Abstract
HIV infection and its outcome is complex because there is great heterogeneity not only in clinical presentation, incomplete clinical information of markers of immunodeficiency and in measurements of viral loads. Also, there many gene variants that control not only viral replication but immune responses to the virus; it has been difficult to study the role of the many AIDS restricting genes (ARGs) because their influence vary depending on the ethnicity of the populations studies and because the cost to follow infected individuals for many years. Nevertheless, at least genes of the major histocompatibility locus (MHC) such as HLA alleles have been informative to classify infected individuals following HIV infection; progression to AIDS and long-term-non-progressors (LTNP). For example, progressors could be defined as up to 5 years, up to 11 years or as we describe in this report up to 15 years from infection, and LTNP could be individuals with normal CD4+ T cell counts for more than 15 years with or without high viral loads. In this review, we emphasized that in the studies of ARGs the HLA alleles are important in LTNP; HLA-B alleles influencing the advantage to pathogens to produce immune defense mediated by CD8+ T cells (cognate immunuity). Our main point we make in this report is that contrary to recent reports claiming that this dominant effect was unlikely due to differences in NK activation through ligands such as HLA-Bw4 motif, we believe that cognate immunity as well as innate immunity conferred by NK cells are involved. The main problem is that HLA-Bw4 alleles can be classified according the aminoacid in position 80. Isoleucine determines LTNP, which is a ligand for 3DS1. Such alleles did not include HLA-B*44. B*13 and B*27 which have threonine at that position. The authors have not considered the fact that in addition to the NK immunoglobulin receptors, NK receptors can be of the lectin like such as NKG2A/HLA-E to influence the HIV infection outcome. HLA-Bw4 as well as HLA-Bw6 alleles can be classified into those with threonine or methionine in the second position of their leader peptides. These leader peptides are ligands for NKG2A in which methionine influences the inhibitory role of NKG2A for killing infected targets. Functional studies have not been done as well as studies of these receptors in infected individuals. However, analyses of the leader peptides of HLA-B alleles in published reports, suggested that threonine in the second position can explain the importance of HLA-B*57, B*13, B*44 as well as certain Bw6 alleles in LNTP. In addition, we analyzed the San Francisco database that was reported and found that the association of HLA-B alleles with LNTP or with progressors can be due to the presence of threonine or methionine in their second position. Therefore, studies of outcome of HIV infection should include not only mechanisms of cognate immunity mediated by peptides and CD8+ T cells but also, NK receptors of two types, NKG2A as well as 3DSI. We propose that the SCID mouse should be used to understand mechanisms mediated by many of the ARGs especially the importance of thymus derived cells as well as NK receptor interactions with their ligands in this experimental animal transplanted with human stem cells, thymus or NK cells obtained from individuals of known HLA genotypes.
I. Introduction
AIDS is not generally considered a genetic disease because there is a great heterogeneity of clinical presentation in part determined by gene variants that control to same extent virus replication and immunity [1-3]. Noteworthy of mentioning is the fact that there is not much information about genes that protect individuals exposed to the virus and do not get infected. Also, it is important to mention that those that get infected develop pathology at different times following the infection, for example those who died from AIDS at 1-5 years from infection, but the definition of such fast progressors should include those where the immunodeficiency and progression to AIDS could go up to 10-11 years or more from the time of the infection. More difficult perhaps, is to define long-term infection since there are patients that progress to immunodeficiency 15-20 or more years after infection and a group of the long-term non progressors that maintain normal CD4+ T cell counts and absence of viremia for more than 20 years without treatment. The heterogeneity of clinical presentation cannot be completely explained, but it is possibly related to the large number of genetic and non-genetic contributing factors: innate, humoral and cell mediated immune responses [4] and the AIDS restricting genes (ARGs). In addition, longitudinal studies have an additional difficulty because some patients are treated with antiretroviral drugs to prevent or diminish the possible progression to AIDS.
Our review will attempt to emphasize that studies of CD8+ T cell immunity should be performed in conjunction with NK receptor/ligand interactions and their variants. We will summarize evidence that HIV infection involves several immune mechanisms resulting in longterm non progression or progression to AIDS in which intrinsic, innate and cognate immunity are important. However, genetic associations with protection from or progression to disease are complex and could vary, related to the ethnicity of the populations studied [5].
II. Immune Functions in HIV
A. Cognate Immunity
Cognate immunity has been extensively studied involving primarily CD4+ and CD8+ T cells interacting with MHC alleles presenting HIV-I peptides on antigen presenting cells [6-12]. Such cells may mediate protection by production of cytotoxic cells or cytokines such as IFN gamma and IL-12 [13]. Macrophages [14], DC and plasmocytoid DCs are type 1 IFN secretion inducers of IL-12. However, several viruses escape MHC class I restricted cytotoxic T lymphocytes (CTL) responses by down-regulating their expression on infected cell surface [15]. In this regard, protection from infection may have genetic basis but it varies in different populations [5]; these associations are complex and difficult to asses because in most cases is difficult to know the date of incidence of infection, the date of presence of viremia, or the date of decrease of CD4 counts. In many cases the studies could be difficult to reproduce because of genetic stratification and population size of case-control studies. MHC studies, in many cases, are incomplete and without strict criteria for population sizes and epidemiological parameters that explain the lack of consistencies of the alleles associated with disease progression [16]. Also, some of the MHC associations maybe present because of genes that are in linkage disequilibrium with HLA alleles, for example TNF [17], that are involved in the regulation of level of HIV-1 viremia since it causes increased viral replication in infected monocytes. This occurs through the activation of the transcription factor NFkB, which binds to the HIV-1 long terminal repeat, causing increased levels of HIV-1 transcription [18-20].
HLA Alleles, and Clinical Outcome of HIV-1 Infection
HLA alleles of class I (A, B and C) and II (DR, DQ and DP) have a large variation between individuals and populations, which could provide recognition of virus agents to which they have been exposed [4]. Since HIV infects immune cells to produce proliferation, spread and CD4+ T lymphocyte damage, the HLA alleles could influence the time from infection to AIDS progression [21]. For example, HLA-Cw4, a ligand for KIR 2DL1 influences the time to develop AIDS, [22]. These findings could have been due to genes in random association with HLA-B and C alleles such as TNF [17]. The contribution of HLA-B*35 alleles has also been postulated [23]. More importantly, HLA influence was observed in relation to HIV-1 subtypes where the HLA recognition motifs are associated to AIDS survival. Two HLA alleles, B*27 and B*57, have been reproducibly associated with long-term non-progression of AIDS [3, 24]. A controversial subject is the role of zygosity of HLA-A alleles in the progression to AIDS. For example HLA class I homozygotes for several alleles progress to AIDS faster than those heterozygous [23, 24]. Further, groups of alleles or supertypes grouped according the B pocket showed association with clinical outcome [16, 25]. The role of zygosity for HLA-Bw6 and HLA-Bw4 has been controversial, Bw6 was found associated with AIDS in Caucasians [25, 26] and Chinese [27] while the Bw4/Bw4 was associated with LTNP in Caucasians but not in Africans [28]. Therefore, genetic markers, ARGs result from different evolutionary effects that would explain that ethnicity and protection from AIDS and genetic effects on clinical outcome vary among populations [4, 5].
