Abstract
The balance between dynamic and stable actin filaments is essential for the regulation of cellular functions including the determination of cell shape and polarity, cell migration, and cytokinesis. Proteins that regulate polymerization at the filament ends and filament stability confer specificity to actin filament structure and cellular function. The dynamics of the barbed, fast-growing end of the filament are controlled in space and time by both positive and negative regulators of actin polymerization. Capping proteins inhibit the addition and loss of subunits, whereas other proteins, including formins, bind at the barbed end and allow filament growth. In this work, we show that tropomyosin regulates dynamics at the barbed end. Tropomyosin binds to constructs of FRL1 and mDia2 that contain the FH2 domain and modulates formin-dependent capping of the barbed end by relieving inhibition of elongation by FRL1- FH1FH2, mDia1-FH2, and mDia2-FH2 in an isoform-dependent fashion. In this role, tropomyosin functions as an activator of formin. Tropomyosin also inhibits the binding of FRL1-FH1FH2 to the sides of actin filaments independent of the isoform. In contrast, tropomyosin does not affect the ability of capping protein to block the barbed end. We suggest that tropomyosin and formin act together to ensure the formation of unbranched actin filaments, protected from severing, that could be capped in stable cellular structures. This role, in addition to its cooperative control of myosin function, establishes tropomyosin as a universal regulator of the multifaceted actin cytoskeleton.
Reversible polymerization of actin is essential for many cellular functions including determination of cell shape and polarity, cell movement, cytokinesis, and intracellular transport. The ends of actin filaments are structurally distinct and differ in the rates of incorporation and dissociation of actin subunits. Elongation at the fast growing, barbed end predominates in cells and is precisely controlled in space and time by positive and negative regulators of actin polymerization. Capping proteins bind and prevent addition and loss of subunits, whereas other proteins, including formins, bind at the barbed end and allow filament growth and depolymerization [reviewed in (1, 2)].
Formins are a widespread family of barbed-end binding proteins that are implicated in the assembly of unbranched actin-containing structures including microvilli and filopodia, stress fibers, and contractile rings. They are required for the formation of actin cables in yeast. Formins prevent the binding of capping protein and modulate the rate of monomer addition to the barbed end [reviewed in (3, 4)].
Formins are classified on the basis of the presence of formin homology domains, FH11 and FH2, which are responsible for most of its effects on actin [reviewed in (5)]. Fragments containing the FH2 domain or FH1FH2 domains form dimers and nucleate de novo filament assembly of monomeric actin. Formin remains associated with the elongating barbed end of the filament, leading to its description as a processive, leaky capper (6-8). Although formins share common mechanisms, isoforms differ in the ability to nucleate polymerization, inhibit elongation, bind to the sides of filaments, and bundle and sever filaments, and in their processivity (9).
The capping protein family of proteins, expressed in virtually all eucaryotic cells, are antagonistic to formins. Capping protein is an αβ heterodimer localized at the ends of filaments where unbranched actin filaments end in stable structures such as Z-discs in muscle and adhering junctions, and at filament ends in lamellipodia [(10); reviewed in (11)]. Capping protein is also associated with actin patches and endosomal vesicles in yeast (12-14). In vertebrates, there are two isoforms of each subunit with tissue-specific expression patterns and functions. Negative regulators of capping protein include the phosphoinositide, PIP2 (15), and proteins that specifically complex with capping proteins such as CARMIL and myotrophin/V-1 (16, 17). No positive regulators have been reported.
Formins and capping protein are often associated with the formation and maintenance of long, unbranched actin filaments [reviewed in (3, 18)] that contain tropomyosin [reviewed in (19)]. Tropomyosins are coiled-coil proteins that associate N-terminus to C-terminus on the actin filament to form a continuous strand that follows the filament helix, one strand on each side of the actin filament [reviewed in (20, 21)]. A large protein family, tropomyosins have cell-and tissue-specific expression patterns and functions [reviewed in (19)]. It is best known for its classic role in the regulation of striated muscle contraction together with troponin and myosin. In addition, tropomyosin physically stabilizes actin filaments and protects them against the action of depolymerizing and severing proteins, such as DNase I, cofilin, and gelsolin as well as the nucleation of branches by the Arp2/3 complex (22-30). Tropomyosin, together with tropomodulin, caps the pointed ends of actin filaments in the sarcomeres of striated muscles and in the erythrocyte membrane cytoskeleton [reviewed in (31)], and can itself depress depolymerization from the pointed end (32-34).
A role for tropomyosin in regulating the barbed end of the filament is less well established. Direct interaction with an actin-binding domain of gelsolin, a barbed end-capping protein, is well documented, as is the ability of tropomyosin to dissociate gelsolin and promote annealing of short actin filaments (35-38). There is circumstantial evidence for an effect of tropomyosin on formin function from studies in Saccharomyces cerevisiae. Actin cables, which are polarized arrays of tropomyosin-containing filaments that are needed for cellular polarization, require both formin (Bni1p) and tropomyosin to form (39-41).
Here, we show that tropomyosin regulates filament dynamics at the barbed end, as it does the pointed end. Because tropomyosin is often present in formin-nucleated filaments in cells, we postulated that it may regulate formin. Tropomyosin binds directly to FRL1 and mDia2 constructs that contain the FH2 domain and relieves the inhibition of barbed-end elongation by formin in an isoform-specific manner. Binding of tropomyosin to actin filaments inhibits the association of FRL1-FH1FH2 with the filament sides. In contrast, the tropomyosin isoforms we studied do not relieve blocking of the barbed end by capping protein. Tropomyosin’s roles in stabilizing the filament from severing, and in regulating the dynamics of the filament ends, alone and in concert with other regulatory proteins, positions it as a universal regulator of the multifaceted actin cytoskeleton.
EXPERIMENTAL PROCEDURES
Protein Purification
Chicken skeletal muscle actin was extracted from acetone powder according to the method of Spudich and Watt (42) and further purified by gel filtration on Sephacryl S-100 HR. Immediately after purification, G-actin (in 2 mM Tris-HCl at pH 8.0, 0.2 mM ATP, 0.2 mM CaCl2, 0.5 mM DTT, and 0.01% NaN3) was quickly frozen in liquid nitrogen at −80 °C or stored at 4 °C and used within 2 weeks. Actin labeled with N-(1-pyrenyl)-iodoacetamide was prepared as described in ref 43, followed by gel filtration on Sephacryl S-100 HR.
Recombinant rat α-tropomyosins, TM5a, TM2, and unacetylated stTM (stTM-rec), were expressed in Escherichia coli BL21(DE3) cells and purified (44). N-acetylated striated muscle α-TM (stTM) was purified from chicken pectoral muscle (45). Recombinant FRL1-FH1FH2 (a.a. res. 449–1094), mDia1-FH2 (a.a. res. 748–1175), and mDia2-FH2 (a.a. res. 612–1034) were expressed in E. coli and purified (46). Recombinant capping proteins (CP) were expressed and purified (47): chicken muscle CPα1β1, mouse CPα1β2, and mouse CPα2β2.
Protein concentrations were determined spectrophotometrically, using absorption coefficients of 22,100 M−1 cm−1, at 280 nM for mDia2-FH2; 25,700 at 280 nm for FRL1-FH1FH2; and 20,160 M−1 cm−1 at 280 nM for mDia1-FH2. Tropomyosin and actin concentrations were calculated by measuring the difference spectrum between pH 12.5 and pH 6 in 6 M guanidine-HCl, using a molar absorption coefficient for tyrosine of 2480 (48).
