Abstract
The genome project of the blacklegged tick, Ixodes scapularis provides sequence data for testing gene function and regulation in this important pathogen vector. We tested Sleeping Beauty (SB), a Tc1/mariner group transposable element, and cationic lipid-based transfection reagents for delivery and genomic integration of transgenes into I. scapularis cell line ISE6. Plasmid DNA and dsRNA were effectively transfected into ISE6 cells and they were successfully transformed to express a red fluorescent protein (DsRed2) and a selectable marker, neomycin phosphotransferase (NEO). Frequency of transformation was estimated as 1 transformant per 5,000–10,000 cells and cultures were incubated 2 to 3 months in medium containing the neomycin analog G418 in order to isolate transformants. Genomic integration of the DsRed2 transgene was confirmed by inverse PCR and sequencing that demonstrated a TA nucleotide pair inserted between SB inverted/direct repeat sequences and tick genomic sequences, indicating that insertion of the DsRed2 gene into the tick cell genome occurred through the activity of SB transposase. RNAi using dsRNA transcribed from the DsRed2 gene silenced expression of red fluorescent protein in transformed ISE6 cells. SB transposition in cell line ISE6 provides an effective means to explore the functional genomics of I. scapularis.
Keywords: Ixodes scapularis, transformation, cell line, RNAi, Sleeping Beauty
1. Introduction
The blacklegged tick, Ixodes scapularis, transmits viruses, bacteria and protozoa (Munderloh et al., 2005), but despite its medical importance, the genetic basis of its vector competence remains poorly defined and methods for genetic analysis and manipulation are limited. This stems partly from the length of the life cycle spanning at least half a year under laboratory conditions, and from difficulties in establishing genetic maps (Ullmann et al., 2003). To better understand the genetic basis of pathogen transmission by I. scapularis, several expressed sequence tag (EST) libraries have been generated (Valenzuela et al., 2002; Ribeiro et al., 2006) and its genome is being sequenced and annotated (Van Zee et al., 2007). The 2.1 Gbp genome contains an abundance of repetitive DNA (approximately 21.5%) complicating assembly (Van Zee et al., 2007). Likewise, regulatory elements (e.g., promoters) have not been identified. The genome project has yielded an abundance of sequences that can be linked with EST data but to exploit these resources, methods for testing gene function and regulation in a native environment are needed. These analyses would be facilitated by experimental methods for transfection and transformation of I. scapularis cells.
Our goal is to develop a genetic manipulation system for I. scapularis cells. I. scapularis cell lines (Munderloh et al., 1994), in lieu of ticks, offer the potential to link genomic data with the genetic basis of tick-pathogen interactions. Line ISE6 has been extensively used to examine I. scapularis cellular interactions with a wide range of tick-associated microorganisms (Munderloh et al., 1999; Mattila et al., 2007). Transposon-based vectors belonging to the Tc1/mariner superfamily are widely used to introduce transgenes into animal cells. Sleeping Beauty (SB) is a broad host range transposon vector that was reconstructed from inactive Tc1/mariner elements in the genome of teleost fish (Ivics et al., 1997). It possesses two transposase binding sites in each inverted repeat, unlike other Tc1 and mariner elements, enhancing transposition (Izsvák et al., 2000). Although the applicability of SB to the transformation of arthropod cells is unknown, SB has been used with considerable success with a wide variety of vertebrate cells including fish, amphibian, bird and mammalian and does not appear to require species-specific factors for transposition. Previously, Rhipicephalus (Boophilus) microplus cell line (BME26) was successfully transfected a using a plasmid that contained a red fluorescent protein reporter gene within the SB transposon and a SB transposase gene outside the transposable element (i.e. cis complementation) (Esteves et al., 2008). The BME26 cells transiently expressed red fluorescent protein but stably transformed cells were not obtained, as a method to select for stable transformants was not applied. In this paper we describe experimental transfection and selection of transformed ISE6 cells using the SB system.
2. Materials and methods
Ixodes scapularis cell line ISE6 was maintained at 34 °C as described (Munderloh et al., 1999).
Transfection frequency was measured using plasmid pT/CAGGS-DsR//CMV-SB having both SB transposon and transposase functions (Sauer et al., 2004). The SB transposase gene (version 10), with the cytomegalovirus (CMV) promoter, resided outside of the transposable element containing the red fluorescent protein reporter gene DsRed2 with a CAGGS promoter (Fig. S1A). Cultures containing 1–5×105 cells/mL were transfected following the manufacturer’s protocol (Qiagen, Valencia, CA, USA) for adherent cells. At selected times after transfection, cultures were examined by fluorescence microscopy and the number of DsRed2 expressing cells per field in 10 randomly selected fields was determined. Data were expressed as the average number of DsRed2 positive cells per cm2 of culture surface ± SD.