B. Innate Immunity
This type of immunity is mediated by a newly discovered endogenously expressed proteins that provide defenses against retroviral infection such as HIV-1 and murine leukemia virus (MLV) [29-31]. These proteins probably prevent the entry of HIV-1 into cells by interaction with CD4, Natural Killer (NK) cells, monocytes and dendritic cells. In addition, several molecules secreted by these cells can protect against such infection, for example IL-12, chemokine receptors CCR4 and CCR5. In this regard, most HIV strains use the CCR5. as a co-receptor and thereby are sensitive to inhibition by the ligands of this receptor; also deletion of CCRS prevents viral entry into cells [2]. HIV-1 infection is potently blocked in rhesus macaques by the cytoplamic component protein tripartite motif 5 (TRIM5). Interestingly, the TRIM5 protein in rhesus macaque is 87% identical to its human homolog. However, human TRIM5 proteins do not restrict HIV-1 [31]. Remarkably, one amino acid change in the protein sequence of human TRIM5 leads to full activity against HIV-1 infection [32, 33]. Even though, several SNPs for human TRIM5 have been identified, none of them corresponded to this specific residue. Differently, the endogenously expressed human cytidine deaminase APOBEC3G potently blocks HIV-1 infection [32]. However, the HIV-I accessory protein vif is able to overcome this restriction allowing productive infection. Finally, the endogenously expressed mouse protein Fvl potently block infection of MLV [29]. Overall, this demonstrates that during evolution, humans developed series of specie-specific blocks for retroviral infection.
In an extensive genetic epidemiological study, SNPs of TRIM5 were identified. In African Americans four alleles exhibited different frequencies in HIV-1 infected and uninfected individuals. SNP2 in the non-coding exon 1 and SNP3 in intron 1 were associated with increased risk of infection. [34]. These finding suggested that any modification of infection susceptibility afforded by particular TRIM5 alleles may be restricted to particular populations or types of exposure. There are population specific effects for AIDS modifying genes [33-36]. For example, the role of TRIM5 polymorphism in Europeans, the TRIM haplotype containing 136Q exhibited increased frequency among HIV-1 infected subjects compared with seronegative exposed individuals [34], which contrasts with the opposite results reported in African Americans [35, 36]. These findings suggest that either the genetic background, non-random association with other genes or the type of exposure may influence susceptibility to HIV-1 infection. However, TRIM5 variants do not influence disease progression. As it will be pointed out below, there are many genes that limit AIDS and such influence may be present in some ethnicities but not in all. This problem stresses the fact that research is needed to determine their frequencies in several ethnicities in order to include sufficient individuals in the calculations of statistical power.
According to estimations there are more than 90% of the genetic and non-genetic influences on AIDS progression that are still undiscovered. Less known is the estimation of these factors influencing genetic susceptibility to be infected by HIV-1. It was calculated that 21.1% of individuals infected who did not develop AIDS for 11 more years did so because they carried one of more protective ARGs. Among those that develop AIDS rapidly, within 5.5 years of HIV-1 infection, the single ARG effects were modest (2.4-8.1%) but the cumulative ARGs associated with rapid progression is high (40.9%). However, the estimates are approximately 10% for both groups including long-term non-progressors and progressors to AIDS. Available databases, although informative, possess problems for analyses, because during the patients follow up some of them are recruited after they have been treated with anti-retroviral drugs during the first 10-11 years from the time of infection. Thus the definition of immunodeficiency is based on CD4+ T cell counts only without consideration of viral loads. Also some patients without treatment maintain normal CD4+ T cell counts beyond 15 years from infection while they may have high viral loads.
2. NK cells
Studies of HIV-1 uninfected persons showed enhanced activity of NK functions despite many years of high-risk exposure demonstrating the importance of NK cells in immunity against HIV-1 infection [37]. NK cells functions involve receptors that interact with HLA ligands. HLA alleles are therefore important in disease progression because target infected cells are more susceptible to NK killing. For example, nef, a product of HIV-1 is known to diminish the levels of HLA-A and HLA-B expression of infected cells [38], whereas HLA-C, which is one ligand for NK receptors, is poorly expressed naturally [39]. By contrast, nef increases the level of HLA-E on the surface of infected cells [40]. Also, HLA-A expression is higher than HLA-B [39] and HLA-B expression is more inducible by IFN alpha than the HLA-A [41]. Therefore, in the environment of the HIV infection there would be more contribution of HLA-B alleles or leader peptides from HLA-B as well as that of HLA-E with HLA-B leader peptides to function in their interaction with NK receptors.
There are two different kinds of NK receptors, immunoglobulin-like such as the inhibitory and activating receptors that include the KIRs and also lectin-like such as the NKG2 receptors. The NKG2A is inhibitory and has as a ligand HLA-E and the NKG2D is an activating-type receptor and has as a ligand the MICA alleles for function, killing of infected cells or cancer cells [42, 43]. Also, some KIRs are expressed on a subset of NK cells with a memory phenotype [44] that suggests that they may regulate T cell as well as NK cell activity. Masking of inhibitory NK receptors on CTLs from HIV infected individuals by monoclonal antibody has produced increases of HIV-1 specific CTL activity [12] suggesting possible involvement of NK cells.
As proposed in this report, HLA-E and its interaction with NKG2A needs to be investigated in both high risk persons exposed to HIV that have or do not have infection or their role in the clinical outcome following HIV-1 infection. However, the MICA alleles, which are ligands for the lectin activating receptor NKG2D, were not associated with HIV-1 clinical progression [17].
It is well known that HLA-E folds to function, depending on the leader peptides from HLA-class I alleles (A, C and B). The critical aminoacid is methionine in second position, whereas threonine is accompanied by poor folding and defective function [45]. The leader peptides with methionine in the second position (VMAPKTVLL and VMAPRTLL) induce higher levels of HLA-E expression than those with threonine at Position 2. Also, they exhibited high affinity for soluble CD4 NKG2A molecules [45-47]. Despite the high expression of HLA-E due to nef, the presence of threonine in the leader peptides of HLA-B alleles render HLA-E to be poorly expressed [40].