Elongation Assays
Elongation of 0.5 μM Mg-G-actin (10% pyrenyl-labeled, in F-buffer: 10 mM Tris at pH 7.4, 0.2 mM ATP, 0.2 mM DTT, 0.01% NaN3, 0.2 mM EGTA, 100 mM NaCl, 2 mM MgCl2) on 1–2 μM F-actin seeds, in the presence or absence of CP, formin, and tropomyosin, was monitored by an increase in pyrene fluorescence, at 25 °C. Mg-G actin was prepared by treating Ca-G-actin with 0.2 mM EGTA and 0.05 mM MgCl2 for 3 min immediately prior to the experiment. For the seeds, 20–28 μM Mg-G-actin was polymerized overnight in 100 mM NaCl and 2 mM MgCl2 to allow spontaneous annealing of filaments before the addition of tropomyosin. Elongation was initiated by the addition of F-buffer (10×) and concentrated (10– 14×) F-actin seeds to the cuvettes containing Mg-G-actin. The F-actin seeds were added using a Microman pipet and gently mixed with a plastic stirrer to minimize shearing. The figure legends include specific experimental details.
Time courses of pyrene fluorescence were measured on a PTI fluorimeter (Lawrenceville, NJ) with the excitation and emission wavelengths of 365 and 386 nm, respectively. Each data set was normalized by setting the final fluorescence increase at steady state, when available, or the value reported following curve fitting, as 100%. Normalized data were fit to the single- or double-exponential growth model, and initial rates of the reaction were calculated as first derivatives at t = 0 (GraphPad Prizm 4).
Actin Filament Sedimentation
A stock solution of 10–30 μM Mg-G-actin was polymerized overnight with 100 mM NaCl and 2 mM MgCl2. F-Actin was diluted with polymerization buffer for a final concentration of 5 μM prior to addition of phalloidin (5 μM), tropomyosin, and/or FRL1-FH1FH2. After 0.5-1 h, the samples (150 μL) were centrifuged for 20 min, at 60,000 rpm, in a TLA-100 rotor (Beckman) at 20 °C. One hundred microliters of supernatant were removed and dried on SpeedVac Plus (Savant). Pellets and supernatants were analyzed using SDS–PAGE (9% gels) according to Laemmli (49) and quantified on a Molecular Dynamics model 300A densitometer.
Circular Dichroism Spectroscopy
Circular dichroism experiments were performed on an Aviv model 215 spectrometer equipped with a 5 sample changer. Spectra of the tropomyosins, TM5a and TM2, and the formins, FRL1-FH1FH2 and mDia2-FH2, and their mixtures were obtained at 0 °C in 100 mM NaCl, 10 mM Tris-HCl, 0.2 mM EGTA, 0.1 mM MgCl2, 0.01% NaN2, and 0.5 mM dithiothreitol at pH 7.4. Data were collected at 0.25 to 0.5 nm intervals, corrected for baselines and smoothed with a polynomial order of 3 and a window of 21 to 11 points, respectively. The concentrations of the proteins ranged from 0.1 to 2 μM. Data were obtained in 0.1 and 1 cm cells. Thermal denaturation curves of 1–2 μM samples were obtained between 0 and 60–70 °C at 0.2 degree intervals with 5 s time averaging. The data were not smoothed. The first derivatives of the denaturation curves were obtained using data smoothed with a polynomial order of 3 and a window of 21 points and fitting the data to a third order polynomial. The helical content of FRL-FH1FH2 was estimated from its CD spectra by fitting it to standards for α-helical, β-pleated sheets, β-turns, and random structures using non-constrained least-squares analysis (50).
RESULTS
Tropomyosin Regulates the Dynamics of the Actin Filament’s Barbed End
We evaluated the influence of muscle and non-muscle tropomyosins on actin filament barbed-end elongation alone, in the presence of three formins that contain the FH2 actin binding domain, FRL1-FH1FH2, mDia1-FH2, and mDia2-FH2 (46, 51), and with three capping protein isoforms, CPα1β1, CPα1β2, and CPα2β2 (47). We compared three tropomyosins, all products of the α-TM gene [TPM1, (19)]. TM5a is a recombinant short, non-muscle isoform; TM2 is a recombinant long, non-muscle isoform; and stTM is a long tropomyosin isolated from skeletal muscle. TM2 and TM5a have the same C-terminal sequence, the end that is oriented toward the barbed end of the actin filament (52). The two long tropomyosins have the same N-terminal sequence that differs from TM5a, but stTM is N-acetylated (19).
We measured the polymerization of the pyrene-actin monomer (10% labeled) on actin filaments polymerized overnight to maximize filament length prior to the addition of tropomyosin because annealing by muscle tropomyosin is inversely related to filament length (38, 53). The initial velocity of elongation (Vo) in the presence of regulatory proteins was calculated and compared to that of F-actin (taken as 100%). Tropomyosin influenced the rate of elongation in an isoform-specific manner. Figure 1 illustrates data from a representative experiment, along with the values relative to actin alone averaged from several experiments. Actin filaments saturated with TM5a and TM2 elongated at a faster rate than actin alone (137 ± 7%, n = 4; 127 ± 14%, n = 4, respectively). The increased rates suggest a specific barbed-end effect that is not the result of new barbed ends formed by severing because TM5a and TM2 had no effect on the rate of elongation of actin filaments in the presence of capping protein (Figure 7) or on gelsolin-capped filaments in which TM5a partially blocked the pointed end (34). Striated muscle TM (stTM) slowed the overall rate of elongation (62 ± 8%, n = 5). In our earlier work using a microscopic assay, we found that rabbit αα-stTM (vs chicken αα-stTM used here) had no significant effect on the rate of barbed-end elongation, but rabbit αβ-stTM reduced the rate (54). The effects of tropomyosin on elongation rate were independent of the time of incubation with F-actin (minutes to 8 h).
Figure 1.

Effect of tropomyosin on elongation at the barbed end of actin filaments. (A) A representative time course of elongation of 0.5 μM Mg-G-actin (10% pyrene labeled) from 2 μM F-actin seeds (1) or F-actin in the presence of saturating amounts of tropomyosin (44): 0.5 μM TM5a (2), 2 μM TM2 (3), and 1 μM stTM (4) in the absence of formin or capping protein, or G-actin alone without F-actin seeds (5) in 100 mM NaCl, 2 mM MgCl2, 10 mM TrisHCl at pH 7.4, 0.2 mM ATP, 0.2 mM EGTA, and 0.2 mM DTT. (B) The initial velocities (Vo) relative to F-actin alone, taken as 100%, with standard error (n = 4–5). The gray curves are the fits to the data.
Figure 7.