We used pT/SVNeo encoding the neomycin phosphotransferase-resistance gene (neo) and SV40 promoter within a SB transposon, together with pCMVSB encoding the SB10 transposase gene and CMV promoter to select for I. scapularis cells with SB mediated transgene integration (Ivics et al., 1997). During prolonged exposure to G418, neomycin analog, only cells that incorporated the neo gene into their genome survived while non-transfected or transiently transfected are eliminated. The helper plasmid expressing SN transposase was either pCMV-SB10 or pT/CAGGS-DsR//CMV-SB. Maps and information about plasmid constructs are available at http://www.cbs.umn.edu/labs/perry/.
Cultures in 6-well plates (35 mm diameter, 1.6 ml per well) were seeded with 1×106 cells per well, with each well receiving 0.4 µg plasmid DNA, 3.2 µl enhancer and 10 µl Effectene Reagent. During 4 to 12 days after transfection the cultures were fed 500 µg G418 per ml of medium to select for resistant cells, and subsequently, the concentration was increased to 1 mg/ml. Plates were incubated in humidified candle jars at 34 °C for 30 to 60 days, and colonies in were visualized and imaged using Dark Reader Transilluminator (Clare Chemical Research, Dolores, CO, USA). The number of colonies indicated efficiency of transformation, and transformants were cloned by transferring colonies to a flask or by serial dilution of cell populations in 96-well plates, using wild-type, untransformed ISE6 cells as feeder layers. Wells containing red fluorescent cells were transferred to flasks two weeks later, and after another 1 – 2 months, wild-type ISE6 cells were eliminated by treatment with 1 mg/ml of G418.
Flow cytometry at a wavelength of 488 nm (FACS Calibur and FACS DiVa, Becton-Dickinson, San Jose, CA, USA) was used to determine cell size and granularity along with the level of red fluorescence to characterize transformed ISE6 cell populations. Data were analyzed using CellQuest Pro software (Becton-Dickinson, Trenton, NJ). Control (untransformed) ISE6 cells were used to adjust for cell size (forward scatter; FSC) and granularity (side scatter; SSC) so that the majority of cells formed a single cloud in the center of the scatter plot. A total of 50,000 events were recorded per sample. Histograms of red fluorescence (FL2) were prepared by selectively gating on the region of the scatter plot containing intact cells and plotting fluorescence against cell numbers. One region of non-fluorescent cells (M1) was established according to the endogenous level of autofluorescence present in wild-type ISE6 cells. Populations of DsRed2 positive cells (M2 and M3) were identified by their fluorescence, and regions were set to correspond with these populations.
Inverse-PCR demonstrated insertion of SB transposons into the genome. Tick genomic sequences flanking the 5’ transposon insertion sites in transformed cells were isolated using a primer with a biotin affinity tag (Fig. S1B). To sequence the 5’ end of the inserted transposon, biotin-labeled primer BPE (CTCCATATATGGGCTATGAACTAATGACCCCGTAATTG) that annealed just downstream from a SpeI restriction endonucleases site, was extended using DNA polymerase (Pfu Ultra, Stratagene). Primer extension products were bound to streptavidin coated magnetic beads (Dynal Dynabeads, Invitrogen, Carlsbad, CA) to separate them from tick genomic DNA and subsequently cleaved from the beads using SpeI and XbaI. Those enzymes digested genomic DNA internally within the transposon, just after the left IR/DR region and within the CMV-1E\enhancer region and also within unknown tick DNA sequences. Restriction enzymes were heat inactivated (65 °C for 10 min) and the digests diluted in ligation mixture (T4 ligase, at 16 °C) to circularize the PCR products. Circularized DNA was PCR amplified using primers specific for the IR/DR-L region of the transposon. The first round PCR used primers 8b (GGCAAGTCAGTTAGGACATCTACTTTGTGC) and 17b (GTTAACAAGAAATTTGTGGAGTAGTTGAAAAACGAG), and second round nested PCR used primers 9b (AATTCCAGTGGGTCAGAAGTTTACATACACTAAG) and 16b (TGAAAAACGAGTTTTAATGACTCCAACTTAAGTG). The PCR products were separated by agarose gel electrophoresis and selected bands were cloned and sequenced. The transgene integration site was determined using the assembled I. scapularis genome sequence. Sequences flanking the inserted SB transposons were used as query sequences in a BLAST search (blastn) of the I. scapularis Genomic Supercontigs hosted at VectorBase (http://www.vectorbase.org/index.php).