HLA-B Ligands for NK Receptors: HLA-Bw4 and HLA-Bw6 Supertypes and the Leader Peptide of HLA-B alleles as Ligands for the NKG2A Receptor
The HLA-B alleles are the most important in HIV viral progression because they restrict infection via CD4/CD8 [48] and they are ligands for NK receptors, KIRs (3DL1 and 3DS1) [26] and lectin receptor (NKG2A) as we propose in this article.
HLA-Bw4 supertype comprising approximately 40% of HLA-B alleles is the ligand for NK receptors encoded by the Killer immunoglobulin receptor (KIR) gene complex on chromosome 19 [49]. This interaction could be due to loss of inhibition of the inhibitory receptor, 3DL1 in effector cells that causes the function of the activating receptor 3DS1. This was found experimentally, that the combination of homozygosity of HLA-Bw4 and KIR3DS1 epistasis influence AIDS progression [23]. Of interest, this interaction did not include the role of B*27 and B*44 in LNTP. The authors suggested that those alleles are involved in a different mechanism of protection from progression. In this report we are proposing a mechanism that needs to be investigated.
All HLA-A and HLA-C alleles encode HLA-E binding peptides with methionine at second position [50]. Importantly, HLA-C alleles are poorly expressed on the surface of cells [39] and HLA-B has higher basal level of expression than HLA-A, and by far more inducible by IFN gamma and alpha than HLA-A gene [41]. Furthermore, HLA-B alleles are the dominant influence to mediate a possible co-evolution of HIV and HLA [44], that we believe should also include HLA-E loaded with class I peptides.
These findings are based on the fact that HLA-B alleles can be classified according to the presence of thr or met at P2 of the leader peptide [41, 43]. Almost all HLA-Bw4 alleles with the exception of HLA-B*.38 encode leader peptides with Thr at P2 and HLA-Bw6 are divided into two groups, those encoding Met at P2 and those encoding Thr at P2 [41, 43, 51] (Table 2).
Table 2.
Comparison of Bw4 and Bw6 frequencies based on the presence of methionine or threonine in second position of HLA-B leader peptides
| aa 2nd position | Caucasians |
African Americans |
Asian Americans |
|---|---|---|---|
| Met | 0.30 | 0.20 | 0.18 |
| Thr | 0.70 | 0.80 | 0.82 |
| Met/Met | 0.09 | 0.04 | 0.04 |
| Met/Thr | 0.42 | 0.32 | 0.27 |
| Thr/Thr | 0.49 | 0.64 | 0.69 |
|
Supertype alleles |
|||
| Bw6 | 0.61 | 0.74 | 0.55 |
| Bw4 | 0.39 | 0.26 | 0.45 |
|
Supertype genotypes |
|||
| Bw6/Bw6 | 0.37 | 0.55 | 0.30 |
| Bw4/Bw4 | 0.15 | 0.07 | 0.20 |
| Bw6/Bw4 | 0.49 | 0.38 | 0.50 |
Statistical Analysis of Influence of Threonine in Second Position in Long Term Non Progressors (LTNP)
Re-analyses of the data published before (Flores et al) (table 3) the number of individuals with two copies of Threonine in the group of controllers was significantly higher than in the progressors. 14/20. (0.70) versus 3/19 (0.15) of the non-controllers p: 0.0006 OR: 12.44. Of interest, the frequency of individuals with TT uninfected was 42/108 (0.39). p: 0.010 OR:3.67.
Table 3.
Frequencies of leader peptides (two copies) of methionine (M) or threonine (T), influence in HIV progression
|
aa 2nd position |
Intermediate Progressors (10-15 years) |
Progressors (0-10 years) |
LTNP | Non-infected controls |
|---|---|---|---|---|
| MM | 7/32 (21.8) | 14/44 (31.8) | 1/22 (4.5) | 24/265 (9.0) |
| MT | 16/32 (50.0) | 19/44 (43.2) | 4/22 (18.2) | 138/265 (52.0) |
| TT | 9/32 (28.1) | 11/44 (25.0) | 17/22 (77.3) | 130/265 (49.0) |
Analyses of the San Francisco cohort reported before also demonstrated that Comparison of LTNP with progressors, either 0-10 years or 0-15 years showed that the presence of TT was significaltly higher in the LTNP than in 0-10 years progressors, 17/22 versus 11/44, p= 0.000005, OR=10.20 and between all progressors (0 to 15 years, 17/22 versus 20/76, p=0.00001, OR= 9.52. Likewise, the frequency of TT was higher in LTNP than in uninfected controls 20/76 versus 130/265, p =0.0004 OR= 2.70. These results should be confirmed using Kaplan-Meier plots to assess more accurately the importance of these findings using outcome of HIV viral infection and need to be studied in relation to the expression of NKG2A receptors on NK cells.
III. Stem Cell Microenvironment in HIV Infection
HIV infection of stem cell microenvironments or niches causes hematopoietic inhibition and hence cytopenias [52-57]. Hematopoietic CD34+ progenitor stem cells are reported to be resistant to HIV-1 infection, in vitro, or in vivo [58, 59]. Those cells that experienced the indirect effects of HIV-1 infection exhibit inhibition of their multilineage hematopoiesis as determined by colony forming activity ex vivo [58, 60-62]. It is reported that the hematopoietic stem cell microenvironment is damaged due to the indirect effects of HIV-1 infection of the thymocytes on the CD34+ progenitor stem cells but in a reversible manner, in the human fetal Thymus/Liver conjoint hematopoietic organ of the transplanted chimeric severe combined immunodeficiency mouse (SCID-hu) model system [60, 62]. It is therefore highly plausible that this implanted human organ in the SCID-hu mouse, which serves as a niche, not only for thymocyte expansion but also supports hematopoiesis, suffers niche dysfunction due to HIV-1 infection. Continued presence of the CD34+ progenitor stem cells in the infected niche seem to suffer due to exacerbation resulting from persistent virus mediated niche disruption via infection of thymocytes and consequent interactions and signaling network of the hubs. In this microenvironment it is possible that several ARGs could produce different outcomes. This is evident from our previous observation that CCR4 and CCR5- tropic HIV-1 produce variable kinetics of inhibition of hematopoiesis [60].