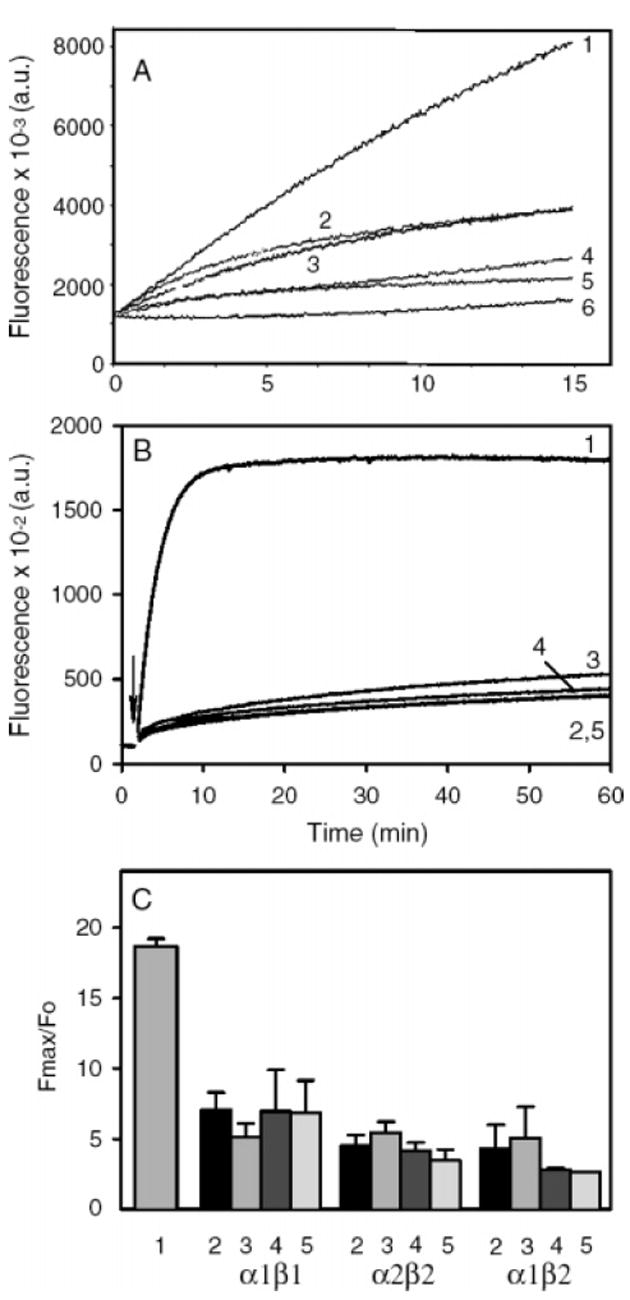
Tropomyosin does not unblock filaments with capping protein at the barbed end. (A) Spectrin-actin seeds were used to nucleate polymerization of pyrene-labeled monomeric actin (5% labeled). The seeds were prepared as previously described (15, 78), by incubating spectrin-actin with F-actin overnight to give F-actin seeds ~200 actins long, capped at their pointed ends by spectrin (1.7 nM spectrin-actin seeds and 0.5 μM F-actin). stTM was added to the F-actin to 2 μM before addition of the spectrin-actin and incubated overnight. The capping protein (α1β2) was added to the filaments just prior to the initiation of polymerization. The F-actin seeds were diluted 1:10 into Mg-G-actin (2 μM, 5% pyrene labeled) to seed polymerization in 100 mM KCl, 2 mM MgCl2, 10 Mm TrisHCl at pH 7.5, 0.2 mM ATP, 0.5 mM DTT, 0.2 mM CaCl2, and 1 mM EGTA. When present, tropomyosin concentration was 2 μM to ensure saturation of any polymer formed. Curves: 1, F-actin seeds; 2, F-actin seeds and 1.5 nM CP; 3, F-actin seeds, stTM, and 1.5 nM CP; 4, F-actin seeds and 4 nM CP; 5, F-actin seeds, stTM, and 4 nM CP; 6, G-actin alone. (B, C) Elongation experiments were carried out as described in Figures 1 and 2, and Experimental Procedures, at a final concentration of 1.5 nM CP. (B) Data from a representative data set with CPα2β2: 1, actin alone; 2, CP and no tropomyosin; 3, CP + TM5a; 4, CP + TM2; 5, CP + stTM. (C) Fmax (at 60 min) relative to the initial fluorescence (Fo) for actin alone, and the three capping proteins with the three tropomyosin isoforms, mean with standard error for 3 experiments with CP alone and TM5a, two to three experiments with TM2, and one to two experiments with stTM. The numbering corresponds to that in B.
Tropomyosin Regulates Capping of the Actin Filament by Formin
The three formins inhibited barbed-end elongation to different extents, as previously reported (5, 46). mDia2-FH2 was the most effective (13 ± 1%, relative to actin alone, n = 4) (Figure 2C and D). The kinetics of the fluorescence increase in the presence of FRL1-FH1FH2 (Figure 2A) and mDia1-FH2 (Figure 2E) closely followed a single-exponential reaction model. However, the best or only fits for the curves in the presence of mDia2-FH2 (Figure 2C) were obtained using a two-phase exponential model that describes two independent, simultaneous reactions. The result suggests the presence of two populations of filament ends elongating at different rates.
Figure 2.
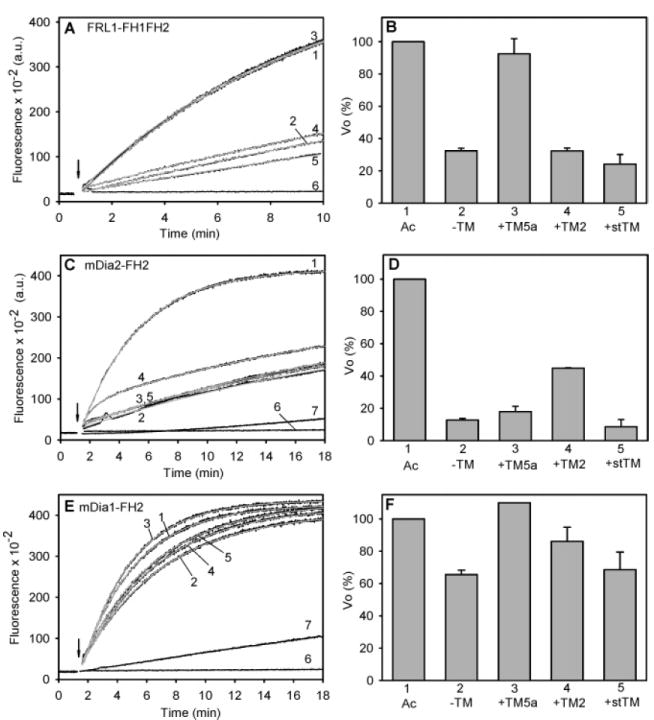
Effect of tropomyosin on elongation at the barbed end of actin filaments in the presence of formin. (A, C, E) Representative time courses. (B, D, F) Relative initial rates of elongation. (A, B) 25 nM FRL1-FH1FH2, 1 μM F-actin seeds; n = 4. (C, D) 50 nM mDia2-FH2, 2 μM F-actin seeds; no TM, n = 4; TM5a, n = 4; TM2, n = 2; stTM, n = 3. (E, F) 25 nM mDia1-FH2, 2 μM F-actin seeds; noTM, n = 3; TM5a, n = 1; TM2, n = 2; stTM, n = 2. The tropomyosin concentrations and ionic conditions are the same as those in Figure 1. Curves: 1, F-actin alone (no formin or tropomyosin); 2, F-actin with formin, no tropomyosin; 3, formin and TM5a; 4, formin and TM2; 5, formin and stTM; 6, G-actin alone, no F-actin seeds; and 7, G-actin with formin, no F-actin seeds. FRL1-FH1FH2 had no effect with G-actin alone and is not shown. The gray curves are the fits to the data. Ac: F-actin, without tropomyosin or formin (from curve 1).
