzed for Ash1 mRNA by Northern blot. (c) She3p and Myo4p form a RNase-insensitive complex. Cells were HA-tagged at MYO4 and c-myc tagged at SHE3. Extracts were prepared from MYO4-HA cells and MYO4-HA/SHE3-MYC cells and incubated with anti-myc antibodies coupled to protein G Sepharose or protein G Sepharose alone as a control. Extracts prepared from MYO4-HA/SHE3-MYC cells were either mock treated or incubated with RNase A. The immunoprecipitates were analyzed for the amount of Myo4p by Western blot.
We next investigated whether the association between Ash1 mRNA and the three She proteins depended on the presence of any of the other She proteins. She2p, She3p, and Myo4p were immunoprecipitated from strains that contained a deletion of a single SHE gene, and the immunoprecipitates were probed for Ash1 mRNA. She2p association with Ash1 mRNA was not affected by deletion of any of the four SHE genes (Fig. 4b). However, in she2Δ cells, both She3p and Myo4p displayed greatly reduced association with Ash1 mRNA. Furthermore, Myo4p association with Ash1 mRNA was significantly reduced in she3Δ cells. However, She3p still associated with Ash1 mRNA in myo4Δ cells. These results reveal that the She2p interaction with Ash1 mRNA is independent of the other She proteins, whereas both Myo4p and She3p require She2p to associate with Ash1 mRNA. Furthermore, Myo4p depends on She3p for its association with Ash1 mRNA, whereas the converse is not true.
Finally, we examined interactions between She2p, She3p, and Myo4p in strains in which two of three proteins were tagged with either the c-myc or HA epitope. After immunoprecipitating one protein with anti-myc antibodies coupled to protein G Sepharose, the presence of the second protein was evaluated by immunoblotting with anti-HA antibodies (Fig. 4c). Myo4p was clearly detected in immunoprecipitates of She3p, and this interaction was not diminished in ash1Δ cells, by excess RNase A treatment or in she2Δ cells, indicating that the association between She3p and Myo4p does not require Ash1 mRNA or She2p (data not shown and Fig. 4c). On the other hand, whereas a small amount of Myo4p was detected in immunoprecipitates of She2p, this interaction was reduced in ash1Δ cells (data not shown). Thus, She3p and Myo4p interact in the absence of Ash1 mRNA, whereas She2p and Myo4p form a weaker, Ash1 mRNA-dependent association.
Discussion
By using three different techniques, we show that She2p, She3p, and Myo4p associate with Ash1 mRNA. First, each of these proteins colocalized with a U1Atag-ASH1 RNA GFP particle in vivo. Second, immunoprecipitation of U1Atag-ASH1 RNA brought down greater amounts of these proteins when compared with U1A-ADH2 RNA immunoprecipitation. Finally, Ash1 mRNA coimmunoprecipitated with each of these three She proteins from wild-type cells. In contrast, we find no evidence that Bni1p and She4p associate with Ash1 mRNA. Both Bni1p (6) and She4p (15) affect the polarity and organization of actin cytoskeleton. Because Ash1 mRNA localization requires an intact actin cytoskeleton (4, 5), it is likely that these two proteins affect Ash1 mRNA localization indirectly through their effects on actin.
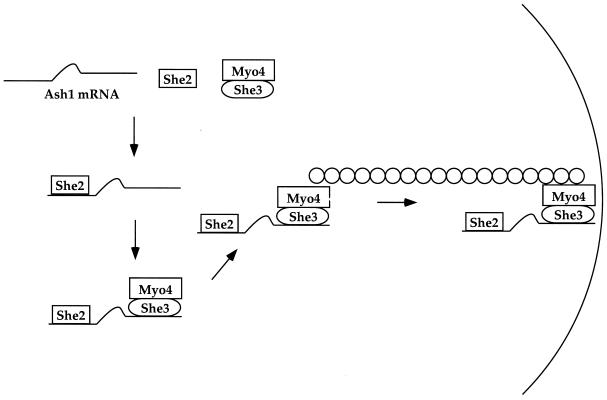
Deletion analyses, in combination with the biochemical techniques described above, suggest a model of how She2p, She3p, and Myo4p assemble onto Ash1 mRNA (Fig. 5). Of these three proteins, we believe that She2p binds first, because its association with Ash1 mRNA does not require any of the other She proteins. In contrast, She3p and Myo4p require She2p for their association with Ash1 mRNA, indicating that they bind subsequent to She2p. The interaction between Myo4p and Ash1 mRNA requires She3p, but She3p does not require Myo4p to associate with Ash1 mRNA. This result indicates that Myo4p binds to Ash1 mRNA via She3p. Furthermore, She3p/Myo4p appears to bind as a preexisting complex to Ash1 mRNA, because these two proteins coimmunoprecipitate in the absence of Ash1 mRNA. In summary, She2p binds Ash1 mRNA, which allows a She3p/Myo4p complex to dock. Once bound, Myo4p transports Ash1 mRNA to the distal tip of the bud.
Figure 5.