Less understood is the role of different lymphoid organs in regards to the diminution of T cells in the gastrointestinal niche which is involved in HIV infection clinical outcome especially the genetic markers that contribute to AIDS progression. The intestinal mucosal immune system is an important target of HIV-1 infection and contributes to disease progression. In addition distinct gene expression profiles correlate with clinical outcome [63]. In this regard we wonder if the variable time of outcome of AIDS following HIV infection is related to several innate unknown mechanisms operating in the host. Independent of these is the fact that patients with HIV do not get diagnosed immediately after infection and the time of diagnosis could be variable taking sometimes several years. For example, we had access to the San Francisco database [46] and several individuals recruited for follow-up were first evaluated from 0 to 11 years and therefore the definition of long-term non-progression can only be analyzed after years of the date of known infection. In addition, some of such patients were censored after they had begun treatment. Only in those individuals followed from the time of infection it would be possible to demonstrate the role of genetic markers in viremia, and their participation in producing decreased CD4 counts and/or conversion to AIDS.
The transcription factors such as STAT5A are involved in stem cell self-renewal that precedes multilineage differentiation of CD34+ progenitor stem cells [64-69]. The proto-oncogene of myeloproliferative leukemia also known as thrombopoietin (Tpo) receptor proto-oncogene, c-mpl, is known to promote multi-lineage pluripotent stem cell differentiation of the CD34+ progenitor cells [70-73]. Both STAT5 and c-mpl are important target genes for control and enhancement of stem cell self-renewal and multi-lineage differentiation to reduce or prevent cytopenias induced during HIV infection [74]. We wonder if some individuals have highly regulated expression of STAT5A/B and c-mpl genes in progenitor cells and if in such individuals CD4 cells are generated in higher numbers than in those without such a regulated niche.
IV. Discussion
It is necessary to investigate the role of NK cells in the innate immunity against HIV infection. It is now clear that there are at least three kinds of immune mechanisms involved in the control of viral infections including that of HIV-1: A) intrinsic innate immunity mediated by a group of major defenses against infection by retroviruses, Fv1 and TRIM5 inhibitors, proteins that target incoming retroviral capsids and the APOBEC3 class of cytidine deaminases that hypermutate and destabilize retroviral genomes. These are probably involved in the prevention of HIV-1 into cells and constitute the first level of protection from infection by specific cells, such as dendritic cells, including plasmocytoid dendritic cells. Of course many proteins such as cytokines, IL 12, chemokines, and chemokine receptors CCR5 and CXCR4 are important. B) Genetic markers of immune effectors that are important in the outcome of HIV-1 infection; Natural killer, NK, receptors of two kinds, Ig-like such as KIRs and lectin-like such as NKG2 are involved. These two kinds of genetic markers have been described to be involved in the outcome of the HIV-1 infection towards long term non-progression and AIDS. In this regard, two reports have described the role of HLA-Bw4/Bw4 as a marker for LTNP and related to that a subgroup of Bw4, isoleucine in position 80 of Bw4 interacting with a KIR receptor, 3DS1 was associated with LTNP [25, 26]. In unpublished work we have re-analyzed the data published before involving the importance of Bw4 in LTNP, discovered that methionine, Met/Met, in the second position of the leader peptide of HLA-B alleles is a marker for progression to AIDS [47]. NK receptors may be involved in the control of viremia; CD94/NKG2A inhibitory receptors [75-77] and the NKG2D stimulatory receptors not only present in NK cells but also in human CD8 lymphocytes [39, 78]. Therefore, NK and CTL activating and inhibitory signals can be provided by NKG2 receptor interaction with MICA and by CD94/NKG2A interaction with HLA-E molecules with the appropriate assembly of peptides [44]. However, the role of NKG2D/MICA in the progression to AIDS was not found [17]. The CD94/NKG2A-HLA-E pathway has not been studied related to HIV-1 infection outcome. It is important to mention that HLA leader peptide sequences with Met at P2 induce significant levels of HLA-E expression compared to those with Thr at P2, the latter failed to confer protection from NK lysis and the HLA-E/peptide complexes exhibited high affinity for soluble CD94/NKG2 molecules [45, 47]. Also, although HLA-A and C alleles encode HLA-E binding peptides with Met at P2, HLA-C alleles are poorly expressed on the surface of cells [39] and nef of HIV-1 down-regulates the cell surface expression of HLA-B and A but not C or HLA-E [40]. Furthermore, HLA-A alleles are rarely used to restrict CTL epitopes [48, 79]. In sum, in the environment of HIV-1 infected cells the HLA-leader peptides have more contribution to generate HLA-E binding peptides. This is supported by the reduced HLA-E poor expression by cells transfected with HLA-B*51 or B*58 [80, 81]. Therefore, it is possible that NK receptors are involved together with the recognition of CD8 T cell responses against the human immunodeficiency virus (HIV-1). Since HIV-1 infection modulates the expression of IFNs that the cellular environment, niche, there would render greater contribution of HLA-B leader sequences to generate HLA-E binding peptides to interact with the NKG2A receptor as well as interaction with the cognate immune system [48]. Based on this scenario, we believe that the presence of Met at P2 produces inhibition of killing favoring HIV progression whereas, Thr at P2 produces loss of inhibition of killing favoring activating receptors of NK cells that negatively influence HIV infection outcome.
Therefore it is important to emphasize that among the multiple genes that limit the progression to AIDS [4] HLA-E molecules loaded with either methionine of threonine in the second position of leader peptides should be studied not only in regard to their influence in infection outcome but also in the innate protection from HIV-1 infection. Furthermore, we will also use a SCID-hu model where a combination of TRIM-5a, CCR5, with threonine in the second position of the HLA-B leader peptide will protect transplants from infection. C) Cognate immunity. There is a large literature of this subject related to viremia progression and future investigations are needed in this respect to identify genes of protection from infection by HIV. These should include genetic markers involved in the role of CD4/CD8 in HIV-1 infection together with those genetic factors involved in mechanisms of innate immunity against HIV-1 viral infection or disease progression, such as those involving NK receptors interacting with their ligands. This mechanism also explains the slow progression to AIDS in a subgroup of patients that maintain normal CD4+ T cell counts but demonstrate viremia even after 15 years of infection [46].
In unpublished studies by Flores, et al [46] previous published results were reanalyzed [25] with 39 individuals of known immune status that included controllers of viremia with a short follow up (not longer than 2 years) The HLA-B alleles were grouped according the Bw4 or Bw6 public specificity encoding epitopes determined by residues 79-83 at the carboxyl-terminal end of the alpha 1 helix (Table 1). Some of the Bw6 alleles have threonine in the second position of their leader peptide, for example HLA-B*1801, B*4101, B*35 alleles, B*40 alleles, and B* 15 alleles. These results were corroborated analyzing 98 HIV infected individuals with follow-up of viral loads and CD4+ T cell counts with progression or lack of progression to immunodeficiency. TT was associated with long-term progressors and with long-term non-progressors that maintain normal CD4 counts beyond 15 years from the date of infection.
Table 1.