Tropomyosin relieved the inhibition by formins in an isoform-specific manner. The inhibition of barbed-end elongation by FRL1-FH1FH2 was almost completely relieved by TM5a, whereas TM2 and stTM had little effect (Figure 2A and B). In contrast, inhibition by mDia2-FH2 was partially relieved by TM2 but was little affected by TM5a or stTM (Figure 2C and D). The inhibition by mDia1-FH2 was less, but it was relieved by TM5a and to a lesser extent by TM2 (Figure 2E and F). In no case did tropomyosin relieve inhibition by formin to the level of actin with tropomyosin alone (compare with Figure 1). The results were independent of the time of incubation or order of addition of formin and tropomyosin to the pre-assembled filaments.
FRL1-FH1FH2 Inhibits ActiVation of Barbed-End Elongation by TM5a
We measured the rate of barbed-end elongation as a function of tropomyosin concentration to understand the relationship between the relief of inhibition by formins and assembly of a tropomyosin-actin filament. The ability of TM2 to relieve inhibition by mDia2-FH2 and the activation by TM2 alone corresponded to saturation of the actin filament (results not shown.).
In contrast, the dependence of activation and relief of inhibition by FRL1-FH1FH2 on TM5a concentration were more complicated (Figure 3). At saturation, TM5a activated the rate of barbed-end elongation ~1.4-fold compared to actin alone (Figure 1). The unanticipated finding was that activation by TM5a was greatest at substoichiometric levels and reached a minimum at saturation, as illustrated in a representative experiment in Figures 3. The activation at 0.05 μM was 540 ± 76% (680%, 530%, and 420% in three experiments), and at 0.1 μM TM5a, the activation was 370 ± 160% (530% and 200% in two experiments) when F-actin was less than 10% saturated with TM5a (i.e., almost undetectable; Figure 3D) compared to 137 ± 7% (n = 3) at saturation. The variability in the magnitude of activation may be because the number of free barbed ends in the filaments used to nucleate polymerization differs from one experiment to the next.
Figure 3.
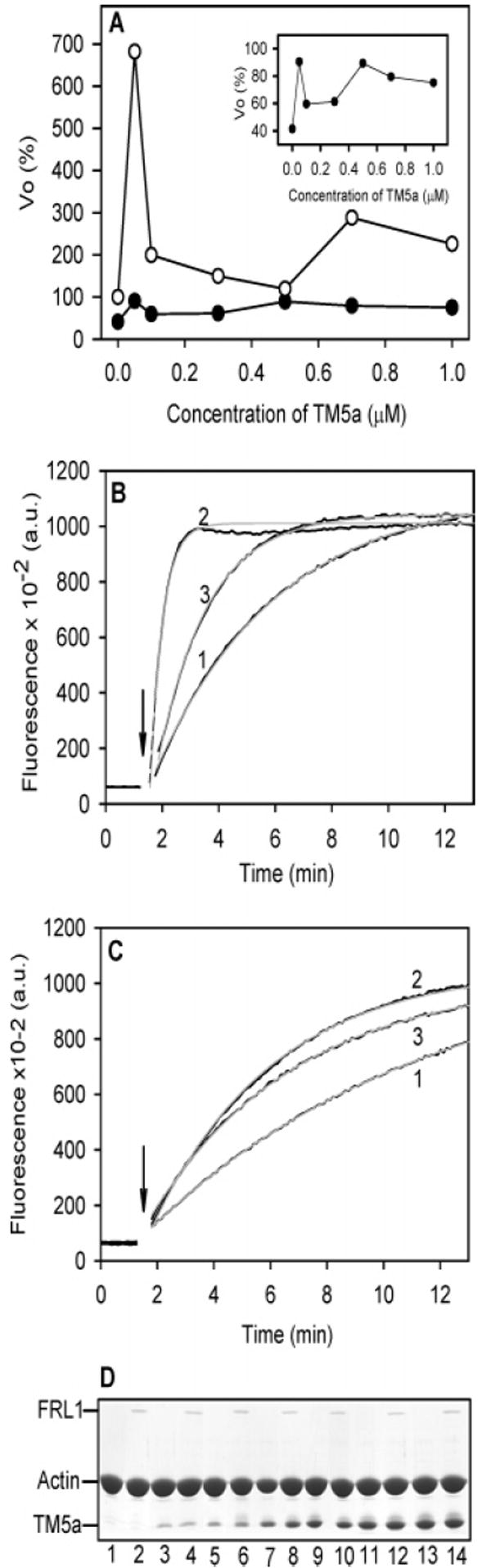
Relief of FRL1-FH1FH2 inhibition of barbed-end elongation by TM5a does not require saturation of the actin filaments with tropomyosin. The results from a representative experiment are shown; 50 nM FRL1-FH1FH2, 2 μM F-actin seeds, TM5a, varied. (A) The Vo of barbed-end elongation as a function of TM5a concentration in the absence of FRL1-FH1FH2 (○) and in the presence of 50 nM FRL1-FH1FH2 (●). The panel shows the data with FRL1-FH1FH2 on an expanded scale. (B) The time course of increase in pyrene fluorescence in the absence of FRL1-FH1FH2 in the absence of TM5a (1), a subsaturating concentration, 0.05 μM TM5a (2), and a saturating concentration, 1 μM TM5a (3). (C) The time course of increase in pyrene fluorescence in the presence of 50 nM FRL1-FH1FH2 in the absence of TM5a (1), a subsaturating concentration, 0.05 μM TM5a (2), and a saturating concentration, 1 μM TM5a (3). The gray curves are the fits to the data. (D) SDS–PAGE gels of the pellets following centrifugation at the end of the polymerization experiment. Even numbers, with FRL1-FH1FH2; odd numbers, no FRL1. TM5a concentration: 1 and 2, no TM5a; 3 and 4, 0.05 μM; 5 and 6, 0.1 μM; 7 and 8, 0.3 μM; 9 and 10, 0.5 μM; 11 and 12, 0.7 μM; 13 and 14, 1.0 μM. The ionic conditions are the same as those in Figure 1.
The presence of FRL1-FH1FH2 inhibited the activation of barbed-end elongation at the lowest TM5a concentration, whereas TM5a relieved the capping activity of the formin. In the presence of FRL1-FH1FH2, the rate of barbed-end elongation was similar at all TM5a concentrations. The amount of FRL1-FH1FH2 in the pellet was independent of tropomyosin (Figure 3D), suggesting that formin remains on the barbed end in the presence of tropomyosin.
The FRL1 concentration and ionic strength in these experiments (0.05 μM FRL1-FH1FH2 and 100 mM NaCl) minimized the formation of filament bundles (51), verified by no observed change in the actin content in the supernatant following low-speed centrifugation (results not shown.). We conclude that TM5a specifically influences the barbed end and that activation may be inhibited by interaction with FRL1-FH1FH2, independent of stoichiometric TM5a binding to actin. The mutual neutralization may take place via direct complex formation because the FRL1-FH1FH2 concentration used (0.05 μM) was sufficient to inhibit activation by 0.05 μM TM5a.