We silenced expression of DsRed2 transgene in transformed cells using RNAi. To make dsRNA, the fluorescent protein genes DsRed2 and GFPuv (Clontech, Mountain View, CA, USA) were cloned into the Litmus 28i vector and transcribed usingthe HiScribeTM RNAi Transcription Kit (New England BioLabs, Beverly, MA, USA).Vectors pDsRed2 and pGFPuv (Beckton Dickson, Palo Alto, CA, USA) were digested with XbaI and EcoRI with XbaI, respectively, cleaving at sites flanking the full-length reporter gene sequences (670 nt). Both digestion products were agarose gel-purified, cloned into the Litmus vector, cleaved with the same enzymes, and transcribed in forward and reverse orientations according to the transcription kit protocol. The RNAs were purified by LiCl precipitation, heat denatured and annealed by slow cooling, and quantified by UV spectrophotometry. Aliquots were electrophoresed on agarose gels to confirm production of full-length double-stranded RNAs. Transformed cultures were transfected with 0.07 nM, 0.7 nM, or 7 nM DsRed2-specific dsRNA per culture, and compared against cultures transfected with 7 nM GFP dsRNA or untreated cultures. Cells were examined by flow cytometry as described above. Changes in fluorescence were determined by comparing the population of cells remaining in the region M3 and the geometric mean fluorescence of cells in the gated region. After addition of dsRNA, cultures were imaged and subsequently examined by flow cytometry as given above. Data were analyzed by comparing the proportion of dsDsRed2-transformed cells within regions M2 and M3 relative to the proportion of autofluorescent cells in M1. Similar comparisons were made for cultures treated with dsGFPuv dsRNA and transfection buffer (no dsRNA).
3. Results
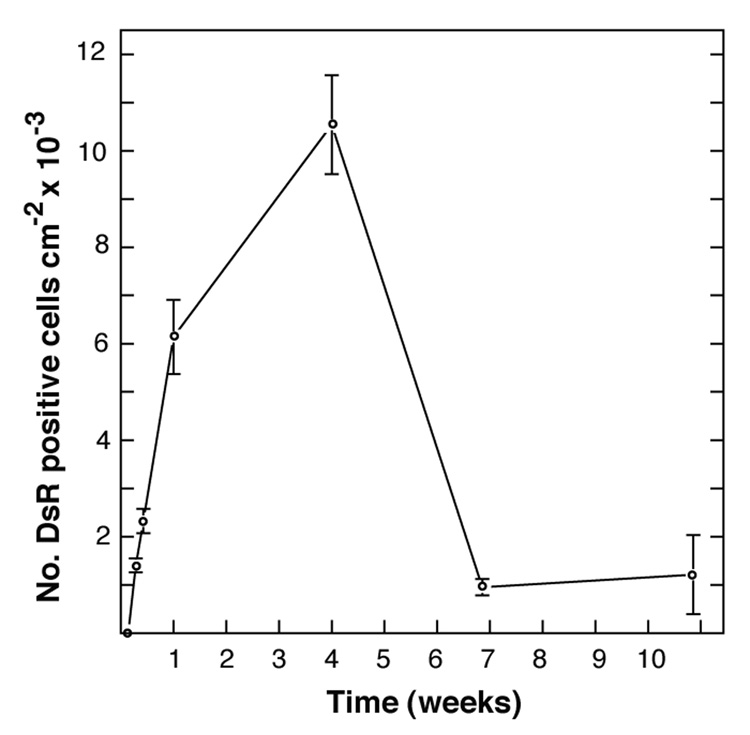
We used pT/CAGGS-DsR//CMV-SB, a single-plasmid SB gene transfer-expression vector (Sauer et al., 2004), to evaluate promoter activity, transgene expression and transformation frequency in transfected ISE6 cells. The ISE6 cell line, developed from embryos of I. scapularis, comprises adherent, non-phagocytic cells (Mattila et al., 2007) with dendritic pseudopodial extensions (Fig. S2). Expression of DsRed2 was first noted by fluorescence microscopy 24 to 48h after transfection, clearly indicating that the CAGGS promoter was functional and the heterologous DsRed2 gene was a suitable reporter gene. Cells incubated with pT/CAGGS-DsR//CMV-SB in the absence of Effectene did not express DsRed2, indicating that the Effectene reagent was essential for mediating transfection. The most rapid increase in cells expressing red fluorescent protein occurred during the first week and by the end of 7 days up to 10% of the cells expressed DsRed2 (Fig. 1). Subsequently, the number of DsRed2 cells per cm2 increased slowly up to 4 weeks post-transfection. The doubling time of ISE6 cells is 4 to 5 days, corresponding to the approximate rate of increase in the number of DsRed2 cells per cm2 during this phase, and suggesting to us that after the first week the increase was a result of cell division rather than an increase in the proportion of cells expressing DsRed2. The decline in cells expressing DsRed2 after 4 weeks indicated the decay of DsRed2 expression in transiently transfected cells. Two months post-transfection, the overall proportion of DsRed2 expressing cells declined to less than 0.1%, and islands of stably transformed cells became visible that grew to comprise >100 cells. This preliminary experiment indicated that transfection of ISE6 cells with pT/CAGGS-DsR//CMV-SB led to stable transformation of a small number of ISE6 cells. This underscored the need for a method to select transformants.