The HLA Bw4 and Bw6 motifs and alleles
| Amino-acid position | Corresponding alleles | |||||
|---|---|---|---|---|---|---|
| 80 | 81 | 82 | 83 | |||
| Bw4 isoleucine |
Ile80 |
Ile |
Ala |
Leu |
Arg |
B*15 (13, 16, 17. 23, 24) B*2702, B*3801, B*4901, B*51, B*5201, B*5301, B*5302, B*57, B*58, B*59 |
| threonine | Thr80 | Thr | Ala | Leu | Arg | B*13 (01, 02, 04), B*3802, B*44 (02, 03, 04, 05, 08, 21) |
| Thr | Leu | Leu | Arg | B*2705, B*2709, B*3701, B*4701 | ||
| Bw6 asparginine |
Asn 80 |
Asp |
Lau |
Arg |
Gly |
B*07 (02, 03, 04, 05, 08, 09, 10, 14) B*08, B*14. |
| B15*(01, 03, 04, 09, 15, 18, 22, 37 45, 48, 13, 16, 17, 23, 24)) B*18, B*35, B*39, B*40, B*41, B*42, B*4501, B*4601, B*48, B*50, B*5401, B*55, B*56, B*67, B*73, B*7801, B*8101, B*82 |
||||||
In bold are alleles whose leader peptide has methionine in the second position and the rest have threonine in the second position [50].
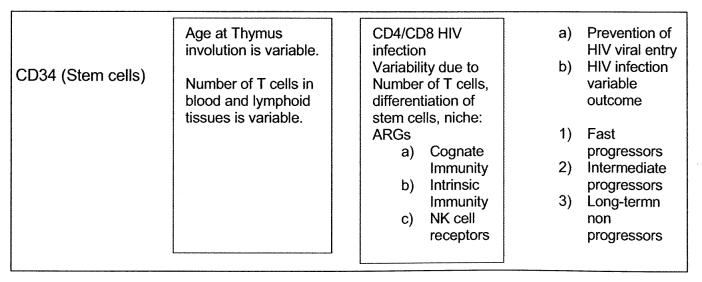
Figure 1, summarizes possible variability of CD34 cells in the population, different age at time of thymic involution.
Figure 1.
Diagram summarizes possible variability of CD34 cells in the population, different age at time of thymic involution.
This hypothesis can be studied as a cross-sectional study using the SCID mouse using peripheral blood and tissues available to investigate the number of CD4, CD8 and NK cells present at the time of short-term progression or long-term non-progression as a first step to study the role of NKG2A/ligand interaction in inhibition from killing, or actual killing, of HIV infected target cells as well as using the SCID-hu mouse model.
V. Future Studies
Most studies of HIV infection outcome reported have described the importance of class I gene products particularly HLA-B. In this regard in a large number of HIV-1 infected individuals from southern Africa, it was reported that HLA-B alleles influence the potential co-evolution of HIV and HLA, providing advantage to pathogen defense mediated by CD8+ T cells. These results were consistent with the findings in B-clade infected Caucasians with non-progression/low viral load or progression/high viral loads [48]. The authors claimed that the dominant effect for HLA-B alleles in HIV infection was unlikely due to differences in NK cell activation through the HLA-Bw4 motif. They mentioned the fact that it was reported that there is epistatic interaction between KIR3DSl and some HLA-Bw4 alleles that mediated protection were entirely independent of this interaction [26]. Since there is a protective role of certain Bw4 alleles, B*2705, B*13 and B*44, as well as those that carry Ile-80, such a protective role should involve the mechanism described of NKG2A with the leader peptide threonine. See diagram. In both reports [26, 48], the authors did not consider the fact that there are two kinds of NK receptors, immunoglobulin receptors such as KIR3DSl and the lectin receptors such as NKG2A. We have described herein that the alleles of Bw4, B*27 and B*57 as well as some Bw6 alleles carry threonine in the second position of their leader peptide that would interact with NKG2A. This mechanism should be studied together with the dominant role of HLA-B alleles influencing HIV specific CD8 T cell responses in the outcome of HIV infection. The use of cross-sectional studies has limitations of statistical power that are corrected when using longitudinal studies and this explains to some extent the major inconsistencies in published reports related to the HIV infection outcome. Perhaps the SCID mouse model could resolve some of these problems but this model also has limitations related to the fact that the experiments are limited to the particular tissues grafted and do not necessarily include the large number of ARGs present in individuals of the population at large [82, 83]. However, the engraftment of CD34+ or even of CD133+ cells give rise to multiple lineages of cells following spontaneous differentiation in vivo that may identify the source of ARGs.
Acknowledgement
Supported by NIH grants HL29583 and HL59838, and also from the Dana Farber Cancer Institute. P.K. is a recipient of NIH grant HL079846.