Formins Containing the FH2 Domain Bind Tropomyosin
We measured binding between formin and tropomyosin using circular dichroism, a method we have used for the analysis of binding between tropomyosin and other proteins (Figure 4) (34, 55-57). When TM5a and FRL1-FH1FH2 were combined in an ~1:1 ratio, the negative ellipticity at 222 nm of the mixture of the proteins increased 9 ± 1% (n = 5) compared to the sum of the curves, evidence for complex formation (Figure 4A). The percent increase in ellipticity was independent of tropomyosin concentration over the range of 0.01 to 0.2 μM, indicating that the binding is relatively tight (Kd < 1 μM). Because TM5a is ~100% helical, we infer that binding of TM5a to FRL1 induces the formation of the α-helix in FRL1-FH1FH2. Although the X-ray structures of FH2 domains are highly helical, the CD spectrum of FRL1-FH1FH2 is at most ~50% helical in the absence of a binding partner. The increase in helical content was lost upon heat denaturation (Figure 4B). TM5a has multiple unfolding transitions that overlap to give one broad unfolding transition at ~41 °C, close to that of FRL1-FH1FH2 (~39.5 °C, Figure 4C). There was no shift in the overall TM of unfolding of the FRL1-FH1FH2-TM5a mixture, compared to that of the unmixed components. However, the unfolding of the complex was more cooperative, as evidenced by the greater peak height of the first derivative of the unfolding curve.
Figure 4.
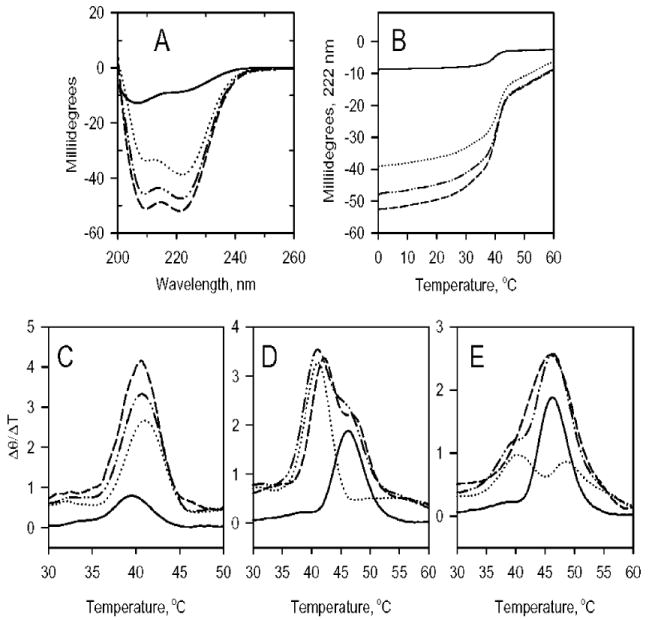
Circular dichroism measurements of the interactions of formin domains with tropomyosin. (A) Spectra of FRL1-FH1FH2 (—); TM5a (····); a 1:1 mixture of FRL1-FH1FH2 and TM5a (- - -); and addition of the curves of unmixed FRL and TM5a (-··-). (B) The change in ellipticity at 222 nm as a function of temperature of the samples in panel A. (C) The first derivatives of the curves in panel B. (D) The first derivative of the unfolding curves of mDia2-FH2 (—); TM5a (····); a 1:1 mixture of mDia2-FH2 and TM5a (- - -); and addition of the curves of the unmixed components (-··-). (E) The first derivatives of the change in ellipticity at 222 nm as a function of temperature of mDia2-FH2 (—); TM2 (····); a 1:1 mixture of mDia2- FH2 and TM2 (- - -); and addition of the curves of the unmixed components (-··-). The TM of the unfolding transition of TM5a increases ~1 °C when bound to mDia2-FH2 (D), and the TM of the first unfolding transition of TM2 is increased by ~2 °C (E). Conditions: 0 °C in 100 mM NaCl, 10 mM Tris-HCl, 0.2 mM EGTA, 0.1 mM MgCl2, 0.01% NaN2, and 0.5 mM dithiothreitol at pH 7.4. The formin and tropomyosin concentrations were 1 μM.
Thermal denaturation experiments show that mDia2-FH2 also binds to TM5a and TM2 and indicate the involvement of the FH2 domain (Figures 4D and E). The mDia2-FH2 domain is more helical and more stable than FRL1-FH1FH2 and unfolded with a TM of ~46 °C. The C-terminal region of TM5a and TM2, encoded by exon 9d, is relatively unstable, and it unfolds during the first major transition of TM2 at ~40 °C (58). When mDia2-FH2 was bound to TM5a (Figure 3D) or TM2 (Figure 3E), the TM of unfolding of the first transitions near 40 °C increased by ~1 to 2 °C, suggesting that the FH2 domain binds to the C-terminal region of tropomyosin.
Even though the CD binding studies indicate that the formin FH2 domain binds the C-terminal domain of tropomyosin, the end oriented toward the barbed end of the filament (52), relief of formin-dependent capping is isoform-specific and is influenced by the rest of the tropomyosin molecule, not just the C-terminus. TM5a relieves FRL1-FH1FH2 inhibition of barbed-end elongation, whereas TM2 affects mDia2, even though the two tropomyosins contain the same C-terminal sequence.
Tropomyosin Dissociates FRL1-FH1FH2 from the Sides of Filaments
Constructs containing the FH2 domain of some formins can bind to the sides of actin filaments and bundle them (51). In the ionic conditions optimal for tropomyosin binding (100 mM NaCl and 2 mM MgCl2), FRL1-FH1FH2 bound poorly to actin (Figure 5). This is consistent with the observation of Harris et al. (59) and others (60) who found that side binding of formins to filamentous actin is sensitive to ionic strength. However, 85–88% of the FRL1-FH1FH2 bound to phalloidin-stabilized actin filaments (Figure 5).
Figure 5.
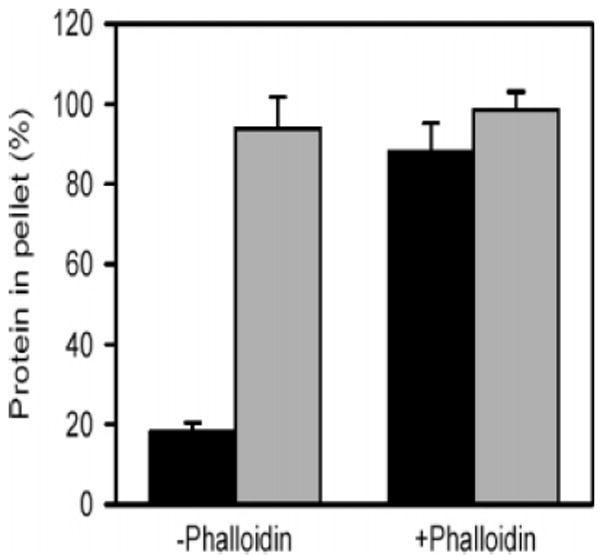
Stabilization of F-actin by phalloidin allows side binding of FRL1-FH1FH2 at near physiological ionic strength. F-actin (5 μM), in the absence or presence of 5 μM phalloidin, was cosedimented with 0.5 μM FRL1-FH1FH2 in 100 mM NaCl, 2 mM MgCl2, 10 mM TrisHCl at pH 7.4, 0.2 mM ATP, 0.2 Mm EGTA, and 0.2 mM DTT. The actin and the formin were quantified in SDS gels of the pellets and supernatants. The percent actin (gray bars) and FRL1-FH1FH2 (black bars) in the pellets are illustrated with standard errors, n = 3.