Fig. 1.
Expression of DsRed2 in pT/CAGGS-DsR//CMV-SB-transfected ISE6 cells over time. Cultures (duplicate 12.5 cm2 flasks) were seeded with 5 × 105 cells/mL one week prior to transfection. At selected times post transfection with pT/CAGGS-DsR//CMV-SB cell layers were examined by fluorescence microscopy and the numbers of DsRed2 positive and negative cells observed in a given field of view were counted. Data points show the means and standard error of the means of 10 microscopic fields. A value of 1 × 104 DsRed2 cells per cm2 was equivalent to 5 × 104 cells / mL.
We used a two-plasmid design (Ivics et al., 1997) (pT/SVNeo and pCMVSB) to select ISE6 transformants stably resistant to the neomycin analog G418. Based on a cell viability assay, the IC50 of G418 for ISE6 cells was approximately 320 µg/ml and the IC95 was 1 mg/ml for ISE6 cells exposed to G418 for a week (data not shown). We initially exposed cells transfected with the neo gene to 0.5 mg G418/ml and gradually increased the dose to 1 mg/ml. Continued addition of G418 to the medium, starting 4–8 days after transfection, gradually eliminated transiently transfected and non-transfected cells. Cytotoxicity (cell detachment) induced by G418 was observed 1 to 2 weeks after its addition and colonies of adherent resistant ISE6 cells were apparent 30 to 60 days post-transfection (Fig. 2A). Colonies varied in size and each well (originally seeded with 106 cells) contained ~200 colonies indicating an approximate efficiency of transformation of 1: 5,000. No colonies of spontaneously resistant ISE6 cells appeared in control cultures incubated with G418, and there was no evidence for endogenous transposase that could mobilize the neo gene within the SB transposon when ISE6 cells were transfected with pT/SVNeo alone.
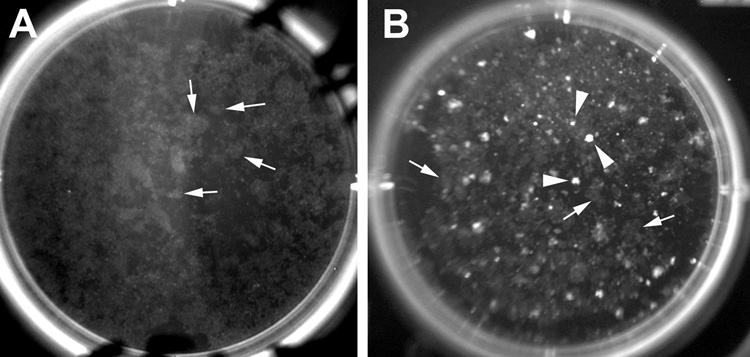
Fig. 2.
Colonies of G418-resistant ISE6 cells in 6-well culture dishes (7 weeks post G418 selection). (A) Colonies of live cells photographed using a transilluminator (Dark Reader) 60 days after co-transfection with pT/SVNeo and pCMV-SB. Note neomycin resistant colonies (grey) (white arrows). At this time colonies of G418-resistant cells had started to merge. (B) Colonies from cells co-transfected with pT/SVNeo and pT/CAGGS-DsR//CMV-SB. Note white colonies that were neomycin-resistant and DsRed2-positive (white arrowheads). No colonies were recovered on control plates (pT/SVNeo alone) (results not shown).
Our goal was to develop a method to select ISE6 transformants expressing specific transgenes. To obtain cells that expressed DsRed2 using G418 selection, we co-transfected ISE6 cells with pT/SVNeo and pT/CAGGS-DsR//CMV-SB. Under G418 selection two different phenotypes arose: G418-resistant cells that did not express DsRed2 (transformants that carried only the neo gene) and G418-resistant cells that expressed DsRed2 (transformants that carried both neo and DsRed2 transgenes) (Fig. 2B). Each well (seeded with 1 × 106 cells) yielded about 100 DsRed2-expressing colonies indicating an efficiency of transformation of 1 per 10,000 cells. After 4 weeks of exposure to G418, no colonies emerged in control cultures not transfected with plasmid DNA or transfected with a plasmid not carrying the neomycin resistance gene (pT/CAGGS-DsR//CMV-SB or pCMV-SB10).