References
- [1].Fauci AS. HIV and AIDS: 20 years of science. Nat. Med. 2003;9:839–843. doi: 10.1038/nm0703-839. [DOI] [PubMed] [Google Scholar]
- [2].O’Brien SJ, Moore J. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol. Rev. 2000;177:99–111. doi: 10.1034/j.1600-065x.2000.17710.x. [DOI] [PubMed] [Google Scholar]
- [3].Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Ann. Rev. Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- [4].O’Brien SJ, Nelson GW. Human genes that limit. AIDS. 2004;36(6):565–74. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- [5].Winkler C, An P, O’Brien SJ. Patterns of ethnic diversity among the genes that influence AIDS. Hum. Mol. Gene. 2004;13(Spec No 1):R9–19. doi: 10.1093/hmg/ddh075. [DOI] [PubMed] [Google Scholar]
- [6].Cao Y, Qin L, Zhang L, Safrit J, Ho DD. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 1995;332(4):201–8. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- [7].Pantaleo G, Menzo S, Vaccarezza M, Graziosi C, Cohen OJ, Demarest JF, Montefiori D, Orenstein JM, Fox C, Schrager LK, et al. Studies in subjects with long-term nonprogressive human immunodeficiency virus infection. N. Engl. J. Med. 1995;332(4):209–16. doi: 10.1056/NEJM199501263320402. [DOI] [PubMed] [Google Scholar]
- [8].HRinaldo C, Huang XL, Fan ZF, Ding M, Beltz L, Logar A, Panicali D, Mazzara G, Liebmann J, Cottrill M, et al. igh levels of anti-human immunodeficiency virus type 1 (HIV-I) memory cytotoxic T-lymphocyte activity and low viral load are associated with lack of disease in HIV-1-infected long-term nonprogressors. J. Virol. 1995;69(9):5838–42. doi: 10.1128/jvi.69.9.5838-5842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Harrer T, Harrer E, Kalams SA, Barbosa P, Trocha A, Johnson RP, Elbeik T, Feinberg MB, Buchbinder SP, Walker BD. Cytotoxic T lymphocytes in asymptomatic long-term nonprogressing HIV-1 infection. Breadth and specificity of the response and relation to in vivo viral quasispecies in a person with prolonged infection and low viral load. J. Immunol. 1996;156(7):2616–23. [PubMed] [Google Scholar]
- [l0].Kalams SA, Buchbinder SP, Rosenberg ES, Billingsley JM, Colbert DS, Jones NG, Shea AK, Trocha AK, Walker BD. Association between virus-specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J. Virol. 1999;73(8):6715–20. doi: 10.1128/jvi.73.8.6715-6720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lisziewicz J, Rosenberg E, Lieberman J, Jessen H, Lopalco L, Siliciano R, Walker B, Lori F. Control of HIV despite the discontinuation of antiretroviral therapy. N. Engl. J. Med. 1999;340(21):1683–4. doi: 10.1056/NEJM199905273402114. [DOI] [PubMed] [Google Scholar]
- [12].De Maria A, Ferraris A, Guastella M, Pilia S, Cantoni C, Polero L, Mingari MC, Bassetti D, Fauci AS, Moretta L. Expression of HLA class I-specific inhibitory natural killer cell receptors in HIV-specific cytolytic T lymphocytes: impairment of specific cytolytic functions. Proc. Natl. Acad. Sci. USA. 1997;94(19):10285–8. doi: 10.1073/pnas.94.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Servet C, Zitvogel L, Hosmalin A. Dendritic Cells in Innate Immune Responses Against HIV. Current Molecular Medicine. 2002;2(8):739–756. doi: 10.2174/1566524023361907. [DOI] [PubMed] [Google Scholar]
- [14].Herbein G, Coaquette A, Perez-Bercoff D, Pancino Hosmalin G. Macrophage Activation and HIV Infection: Can the Trojan Horse Turn into a Fortress? Current Molecular Medicine. 2002;2(8):723–738. doi: 10.2174/1566524023361844. [DOI] [PubMed] [Google Scholar]
- [15].Rappocciolo G, Birch J, Ellis SA. Down-regulation of MHC class I expression by equine herpesvirus-1. J Gen Virol. 2003;84(2):293–300. doi: 10.1099/vir.0.18612-0. [DOI] [PubMed] [Google Scholar]
- [16].Trachtenberg EA, Erlich HA. A review of the role of the human leukocyte antigen (HLA) system as a host immunogenic factor influencing HIV transmission and progression to AIDS. In: Korber BTK, Brander C, Haynes BF, Moore JP, Koup RA, Kuiken C, Walker BD, Watkins DI, editors. HIV Molecular Immunology 2001. Theoretical Biology and Biophysics Group; 2001. pp. 143–160. [Google Scholar]
- [17].Delgado JC, Leung JY, Baena A, Clavijo OP, Vittinghoff E, Buchbinder S, Wolinsky S, Addo M, Walker BD, Yunis EJ, Goldfeld AE. The -1030/-862-linked TNF promoter single-nucleotide polymorphisms are associated with the inability to control HIV-1 viremia. Immunogenetics. 2003;55(7):497–501. doi: 10.1007/s00251-003-0604-7. [DOI] [PubMed] [Google Scholar]
- [18].Folks TM, Clouse KA, Justement J, Rabson A, Duh E, Kehrl JH, Fauci AS. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc. Natl. Acad. Sci. USA. 1989;86(7):2365–8. doi: 10.1073/pnas.86.7.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Marshall WL, Brinkman BM, Ambrose CM, Pesavento PA, Uglialoro AM, Teng E, Finberg RW, Browning JL, Goldfeld AE. Signaling through the lymphotoxin-beta receptor stimulates HIV-1 replication alone and in cooperation with soluble or membrane-bound TNF-alpha. J. Immunol. 1999;162(10):6016–23. [PubMed] [Google Scholar]
- [20].Poli G, Kinter A, Justement JS, Kehrl JH, Bressler P, Stanley S, Fauci AS. Tumor necrosis factor alpha functions in an autocrine manner in the induction of human immunodeficiency virus expression. Proc. Natl. Acad. Sci. USA. 1990;87(2):782–5. doi: 10.1073/pnas.87.2.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gao X, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 2001;344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- [22].O’Brien SJ, Gao X, Carrington M. HLA and AIDS: A cautionary tale. Trends Mol. Med. 2002;7:379–381. doi: 10.1016/s1471-4914(01)02131-1. [DOI] [PubMed] [Google Scholar]
- [23].Carrington M, et al. HLA and HIV-1: Heterozygote advantage and B*35- Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- [24].Tang JM, et al. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type I infection AIDS Res. Hum 1999, Retroviruses 15317–324. [DOI] [PubMed] [Google Scholar]
- [25].Flores-Villanueva PO, et al. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc. Natl. Acad. Sci. USA. 2001;98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O’Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nut. Genet. 2002;4:429–34. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- [27].Qing M, Li T, Han Y, Qiu Z, Jiao Y. Accelerating effect of human leukocyte antigen-Bw6 homozygosity on disease progression in Chinese HIV-1-infected patients. J. Acquir. Immune Defic. Syndr. 2006;41(2):137–9. doi: 10.1097/01.qai.0000195607.25262.92. [DOI] [PubMed] [Google Scholar]