Tropomyosin inhibited the side binding of FRL1-FH1FH2 to phalloidin-stabilized actin filaments, independent of the tropomyosin isoform (Figure 6). Filaments saturated with tropomyosin reduced formin binding by about 50%. Recombinant striated muscle TM, a form that lacks the N-terminal acetyl group required for high affinity binding (61, 62), did not inhibit FRL1-FH1FH2 binding and serves as a control to show that inhibition of side binding of FRL1-FH1FH2 requires tropomyosin binding to the actin filament. The lack of isoform specificity suggests that tropomyosin’s inhibition of side binding is via a general mechanism, comparable to its inhibition of cross-linking and severing by cofilin, gelsolin, and related proteins (24-26, 28) and the inhibition of nucleation of branches of filaments by the Arp2/3 complex (63).
Figure 6.

Tropomyosin binding to F-actin partially dissociates FRL1-FH1FH2 from the filament side independent of the isoform. Phalloidin-stabilized F-actin (5 μM) was cosedimented with 0.5 μM FRL1-FH1FH2 and 1.3 μM TM5a, TM2, stTM, or recombinant, unacetylated stTM expressed in E. coli, stTM(rec). The proteins in the pellets were quantified following SDS–PAGE. (A) A gel from a representative experiment. (B) FRL1-FH1FH2/actin ratio in the pellet; the value in the absence of TM was set at 100%. (C) TM/actin ratio in the pellet with arbitrary units. A value of 0.35–0.4 represents saturation. stTM(rec) has low affinity for actin and is used here as a control to show that dissociation from the sides of the filaments requires binding of tropomyosin. The bar graphs show standard error, n = 3.
Tropomyosin Does Not Unblock Filaments with Capping Protein at the Barbed End
Stable actin filaments with capping protein at the barbed end, such as those in adhering junctions or at the Z-disc or dense bodies in muscle, contain tropomyosin. We tested the ability of tropomyosin to regulate barbed-end capping by three recombinant CapZ isoforms: α1β1 (β1 is muscle-specific; CapZ localized at the Z-line), α1β2 and α2β2 (β2 is widely expressed) (47, 64, 65). Actin polymerization was nucleated by actin filaments with bound capping protein and tropomyosin. We selected conditions in which capping in the absence of tropomyosin was incomplete. Tropomyosin neither relieved the inhibition of elongation by capping protein as it did formin-capped filaments (above) nor improved capping activity (Figure 7). In all cases, the amount of polymer formed was well below that of the actin and similar to that with capping protein alone, indicating no significant effect of tropomyosin on capping protein function. The results of binding measurements using circular dichroism were negative (results not shown).
DISCUSSION
We show here for the first time that tropomyosin regulates the dynamics of the barbed, plus end of the actin filament, alone, by uncoupling the capping function of formins and by inhibiting the binding of FRL1 to the sides of filaments. Tropomyosin’s role as a universal regulator of the actin filament now extends to both ends of the filament, in addition to overall filament stiffening and stabilization and the allosteric regulation of the actin filament in contractile function via its cooperative regulation of myosin and with troponin [reviewed in (19-21)].
Formins are leaky, processive cappers [reviewed in (3, 4)]. The binding of the mDia1-FH2 dimer to the barbed end makes the actin filament more flexible over a long range and the entire filament more dynamic (60, 66), just as binding of gelsolin at the filament end cooperatively influences filament structure and dynamics (67). Assuming that the FH2 domains of other formins have similar allosteric effects, the stabilization of the filament by tropomyosin may antagonize the effect of formin and reduce its ability to block the barbed end, thereby making it a leakier but still processive capper.
An effect of tropomyosin itself on the barbed end is illustrated by the marked stimulation of elongation at substoichiometric TM5a and its neutralization by FRL1-FH1FH2. The results infer that TM5a has a specific influence on the barbed end where the first molecules may bind the filament. As tropomyosin cooperatively saturates the filament and stabilizes the structure, stimulation of barbed-end elongation is dampened. The binding of FRL1-FH1FH2 at the barbed end also depresses the effect of substoichiometric TM5a, presumably because the cap formed by the FH2 domain with the two terminal actin subunits (68) impedes the addition of actin subunits. Tropomyosin, by binding to the FH2 domain, relieves capping by formin. In this scenario, tropomyosin is an activator. Although there is no specific structural information available, we postulate that the flexible hinge near the C-terminus of tropomyosin (69-71) allows interaction of the C-terminal chains with helical regions of the FH2 domain, possibly forming a coiled coil as they do with the N-terminus of tropomyosin and troponin T (69, 71).
Tropomyosin has no observed effect on capping protein function, and we have no evidence for direct interaction between the proteins. Formins compete with CapZ proteins for the barbed end, but they should operate by distinct mechanisms because they have different structures and different proposed interactions with the filament end (68, 72-74). Tropomyosin binds to the G2 domain of gelsolin (37), but it has no marked effect on gelsolin capping of long filaments (34). The cartoons in Figure 8 illustrate the different relationships of tropomyosin to the formin FH2 domain and capping protein at the barbed end of the actin filament (based on models in refs 68 and 74). Tropomyosin binds along the sides of the actin filament and, at the barbed end, binds to the formin FH2 domain but not capping protein.
Figure 8.
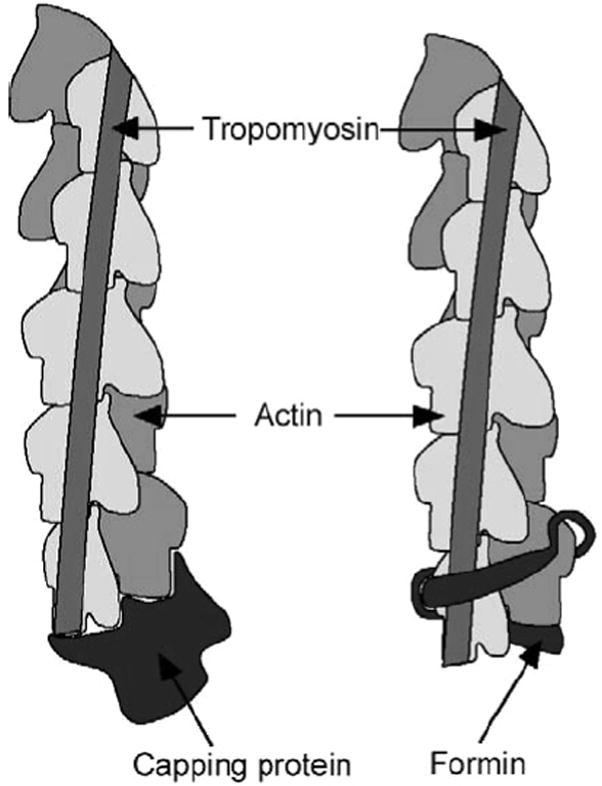
Regulatory proteins at the barbed end of the actin filament. A cartoon of the barbed end of the actin filament shows the formin FH2 domain based on ref 68 or the capping protein based on ref 74. Tropomyosin binds along the actin filament (not precisely positioned in the cartoon; the tropomyosin on only one side of the filament is shown) such that the C-terminal end interacts with formin-FH2, regulating its assembly function but with no effect on the capping protein.