A model of Ash1 mRNA localization. She2p binds Ash1 mRNA which then allows the She3p/Myo4p complex to bind. She3p mediates the interaction between Ash1 mRNA and Myo4p. Myo4p transports the Ash1 ribonucleoprotein complex along actin filaments to the distal tip of the daughter cells.
Our results indicate that She2p is a key protein in the assembly of the Ash1 ribonucleoprotein complex. Although She2p is needed to dock She3p/Myo4p onto the mRNA, the mechanism is unknown, because our results do not reveal a direct interaction between these proteins; rather they appear to require Ash1 mRNA to facilitate their association. One possibility is that She2p binding to Ash1 mRNA induces a change in the RNA that enables She3p/Myo4p to associate. Alternatively, Ash1 mRNA may induce a conformational change in She2p, allowing it to recruit She3p/Myo4p.
Our result also shows that She3p acts as an adapter protein that directly binds to the myosin motor (Myo4p) and links it to Ash1 mRNA. Munchow et al. (9) also found that She3p is required for the interaction between Myo4p and Ash1 mRNA; however, they did not show that She3p and Myo4p associate to form a stable complex. The only other known adapter that links a motor to its cargo is dynactin, a 20S multisubunit complex that interacts with cytoplasmic dynein (16–18). Dynactin is important for docking dynein onto various types of cargo, including the Golgi apparatus (19), lysosomes (20), and kinetochores (21). Similarly, it will be interesting to establish whether She3p serves as a general Myo4p adapter for various cargo besides Ash1 mRNA. In support of this idea, both She3p and Myo4p colocalize to the bud before ASH1 expression (22). This result suggests that She3p/Myo4p transports other cargo to the growing bud, although she3Δ and myo4Δ strains have, as yet, no detectable phenotype other that Ash1 mRNA mislocalization. The cargo transported by She3p/Myo4p, however, may not be essential for growth, as is the case for Ash1 mRNA. Further analysis of how She3p interacts with Myo4p and how She3p/Myo4p binds the Ash1 ribonucleoprotein complex provides an excellent opportunity for understanding how a motor protein recognizes its cargo.
Acknowledgments
We thank Michael Rosbash and Bruno Charpentier for suggesting the use of U1A in our experiments. We thank Ira Herskowitz, Anita Sil, Kenji Irie, and Catherine Takizawa for helpful discussions. This work was supported in part by National Institutes of Health Grant 38496 (R.D.V.) and a grant from the Jane Coffin Childs Memorial Fund for Medical Research Fellowship (P.A.T.).
Abbreviations
- GFP
green fluorescent protein
- UTR
untranslated region
- HA
hemagglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080585897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080585897
References
- 1.Wilhelm J E, Vale R D. J Cell Biol. 1993;123:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.St Johnston D. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 3.Hazelrigg T. Cell. 1998;95:451–460. doi: 10.1016/s0092-8674(00)81613-x. [DOI] [PubMed] [Google Scholar]
- 4.Takizawa P A, Sil A, Swedlow J R, Herskowitz I, Vale R D. Nature (London) 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 5.Long R M, Singer R H, Meng X, Gonzalez I, Nasmyth K, Jansen R P. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 6.Evangelista M, Blundell K, Longtine M S, Chow C J, Adames N, Pringle J R, Peter M, Boone C. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 7.Haarer B K, Petzold A, Lillie S H, Brown S S. J Cell Sci. 1994;107:1055–1064. doi: 10.1242/jcs.107.4.1055. [DOI] [PubMed] [Google Scholar]
- 8.Bertrand E, Chartrand P, Schaefer M, Shenoy S M, Singer R H, Long R M. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 9.Munchow S, Sauter C, Jansen R P. J Cell Sci. 1999;112:1511–1518. doi: 10.1242/jcs.112.10.1511. [DOI] [PubMed] [Google Scholar]
- 10.Allain F H, Gubser C C, Howe P W, Nagai K, Neuhaus D, Varani G. Nature (London) 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- 11.Longtine M S, McKenzie A, III, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 12.Beach D L, Salmon E D, Bloom K. Curr Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- 13.Chartrand P, Meng X H, Singer R H, Long R M. Curr Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez I, Buonomo S B, Nasmyth K, von Ahsen U. Curr Biol. 1999;9:337–340. doi: 10.1016/s0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]
- 15.Wendland B, McCaffery J M, Xiao Q, Emr S D. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill S R, Schroer T A, Szilak I, Steuer E R, Sheetz M P, Cleveland D W. J Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karki S, Holzbaur E L. J Biol Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- 18.Vaughan K T, Vallee R B. J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holleran E A, Tokito M K, Karki S, Holzbaur E L. J Cell Biol. 1996;135:1815–1829. doi: 10.1083/jcb.135.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burkhardt J K, Echeverri C J, Nilsson T, Vallee R B. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Echeverri C J, Paschal B M, Vaughan K T, Vallee R B. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansen R P, Dowzer C, Michaelis C, Galova M, Nasmyth K. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]