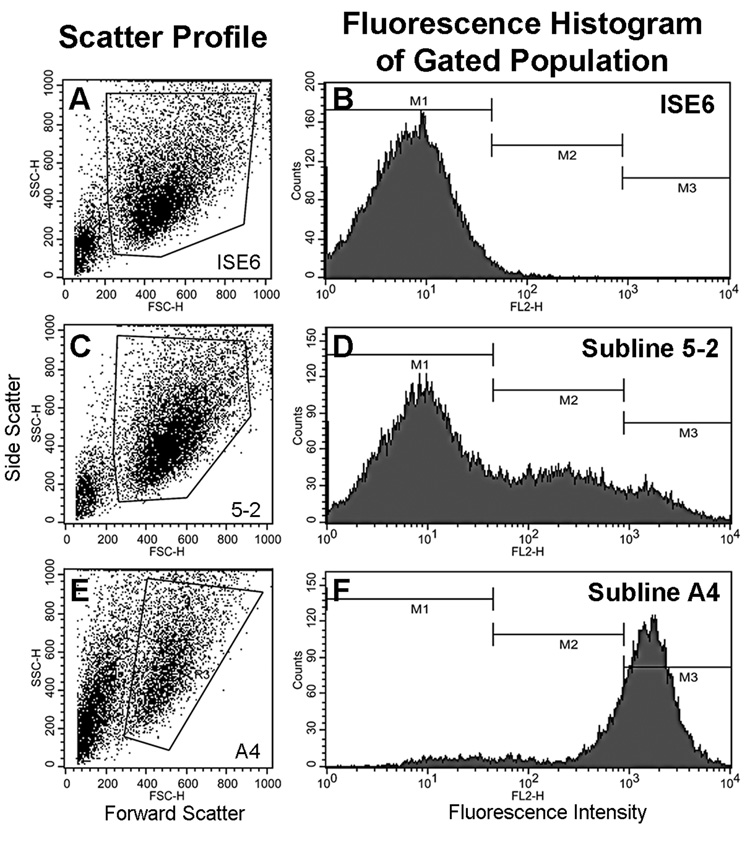
Subline 5-2 derived from a G418-resistant culture interspersed with DsRed2-positive colonies of different sizes and fluorescence was characterized by flow cytometry 70 days post transfection with pTSVNeo and pT/CAGGS-DsR//CMV-SB. Cell size or granularity (Figure 3 A, C and E, scatter profiles) were unchanged, but fluorescence histogram data revealed a single peak of autofluorescent cells (Figure 3B, M1) in wild-type ISE6 (neo−DsRed2−), whereas the 5-2 subline had three populations: non-DsRed2 expressing neo-resistant cells (M1; neo+ DsRed2−) comprised ~64% of the cells, intermediate-level DsRed2 expressing cells (M2) constituted ~27% of cells, and the remaining 9% of the population were highly fluorescent (M3) (neo+, DsRed2+; Figure 3D) (Table S1). Inverse PCR and analysis of tick genomic DNA sequences flanking the integrated transposons (Fig. S1C) demonstrated that the DsRed2 gene had been spliced into different locations within the ISE6 genome (Fig. S1B). This conclusion is supported by multiple bands observed after gel electrophoretic separation of inverse PCR products (data not shown) amplified from genomic DNA of subline 5-2 which constitutes a heterogeneous population of transformed cells. Three distinct PCR product bands were cloned and included the predicted IR/DR-L sequences as well as the TA dinucleotide derived from the vector DNA originally flanking the transposon. In all three sequences, TA dinucleotides were found at the 5’ insertion site indicating Sleeping Beauty transposase had mediated the integrations. The transgene integration site was investigated using the first publicly available genome assembly of I. scapularis (Wikel strain) during 07–2008. Sequence 1 was 96.36% identical (2e–73) to a 165 bp region of scaffold DS664373 (254,775 bp) from basepairs 212,566–212,730. Sequence 2 yielded significant BLAST hits (>88% identity, ≤ 7e-52) to 23 different scaffolds, and sequence 3 had 91.04% identity (1e–168) with a 477 bp region of scaffold DS965390 (1,200 bp) from basepairs 721–1,197.
Fig. 3.