- [28].Kaslow RA, Tand J, Dorak MT, Tang s, Musonda R, Karita E, Wilson C, allen S.Homozigosity for HLA-Bw4 is not associated with protection of HIV-1 infected persons in African ancestry Conference retroviruses opportunistic inject 20029Feb 2428: abstract # 320-W. [Google Scholar]
- [29].Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fvl. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- [30].Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- [31].Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- [32].Li Y, Li X, Stremlau M, Lee M, Sodroski J. Removal of arginine 332 allows human TRIM5alpha to bind human immunodeficiency virus capsids and to restrict infection. J Virol. 2006;80:6738–6744. doi: 10.1128/JVI.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- [34].Javanbakht H, An P, Gold B, Petersen DC, O’Huigin C, Nelson GW, O’Brien SJ, Kirk GD, Detels R, Buchbinder S, Donfield S, Shulenin S, Song B, Perron MJ, Stremlau M, Sodroski J, Dean M, Winkler C. Effects of human TRIM5alpha polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology. 2006;354(1):15–27. doi: 10.1016/j.virol.2006.06.031. [DOI] [PubMed] [Google Scholar]
- [35].Winkler CA, O’Brien SJ. In: AIDS restriction genes in human ethnic groups: An assessment. in AIDS in Africa. 2nd Essex M, Mboup S, Kanki PJ, Marlink R, Tlou SD, editors. Kluwer Academic; New York: 2002. [Google Scholar]
- [36].Speelmon EC, Livingston-Rosanoff D, Li SS, Vu Q, Bui J, Geraghty DE, Zhao LP, McElrath MJ. Genetic association of the antiviral restriction factor TRIM5alpha with human immunodeficiency virus type 1 infection. J. Virol. 2006;80(5):2463–71. doi: 10.1128/JVI.80.5.2463-2471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Scott-Algara D, Truong LX, Versmisse P, David A, Luong TT, Nguyen NV, Theodorou I, Barre-Sinoussi F, Pancino G. Cutting edge: increased NK cell activity in HIV-1-exposed but uninfected Vietnamese intravascular drug users. J. Immunol. 2003;171(11):5663–7. doi: 10.4049/jimmunol.171.11.5663. [DOI] [PubMed] [Google Scholar]
- [38].Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391(6665):397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- [39].McCutcheon JA, Gumperz J, Smith KD, Lutz CT, Parham P. Low HLA-C expression at cell surfaces correlates with increased turnover of heavy chain mRNA. J. Exp. Med. 1995;181(6):2085–95. doi: 10.1084/jem.181.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10(6):661–71. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- [41].Liu K, Kao KJ.Mechanisms for genetically predetermined differential quantitative expression of HLA-A and -B antigens Hum. Immunol 2000. Aug; 618799–807. [DOI] [PubMed] [Google Scholar]
- [42].Katsuyama Y, Ota M, Ando H, Saito S, Mizuki N, Kera J, Bahram S, Nose Y, Inoko H. Sequencing based typing for genetic polymorphisms in exons, 2, 3 and 4 of the MICA gene. Tissue Antigens. 1999;54(2):178–84. doi: 10.1034/j.1399-0039.1999.540209.x. [DOI] [PubMed] [Google Scholar]
- [43].Natarajan K, Dimasi N, Wang J, Mariuzza RA, Margulies DH. Structure and function of natural killer cell receptors: multiple molecular solutions to self, nonself discrimination. Annu. Rev. Immunol. 2002;20:853–85. doi: 10.1146/annurev.immunol.20.100301.064812. [DOI] [PubMed] [Google Scholar]
- [44].Phillips JH, Gumperz JE, Parham P, Lanier LL. Superantigen-dependent, cell-mediated cytotoxicity inhibited by MHC class I receptors on T lymphocytes. Science. 1995;268(5209):403–5. doi: 10.1126/science.7716542. [DOI] [PubMed] [Google Scholar]
- [45].Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J. Exp. Med. 1998;187(5):813–8. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Flores-Villanueva, Yunis E, Buchbinder S, Vittinghoff E, walker B.Association of two copies of HLA-B alleles encoding HLA-E binding peptides with Threonine at postion 2 with control of viremia and progression to AIDS. Unpublished.
- [47].Brooks AG, Borrego F, Posch PE, Patamawenu A, Scorzelli CJ, Ulbrecht M, Weiss EH, Coligan JE. Specific recognition of HLA-E, but not classical, HLA class I molecules by soluble CD94/NKG2A and NK cells. J. Immunol. 1999;162(1):305–13. [PubMed] [Google Scholar]
- [48].Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432(7018):769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- [49].Lanier LL. Natural killer cells: from no receptors to too many. Immunity. 1997;(4):371–8. doi: 10.1016/s1074-7613(00)80280-0. [DOI] [PubMed] [Google Scholar]
- [50].Miller JD, Weber DA, Ibegbu C, Pohl J, Altman JD, Jensen PE. Analysis of HLA-E peptide-binding specificity and contact residues in bound peptide required for recognition by CD94/NKG2. J. Immunol. 2003;171(3):1369–75. doi: 10.4049/jimmunol.171.3.1369. [DOI] [PubMed] [Google Scholar]
- [51]. WWW.anthonynolan.com/HIGseq/pep
- [52].Koka PS, Reddy ST. Cytopenias in HIV infection: Mechanisms and alleviation of hematopoietic inhibition. Curr. HIV Res. 2004;2:275–282. doi: 10.2174/1570162043351282. [DOI] [PubMed] [Google Scholar]
- [53].Miles SA, Mitsuyasu RT, Moreno J, Baldwin G, Alton NK, Souza L, Glaspy JA. Combined therapy with recombinant granulocyte colony-stimulating factor and erythropoietin decreases hematologic toxicity from zidovudine. Blood. 1991;77:2109–2117. [PubMed] [Google Scholar]
- [54].Miles SA, Lee S, Hutlin L, Zsebo KM, Mitsuyasu RT. Potential use of Human stem cell factor as adjunctive therapy for Human immunodeficiency virus-related cytopenias. Blood. 1991;78:3200–3208. [PubMed] [Google Scholar]
- [55].Ratner L. Human immunodeficiency virus-associated autoimmune thrombocytopenic purpua: A review. Am. J. Med. 1989;86:194–198. doi: 10.1016/0002-9343(89)90268-4. [DOI] [PubMed] [Google Scholar]
- [56].Fauci AS. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- [57].Harbol AW, Liesveld JL, Simpson-Haidaris PJ, Abboud CN. Mechanisms of cytopenia in human immunodeficiency virus infection. Blood Rev. 1994;8:241–251. doi: 10.1016/0268-960x(94)90112-0. [DOI] [PubMed] [Google Scholar]