The influence of tropomyosin on the side binding of FRL1-FH1FH2 (and presumably other formins) occurs by a general mechanism, without tropomyosin isoform specificity. In this regard, it is similar to the effect of tropomyosin on the side binding, severing, and bundling by other actin binding proteins, including DNase I, gelsolin, the Arp2/3 complex, and cofilin (23-26, 28). Tropomyosin and formin acting together would ensure the formation of a stable, straight filament, protected from severing that could eventually be capped by a CapZ family protein in a stable cellular structure such as an adhering junction or a Z-disk in striated muscle. We suggest that tropomyosin may have a similar role in modulating the formation of stable actin filaments in concert with Ena/VASP family proteins (75-77).
Acknowledgments
We thank Elizabeth Harris for the preparation of the formin constructs and Dr. Nandini Bhattacharya for the capping proteins.
Footnotes
This work was supported by NIH Grants GM36326 and GM63257 to S.E.H.D., GM38542 to J.A.C., and GM69818 to H.N.H.
Abbreviations: FH, formin homology domain; TM, tropomyosin; CP, capping protein.
References
- 1.dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol ReV. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson-Dykstra S, Higgs HN, Harris ES. Actin dynamics: growth from dendritic branches. Curr Biol. 2005;15:R346–357. doi: 10.1016/j.cub.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 3.Faix J, Grosse R. Staying in shape with formins. DeV Cell. 2006;10:693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Kovar DR. Cell polarity: formin on the move. Curr Biol. 2006;16:R535–R538. doi: 10.1016/j.cub.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 5.Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe N, Higashida C. Formins: processive cappers of growing actin filaments. Exp Cell Res. 2004;301:16–22. doi: 10.1016/j.yexcr.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 9.Michelot A, Derivery E, Paterski-Boujemaa R, Guerin C, Huang S, Parcy F, Staiger CJ, Blanchoin L. A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr Biol. 2006;16:1924–1930. doi: 10.1016/j.cub.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 10.Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 11.Wear MA, Cooper JA. Capping protein: new insights into mechanism and regulation. Trends Biochem Sci. 2004;29:418–428. doi: 10.1016/j.tibs.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Waddle JA, Karpova TS, Waterston RH, Cooper JA. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K, Yamashita A, Wear MA, Maeda Y, Cooper JA. Capping protein binding to actin in yeast: biochemical mechanism and physiological relevance. J Cell Biol. 2004;164:567–580. doi: 10.1083/jcb.200308061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu ReV Cell DeV Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 15.Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uruno T, Remmert K, Hammer JA., III CARMIL is a potent capping protein antagonist: identification of a conserved CARMIL domain that inhibits the activity of capping protein and uncaps capped actin filaments. J Biol Chem. 2006;281:10635–10650. doi: 10.1074/jbc.M513186200. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharya N, Ghosh S, Sept D, Cooper JA. Binding of myotrophin/V-1 to actin-capping protein: implications for how capping protein binds to the filament barbed end. J Biol Chem. 2006;281:31021–31030. doi: 10.1074/jbc.M606278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 19.Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Perry SV. Vertebrate tropomyosin: distribution, properties and function. J Muscle Res Cell Motil. 2001;22:5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- 21.Brown JH, Cohen C. Regulation of muscle contraction by tropomyosin and troponin: how structure illuminates function. Adv Protein Chem. 2005;71:121–159. doi: 10.1016/S0065-3233(04)71004-9. [DOI] [PubMed] [Google Scholar]
- 22.Ishiwata S, Fujime S. Effect of calcium ions on the flexibility of reconstituted thin filaments of muscle studied by quasielastic scattering of laser light. J Mol Biol. 1972;68:511–522. doi: 10.1016/0022-2836(72)90103-9. [DOI] [PubMed] [Google Scholar]
- 23.Hitchcock SE, Carisson L, Lindberg U. Depolymerization of F-actin by deoxyribonuclease I. Cell. 1976;7:531–542. doi: 10.1016/0092-8674(76)90203-8. [DOI] [PubMed] [Google Scholar]
- 24.Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi R, Nonomura Y, Okano A, Tashima Y. Purification and some properties of porcine kidney tropomyosin. J Biochem (Tokyo) 1983;94:171–179. doi: 10.1093/oxfordjournals.jbchem.a134327. [DOI] [PubMed] [Google Scholar]
- 26.Pruliere G, d’Albis A, der Terrossian E. Effect of tropomyosin on the interactions of actin with actin-binding proteins isolated from pig platelets. Eur J Biochem. 1986;159:535–547. doi: 10.1111/j.1432-1033.1986.tb09920.x. [DOI] [PubMed] [Google Scholar]
- 27.Ludescher RD, Liu Z. Characterization of skeletal muscle actin labeled with the triplet probe erythrosin-5-iodoacetamide. Photochem Photobiol. 1993;58:858–866. doi: 10.1111/j.1751-1097.1993.tb04984.x. [DOI] [PubMed] [Google Scholar]
- 28.DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE. Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J Cell Sci. 2002;115:4649–4660. doi: 10.1242/jcs.00147. [DOI] [PubMed] [Google Scholar]
- 29.Ono S, Ono K. Tropomyosin inhibits ADF/cofilindependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu R, Ono S. Dual roles of tropomyosin as an F-actin stabilizer and a regulator of muscle contraction in Caenorhabditis elegans body wall muscle. Cell Motil Cytoskeleton. 2006;63:659–672. doi: 10.1002/cm.20152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer RS, Fowler VM. Tropomodulins: life at the slow end. Trends Cell Biol. 2003;13:593–601. doi: 10.1016/j.tcb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Broschat KO, Weber A, Burgess DR. Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry. 1989;28:8501–8506. doi: 10.1021/bi00447a035. [DOI] [PubMed] [Google Scholar]
- 33.Broschat KO. Tropomyosin prevents depolymerization of actin filaments from the pointed end. J Biol Chem. 1990;265:21323–21329. [PubMed] [Google Scholar]
- 34.Kostyukova AS, Hitchcock-DeGregori SE. Effect of the structure of the N terminus of tropomyosin on tropomodulin function. J Biol Chem. 2004;279:5066–5071. doi: 10.1074/jbc.M311186200. [DOI] [PubMed] [Google Scholar]
- 35.Ishikawa R, Yamashiro S, Matsumura F. Annealing of gelsolin-severed actin fragments by tropomyosin in the presence of Ca2+. Potentiation of the annealing process by caldesmon. J Biol Chem. 1989;264:16764–16770. [PubMed] [Google Scholar]
- 36.Koepf EK, Burtnick LD. Interaction of plasma gelsolin with tropomyosin. FEBS Lett. 1992;309:56–58. doi: 10.1016/0014-5793(92)80738-3. [DOI] [PubMed] [Google Scholar]
- 37.Maciver SK, Ternent D, McLaughlin PJ. Domain 2 of gelsolin binds directly to tropomyosin. FEBS Lett. 2000;473:71–75. doi: 10.1016/s0014-5793(00)01507-6. [DOI] [PubMed] [Google Scholar]
- 38.Nyakern-Meazza M, Narayan K, Schutt CE, Lindberg U. Tropomyosin and gelsolin cooperate in controlling the microfilament system. J Biol Chem. 2002;277:28774–28779. doi: 10.1074/jbc.M203360200. [DOI] [PubMed] [Google Scholar]