Flow cytometry of untransformed ISE6 and sublines 5-2 and A4 containing transformed cells expressing the red fluorescent protein DsRed2. Scatter profiles display the selective gating of events to exclude debris and aggregated cells from the fluorescence histograms. Region M1 includes autofluorescent cells, region M2 contains cells of low and intermediate fluorescence and region M3 includes the highly fluorescent cells.
We developed subline A4 comprised of DsRed2 expressing cells by serial dilution of the 5-2 subline. A4 tended to form clusters and was more fragile during resuspension and cytomtery procedures. After 5 serial transfers the proportion of cells expressing red fluorescent protein was 95% with 73% of the cells in the M3 region (Fig. 3F, Table S1).
We analyzed the ability of DsRed2 dsRNA to silence expression of red fluorescent proteins by transformed cells. Initially, when dsRNA (up to 30 nM) alone was added to complete culture medium, DsRed2 expression was reduced but not eliminated, and results were inconsistent between experiments (data not shown). As we had successfully used a cationic lipid-based transfection reagent to deliver plasmids into ISE6 cells, we chose RNAiFect Transfection Reagent to deliver dsRNA into A4 cells. The concentration of dsRNA required to maximally silence a target gene while minimally affecting expression of non-target genes varies from gene to gene, and there is little information describing the effect of dsRNA concentration on RNAi-induced gene silencing in ticks. We therefore evaluated different concentrations of dsRNA (0.07, 0.7 and 7 nM) on DsRed2 expression during a period of 21 days. Neither concentration produced an obvious effect on cell growth (Fig. S3, left two columns), but there was a measurable difference in the overall red fluorescence in A4 cells treated with 0.07 vs. 0.7 nM of dsRNA, as determined by flow cytometry. No further reduction in fluorescence was seen with 7 nM dsRNA (Fig. S3, right column), suggesting that higher concentrations of dsRNA do not enhance RNAi once a saturation threshold has been met.
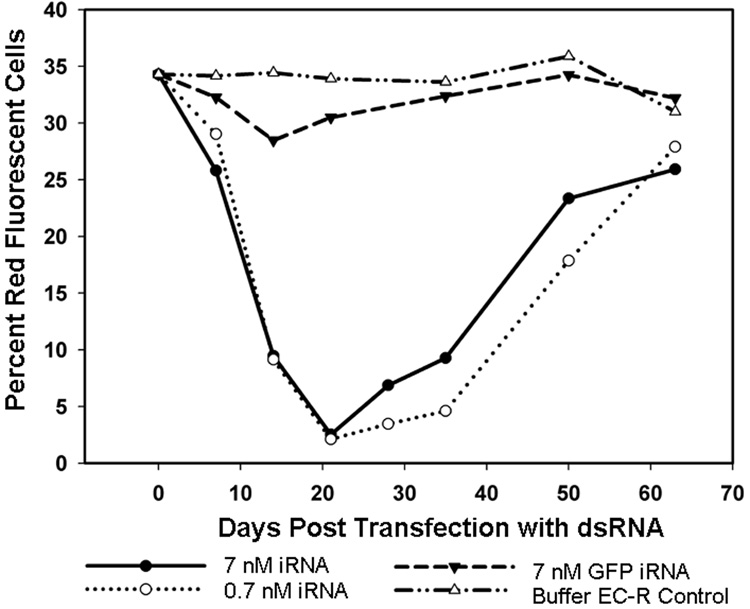
To examine the effect of dsRNA on DsRed2 expression over time, subline 5-2 was transfected with DsRed2-specific dsRNA (0.7 and 7 nM), while controls received GFPuv-specific dsRNA (7 nM) or transfection-buffer (EC-R) alone without dsRNA. Cultures were examined by flow cytometry to determine the proportion of cells that expressed DsRed2. In dsRNA transfected DsRed2-cultures, red fluorescence declined within seven days of transfection as determined by the leftward shift in the fluorescence profile (data not shown). The lowest levels of fluorescence were noted on day 21 post-transfection (PT, Fig. 4), with 2.5% of cells retaining sufficient fluorescence to be placed within histogram regions M2 and M3. Approximately 84% fewer cells retained fluorescence in the DsRed2-dsRNA transfected cultures relative to the proportion of fluorescent cells in controls receiving buffer alone. Early indications of recovery were seen 28 days PT, and by 63 days PT, fluorescence levels were similar to control cultures. The fluorescence of cells transfected with GFPuv-dsRNA declined slightly in the early time points PT, suggesting transfection with GFPuv-dsRNA may have had negative effects on non-target processes within the cells when used at this concentration. DsRed2 expression was unchanged in buffer-only transfection controls. Cellular characteristics such as morphology and adherence, as well as parameters assessed by flow cytometry (cell size and granularity) were comparable in all cultures.