- [58].Shen H, Cheng T, Preffer FI, Dombkowski D, Tomasson MH, Golan DE, Yang O, Hofmann W, Sodroski JG, Luster AD, Scadden DT. Intrinsic Human immunodeficiency virus type 1 resistance of hematopoietic stem cells despite coreceptor expression. J. Virol. 1999;73:728–737. doi: 10.1128/jvi.73.1.728-737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Koka PS, Jamieson BD, Brooks DG, Zack JA. Human immunodeficiency virus type-1 induced hematopoietic inhibition is independent of productive infection of progenitor cells in vivo. J. Virol. 1999;73:9089–9097. doi: 10.1128/jvi.73.11.9089-9097.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Koka PS, Fraser JK, Bryson Y, Bristol GC, Aldrovandi GM, Daar ES, Zack JA. Human immunodeficiency virus type 1 inhibits multilineage hematopoiesis in vivo. J. Virol. 1998;72:5121–5127. doi: 10.1128/jvi.72.6.5121-5127.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jenkins M, Hanley MB, Moreno MB, Wieder E, McCune JM. Human immunodeficiency virus-1 infection interrupts thymopoiesis and multilineage hematopoiesis in vivo. Blood. 1998;91:2672–2678. [PubMed] [Google Scholar]
- [62].Koka PS, Kitchen CM, Reddy ST. Targeting c-Mpl for revival of human immunodeficiency virus type 1-induced hematopoietic inhibition when CD34+ progenitor cells are re-engrafted into a fresh stromal microenvironment in vivo. J. Virol. 2004;78:11385–11392. doi: 10.1128/JVI.78.20.11385-11392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sankaran S, Guadalupe M, Reay E, George MD, Flamm J, Prindiville T, Dandekar S. Gut mucosal T cell responses and gene expression correlate with protection against disease in long-term HIV-1-infected nonprogressors. Proc. Natl. Acad. Sci. USA. 2005;102(28):9860–5. doi: 10.1073/pnas.0503463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Stier S, Cheng T, Dombkowski D, Carlesso N, Scadden DT. Notchl activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood. 2002;99:2369–2378. doi: 10.1182/blood.v99.7.2369. [DOI] [PubMed] [Google Scholar]
- [65].Pestina TI, Jackson CW. Differential role of Stat5 isoforms in effecting hematopoietic recovery induced by Mpl-ligand in lethally myelosuppressed mice. Exp. Hematol. 2003;31:1198–1205. doi: 10.1016/j.exphem.2003.09.002. [DOI] [PubMed] [Google Scholar]
- [66].Schulze H, Ballmaier M, Welte K, Germeshausen M. Thrombopoietin induces the generation of distinct Statl, Stat3, Stat5a and Stat5b homo- and heterodimeric complexes with different kinetics in human platelets. Exp. Hematol. 2000;28:294–304. doi: 10.1016/s0301-472x(99)00154-x. [DOI] [PubMed] [Google Scholar]
- [67].Zeng H, Masuko M, Jin L, Neff T, Otto KG, Blau CA. Receptor specificity in the self-renewal and differentiation of primary multipotential hematopoietic cells. Blood. 2001;98:328–334. doi: 10.1182/blood.v98.2.328. [DOI] [PubMed] [Google Scholar]
- [68].Bradley HL, Couldrey C, Bunting KD. Hematopoietic-repopulating defects from STAT5-deficient bone marrow are not fully accounted for by loss of thrombopoietin responsiveness. Blood. 2004;103:2965–2972. doi: 10.1182/blood-2003-08-2963. [DOI] [PubMed] [Google Scholar]
- [69].Goncalves F, Lacout C, Villeval JL, Wendling F, Vainchenker W, Dumenil D. Thrombopoetin does not induce lineage-restricted commitment of Mpl-R expressing pluripotent progenitors but permits their complete erythroid and megakaryocytic differentiation. Blood. 1997;89:3544–3553. [PubMed] [Google Scholar]
- [70].Kaushansky K. Thrombopoietin and the hematopoietic stem cell. Blood. 1998;92:l–3. [PubMed] [Google Scholar]
- [71].Kaushansky K, Lin N, Grossman A, Humes J, Sprugel KH, Broudy VC. Thrombopoietin expands erythroid, granulocyte-macrophage, and megakaryocytic progenitor cells in normal and myelosuppressed mice. Exp. Hematol. 1996;24:265–269. [PubMed] [Google Scholar]
- [72].Silvestris F, Cafforio P, Tucci M, Dammacco F. Negative regulation of erythroblast maturation by Fas-L+/TRAIL+ highly malignant plasma cells: a major pathogenic mechanism of anemia in multiple myeloma. Blood. 2002;99:1305–1313. doi: 10.1182/blood.v99.4.1305. [DOI] [PubMed] [Google Scholar]
- [73].Solar GP, Kerr WG, Zeigler FC, Hess D, Donahue C, de Sauvage FJ, Eaton DL. Role of c-mpl in early hematopoiesis. Blood. 1998;92:4–10. [PubMed] [Google Scholar]
- [74].Schuringa JJ, Chung KY, Morrone G, Moore MAS. Constitutive activation of STAT5A promotes human hematopoietic stem cell self-renewal and erythroid differentiation. J. Exp. Med. 2004;200:623–635. doi: 10.1084/jem.20041024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mingari MC, Moretta A, Moretta L. Regulation of KIR expression in human T cells: a safety mechanism that may impair protective T-cell responses. Immunol. Today. 1998;19(4):153–7. doi: 10.1016/s0167-5699(97)01236-x. [DOI] [PubMed] [Google Scholar]
- [76].Ponte M, Bertone S, Vitale C, Tradori-Cappai A, Bellomo R, Castriconi R, Moretta L, Mingari MC. Cytokine-induced expression of killer inhibitory receptors in human T lymphocytes. Eur. Cytokine Netw. 1998;9(3 Suppl):69–72. [PubMed] [Google Scholar]
- [77].Mingari MC, Ponte M, Bertone S, Schiavetti F, Vitale C, Bellomo R, Moretta A, Moretta L.HLA class I-specific inhibitory receptors in human T lymphocytes: interleukin 15-induced expression of CD94/NKG2A in superantigenor alloantigen-activated CD8+ T cells Proc. Natl. Acad. Sci. USA 1998. Feb 3;9531172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Fodil N, Pellet P, Laloux L, Hauptmann G, Theodorou I, Bahram S. MICA haplotypic diversity. Immunogenetics. 1999;49(6):557–60. doi: 10.1007/s002510050536. [DOI] [PubMed] [Google Scholar]
- [79].Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty DE. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA. 1998;95(9):5199–204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Llano M, Lee N, Navarro F, Garcia P, Albar JP, Geraghty DE, Lopez-Botet M. HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA-G-derived nonamer. 1998. Eur. J. Immunol. 28(9):2854–63. doi: 10.1002/(SICI)1521-4141(199809)28:09<2854::AID-IMMU2854>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- [81].Andre P, Brunet C, Guia S, Gallais H, Sampol J, Vivier E, Dignat-George F. Differential regulation of killer cell Ig-like receptors and CD94 lectin-like dimers on NK and T lymphocytes from HIV-1-infected individuals. Eur. J. Immunol. 1999;29(4):1076–85. doi: 10.1002/(SICI)1521-4141(199904)29:04<1076::AID-IMMU1076>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- [82].Sundell IB, Koka PS. Chimeric SCID-hu model as a human hematopoietic stem cell host that recapitulates the effects of HIV-1 on bone marrow progenitors in infected patients. J. Stem Cells. 2006;1(4):283–300. [PMC free article] [PubMed] [Google Scholar]
- [83].Melkus MW, Estes JD, Padgett-Thomas A, Gatlin J, Denton PW, Othieno FA, Wege AK, Haase AT, Garcia JV. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-I. Nature Med. 2006;12(11):1316–1322. doi: 10.1038/nm1431. [DOI] [PubMed] [Google Scholar]