- 39.Liu HP, Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57:233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- 40.Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 41.Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2002;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- 42.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 43.Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- 44.Moraczewska J, Nicholson-Flynn K, Hitchcock-DeGregori SE. The ends of tropomyosin are major determinants of actin affinity and myosin subfragment 1-induced binding to F-actin in the open state. Biochemistry. 1999;38:15885–15892. doi: 10.1021/bi991816j. [DOI] [PubMed] [Google Scholar]
- 45.Hitchcock-DeGregori SE, Lewis SF, Chou TM. Tropomyosin lysine reactivities and relationship to coiled-coil structure. Biochemistry. 1985;24:3305–3314. doi: 10.1021/bi00334a035. [DOI] [PubMed] [Google Scholar]
- 46.Harris ES, Li F, Higgs HN. The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem. 2004;279:20076–20087. doi: 10.1074/jbc.M312718200. [DOI] [PubMed] [Google Scholar]
- 47.Schafer DA, Korshunova YO, Schroer TA, Cooper JA. Differential localization and sequence analysis of capping protein beta-subunit isoforms of vertebrates. J Cell Biol. 1994;127:453–465. doi: 10.1083/jcb.127.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 49.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 50.Brahms S, Brahms J. Determination of protein secondary structure in solution by vacuum ultraviolet circular dichroism. J Mol Biol. 1980;138:149–178. doi: 10.1016/0022-2836(80)90282-x. [DOI] [PubMed] [Google Scholar]
- 51.Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 52.Ohtsuki I. Molecular arrangement of troponin-T in the thin filament. J Biochem (Tokyo) 1979;86:491–497. doi: 10.1093/oxfordjournals.jbchem.a132549. [DOI] [PubMed] [Google Scholar]
- 53.Teubner A, Wegner A. The rate of annealing of actin tropomyosin filaments depends strongly on the length of the filaments. Biochim Biophys Acta. 1996;1297:214–218. doi: 10.1016/s0167-4838(96)00111-2. [DOI] [PubMed] [Google Scholar]
- 54.Hitchcock-DeGregori SE, Sampath P, Pollard TD. Tropomyosin inhibits the rate of actin polymerization by stabilizing actin filaments. Biochemistry. 1988;27:9182–9185. doi: 10.1021/bi00426a016. [DOI] [PubMed] [Google Scholar]
- 55.Greenfield NJ. Analysis of circular dichroism data. Methods Enzymol. 2004;383:282–317. doi: 10.1016/S0076-6879(04)83012-X. [DOI] [PubMed] [Google Scholar]
- 56.Greenfield NJ, Kostyukova AS, Hitchcock-DeGregori SE. Structure and tropomyosin binding properties of the N-terminal capping domain of tropomodulin 1. Biophys J. 2005;88:372–383. doi: 10.1529/biophysj.104.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kostyukova AS, Rapp BA, Choy A, Greenfield NJ, Hitchcock-DeGregori SE. Structural requirements of tropomodulin for tropomyosin binding and actin filament capping. Biochemistry. 2005;44:4905–4910. doi: 10.1021/bi047468p. [DOI] [PubMed] [Google Scholar]
- 58.Hammell RL, Hitchcock-DeGregori SE. Mapping the functional domains within the carboxyl terminus of alpha-tropomyosin encoded by the alternatively spliced ninth exon. J Biol Chem. 1996;271:4236–4242. doi: 10.1074/jbc.271.8.4236. [DOI] [PubMed] [Google Scholar]
- 59.Harris ES, Higgs HN. Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 2006;406:190–214. doi: 10.1016/S0076-6879(06)06015-0. [DOI] [PubMed] [Google Scholar]
- 60.Bugyi B, Papp G, Hild G, Lorinczy D, Nevalainen EM, Lappalainen P, Somogyi B, Nyitrai M. Formins regulate actin filament flexibility through long range allosteric interactions. J Biol Chem. 2006;281:10727–10736. doi: 10.1074/jbc.M510252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heald RW, Hitchcock-DeGregori SE. The structure of the amino terminus of tropomyosin is critical for binding to actin in the absence and presence of troponin. J Biol Chem. 1988;263:5254–5259. [PubMed] [Google Scholar]
- 62.Urbancikova M, Hitchcock-DeGregori SE. Requirement of amino-terminal modification for striated muscle alpha-tropomyosin function. J Biol Chem. 1994;269:24310–24315. [PubMed] [Google Scholar]
- 63.Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- 64.Hart MC, Korshunova YO, Cooper JA. Vertebrates have conserved capping protein alpha isoforms with specific expression patterns. Cell Motil Cytoskeleton. 1997;38:120–132. doi: 10.1002/(SICI)1097-0169(1997)38:2<120::AID-CM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 65.Hart MC, Cooper JA. Vertebrate isoforms of actin capping protein beta have distinct functions in vivo. J Cell Biol. 1999;147:1287–1298. doi: 10.1083/jcb.147.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Papp G, Bugyi B, Ujfalusi Z, Barko S, Hild G, Somogyi B, Nyitrai M. Conformational changes in actin filaments induced by formin binding to the barbed end. Biophys J. 2006;91:2564–2572. doi: 10.1529/biophysj.106.087775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prochniewicz E, Zhang Q, Janmey PA, Thomas DD. Cooperativity in F-actin: binding of gelsolin at the barbed end affects structure and dynamics of the whole filament. J Mol Biol. 1996;260:756–766. doi: 10.1006/jmbi.1996.0435. [DOI] [PubMed] [Google Scholar]
- 68.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 69.Greenfield NJ, Palm T, Hitchcock-DeGregori SE. Structure and interactions of the carboxyl terminus of striated muscle alpha-tropomyosin: it is important to be flexible. Biophys J. 2002;83:2754–2766. doi: 10.1016/S0006-3495(02)75285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y, Mui S, Brown JH, Strand J, Reshetnikova L, Tobacman LS, Cohen C. The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site. Proc Natl Acad Sci U S A. 2002;99:7378–7383. doi: 10.1073/pnas.102179999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Greenfield NJ, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT, Hitchcock-Degregori SE. Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J Mol Biol. 2006;364:80–96. doi: 10.1016/j.jmb.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 72.Yamashita A, Maeda K, Maeda Y. Crystal structure of CapZ: structural basis for actin filament barbed end capping. EMBO J. 2003;22:1529–1538. doi: 10.1093/emboj/cdg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 74.Narita A, Takeda S, Yamashita A, Maeda Y. Structural basis of actin filament capping at the barbed-end: a cryo-electron microscopy study. EMBO J. 2006;25:5626–5633. doi: 10.1038/sj.emboj.7601395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu ReV Cell DeV Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 76.Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem. 2005;280:28653–286562. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scott JA, Shewan AM, den Elzen NR, Loureiro JJ, Gertler FB, Yap AS. Ena/VASP proteins can regulate distinct modes of actin organization at cadherin-adhesive contacts. Mol Biol Cell. 2006;17:1085–1095. doi: 10.1091/mbc.E05-07-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Casella JF, Maack DJ, Lin S. Purification and initial characterization of a protein from skeletal muscle that caps the barbed ends of actin filaments. J Biol Chem. 1986;261:10915–10921. [PubMed] [Google Scholar]