Fig. 4.
Effect of dsRNA on expression of DsRed2 over time in ISE6 subline 5-2. Data points indicate the percentage of DsRed2 expressing cells within the gated M2 and M3 populations (see Fig. 3, histogram D).
4. Discussion
Tick cell lines provide tractable models for the development of transfection and transformation methods and have advantages over using ticks for functional genomics. Several I. scapularis cell lines (Munderloh et al., 1994) are available for the identification of transposition systems that are active in tick cells. A variety of transfection and transformation methods and reagents are available, but as we know so little about the molecular genetics of ticks, we adapted a transformation system used with other eukaryotic cells. Transformation systems for insects were developed by testing combinations of expression vectors, transposases, transposons, promoters, reporter genes, and methods for introduction of the DNA for their activity in insects or cell lines (Huynh and Zieler, 1999; Horn et al., 2002), which yielded results that cannot easily be compared. Similarly, vertebrate transformation systems used in different laboratories produced divergent results, and careful in vitro comparisons are still required for making rational choices (Wu et al., 2006). Because mariner transposition systems are widely and successfully used in both prokaryotic and eukaryotic cells (Izsvák et al., 2000; Felsheim et al., 2006), we examined the potential of the SB gene transfer system to function in I. scapularis cells. The SB system, based on a transposase that was reconstructed from an ancestral Tc1/mariner fish element (Ivics et al., 1997), has been used with a wide variety of vertebrate cells (Izsvák et al., 2000) but its functionality in arthropod cells has not been reported. Our results demonstrate that the SB transposition system can be used to transform I. scapularis cells, although transposase activity may have been limited by host-specific factors. BLAST searches against the I. scapularis genome sequences that have been assembled into scaffolds with regions flanking the inserted transposons confirmed that SB transposons were inserted into the I. scapularis genome. These genomic supercontigs have not yet been mapped to I. scapularis chromosomes, representing a prospect for future research.
Interfering RNA has been introduced into ticks for analysis of gene function (de la Fuente et al., 2007) at nanomolar concentrations of dsRNA similar to those used in this study. dsRNA silenced DsRed2 expression in transformed I. scapularis cells for 21 – 28 days before signs of restoration of DsRed2 expression became apparent. That dsRNA is stable for over 3 weeks correlates with our observation that transfecting plasmids that were not translocated to the genome persisted and were active in ISE6 cells for up to 4 weeks, but further studies are needed to confirm this. Our results demonstrate that standard protocols and reagents for RNAi, and cytometric techniques used in better-characterized systems (Agaisse et al., 2005), are useful tools for investigating functional genomics of tick cells in vitro.
The ISE6 line offers a means for functional characterization of tick genes and their promoters in a native cellular environment. Previously, functional analysis of tick genes has relied on the use of bacterial, mammalian or insect cell based expression systems (Alarcon-Chaidez et al., 2003; Chen et al., 2004). The development of transfection and transformation protocols for I. scapularis cell lines is an important first step towards introducing DNA into target cells and identifying conditions required for genomic integration and stable transformation. Furthermore, stable expression of DsRed2 and neomycin phosphotransferase genes indicated that the promoters used to drive these genes were functional in the tick cells. The prolonged survival of cells that transiently expressed the neo gene provided a helper effect that supported the growth of cells that had incorporated the neo gene into their genomes. Established I. scapularis cell lines provide a useful tool to overcome the limitations and difficulties of working with ticks on subjects such as tick-borne pathogens, and this study demonstrates their potential for functional tick genomics. The sequenced I. scapularis genome will help to identify more efficient promoters and transposable elements that can be used instead of those reported here, and will make this model system more applicable to the discovery of new pathways and biological processes important to ticks.
Supplementary Material
Acknowledgments
This research was supported by Public Health Service Grants R01 AI 042792 and AI49424 from the National Institutes of Health to UGM and in part by state funds from the Minnesota Agriculture Experiment Station.
Appendix A. Supplementary materials
Supplementary data associated with this article can be found in the online version at doi:........
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 2005;309:1248–1251. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- Alarcon-Chaidez FJ, Müller-Doblies UU, Wikel SK. Characterization of a recombinant immunomodulatory protein from the salivary glands of Dermacentor andersoni. Parasite Immunol. 2003;25:69–77. doi: 10.1046/j.1365-3024.2003.00609.x. [DOI] [PubMed] [Google Scholar]
- Chen A, Holmes SP, Pietrantonio PV. Molecular cloning and Functional expression of a serotonin receptor from the Southern cattle tick, Boophilus microplus (Acari: Ixodidae) Insect Mol. Biol. 2004;13:45–54. doi: 10.1111/j.1365-2583.2004.00457.x. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Kocan KM, Almazan C, Blouin EF. RNA interference for the study and genetic manipulation of ticks. Trends Parasitol. 2007;23:427–433. doi: 10.1016/j.pt.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Esteves E, Lara FA, Lorenzini DM, Costa GHN, Fukuzawa AH, Pressinotti LN, Silva JRMC, Ferro JA, Kurtti TJ, Munderloh UG, Daffre S. Cellular and molecular characterization of an embryonic cell line (BME26) from the tick Rhipicephalus (Boophilus) microplus. Insect Biochem. Mol. Biol. 2008;38:568–580. doi: 10.1016/j.ibmb.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsheim RF, Herron MJ, Nelson CM, Burkhardt N, Barbet AF, Kurtti TJ, Munderloh UG. Transformation of Anaplasma phagocytophilum. BMC Biotechnol. 2006;6:42. doi: 10.1186/1472-6750-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn C, Schmid BGM, Pogoda FS, Wimmer EA. Fluorescent transformation markers for insect transgenesis. Insect Biochem. Mol. Biol. 2002;32:1221–1235. doi: 10.1016/s0965-1748(02)00085-1. [DOI] [PubMed] [Google Scholar]
- Huynh CQ, Zieler H. Construction of modular and versatile plasmid vectors for the high-level expression of single or multiple genes in insects and insect cell lines. J. Mol. Biol. 1999;288:13–20. doi: 10.1006/jmbi.1999.2674. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH, Izsvák S. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Izsvák S, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- Mattila JT, Munderloh UG, Kurtti TJ. Phagocytosis of the Lyme disease spirochete, Borrelia burgdorferi, by cells from the ticks, Ixodes scapularis and Dermacentor andersoni, infected with an endosymbiont, Rickettsia peacockii. J. Insect Science. 2007;7:58. doi: 10.1673/031.007.5801. (available online: insectscience.org/57.58) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munderloh UG, Liu Y, Wang M, Chen C, Kurtti TJ. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J. Parasitol. 1994;80:533–543. [PubMed] [Google Scholar]
- Munderloh UG, Jauron SD, Fingerle V, Leitritz L, Hayes SF, Hautman JM, Nelson CM, Huberty BW, Kurtti TJ, Ahlstrand GG, Greig B, Mellencamp MA, Goodman JL. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J. Clin. Microbiol. 1999;37:2518–2524. doi: 10.1128/jcm.37.8.2518-2524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munderloh UG, Jauron SD, Kurtti TJ. The tick: a different kind of host for human pathogens. In: Goodman JL, Dennis D, Sonenshine DE, editors. Tick-Borne Diseases of Humans. Washington, D. C.: ASM Press; 2005. pp. 37–64. [Google Scholar]
- Ribeiro JMC, Alarcon-Chaidez FJ, Francischetti IMB, Mans BJ, Mather TN, Valenzuela JG, Wikel SK. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem. Mol. Biol. 2006;36:111–129. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Sauer MG, Ericson ME, Weigel BJ, Herron MJ, Panoskaltsis-Mortari A, Kren BT, Levine BL, Serody JS, June CH, Taylor PA, Blazar BR. A novel system for simultaneous in vivo tracking and biological assessment of leukemia cells and ex vivo generated leukemia-reactive cytotoxic T cells. Cancer Res. 2004;64:3914–3921. doi: 10.1158/0008-5472.CAN-03-3991. [DOI] [PubMed] [Google Scholar]
- Ullmann AJ, Piesman J, Dolan MC, Black WC., IV A preliminary linkage map of the hard tick. Ixodes scapularis. Insect Mol. Biol. 2003;12:201–210. doi: 10.1046/j.1365-2583.2003.00402.x. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IMB, Pham VM, Garfield MK, Mather TN, Ribeiro JMC. Exploring the sialome of the tick Ixodes scapularis. J. Exp. Biol. 2002;205:2843–2864. doi: 10.1242/jeb.205.18.2843. [DOI] [PubMed] [Google Scholar]
- Van Zee JP, Geraci NS, Guerrero FD, Wikel SK, Stuart JJ, Nene V, Hill CA. Tick genomics: The Ixodes genome project and beyond. Int. J. Parasit. 2007;37:1297–1305. doi: 10.1016/j.ijpara.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Wu SC-Y, J, M.Y. J, Coates CJ, Handler AM, Pelczar P, Moisyadi S, Kaminski JM. piggyBac is a flexible and highly active transposon as compared to Sleeping Beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl. Acad. Sci. USA. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.