Abstract
Processive motor proteins are stochastic steppers that perform actual mechanical steps for only a minor fraction of the time they are bound to the filament track. Motors usually work in teams and therefore the question arises whether the stochasticity of stepping can cause mutual interference when motors are mechanically coupled. We used biocompatible surfaces to immobilize processive kinesin-1 motors at controlled surface densities in a mechanically well-defined way. This helped us to study quantitatively how mechanical coupling between motors affects the efficiency of collective microtubule transport. We found that kinesin-1 constructs that lack most of the non-motor sequence slow each other down when collectively transporting a microtubule, depending on the number of interacting motors. This negative interference observed for a motor ensemble can be explained quantitatively by a mathematical model using the known physical properties of individual molecules of kinesin-1. The non-motor extension of kinesin-1 reduces this mutual interference, indicating that loose mechanical coupling between motors is required for efficient transport by ensembles of processive motors.
Keywords: molecular motor, conventional kinesin, gliding assay, microtubule transport, surface immobilization
Introduction
Motor proteins often work in ensembles, either in clusters bound to cargoes or as crosslinkers in highly interconnected filament networks. Single-molecule experiments have shown that motors are stochastic steppers (Svoboda et al, 1993; Mehta et al, 1999; Reck-Peterson et al, 2006). For example, a processive motor such as kinesin-1 (Lawrence et al, 2004) makes, on average, about 100 consecutive steps along a microtubule in the absence of load. The duration of the actual mechanical step is several orders of magnitude shorter than the dwell time between individual steps (Carter & Cross, 2005) during which the motor is firmly attached to the microtubule (Mori et al, 2007). If motors in an ensemble are mechanically coupled, one might expect that each stepping motor experiences a hindering load, because it has to work against its pausing team members during an actual step. It is known that individual kinesins slow down when experiencing a hindering load (Svoboda & Block, 1994; Carter & Cross, 2005). Therefore, the question arises whether mechanical coupling between motors can result in negative effects on motor stepping. The degree of such a negative interference should depend on the extent of mechanical coupling and on the number of mechanically coupled motors interacting with the same microtubule. Such an interference has, however, not yet been reported. By contrast, it has become an accepted view that processive motors immobilized, for example on a glass surface, collectively transport microtubules with a speed that is independent of the concentration of motors used for immobilization (Howard et al, 1989).
The lack of evidence for negative interference between mechanically coupled motors could be understood if the motors stepped synchronously. However, kinesins step stochastically also when collectively propelling a microtubule as a small team of motors (Leduc et al, 2007). Many kinesins have long, non-motor extensions and flexible parts between the segments of these extensions, providing some flexibility between an individual motor and its cargo (Hunt & Howard, 1993; Kerssemakers et al, 2006). This flexibility is thought to be important for the functionality of individual motors (Hunt & Howard, 1993). Whether these long and flexible extensions also provide loose coupling between processive motors of a team to prevent mutual interference remain to be explained.
To investigate this question, we tested whether dimeric kinesin-1 constructs that contain non-motor extensions of differing lengths and that behave identically at the single-molecule level show various microtubule transport behaviours when working as an ensemble in surface-gliding assays. To ensure motor functionality, even of very short motor constructs, and at the same time well-defined mechanical coupling, we used chemically functionalized glass surfaces for motor immobilization.
Results And Discussion
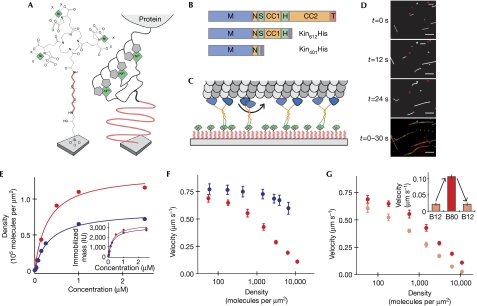
To explore the function of the first extended coiled coil and of the two adjacent possibly flexible regions of kinesin-1 (Fig 1A) for efficient microtubule transport by motor ensembles, we generated two truncated dimeric constructs. The first was a minimal dimeric construct with the amino-terminal 401 amino acids of kinesin (Kin401), containing the motor domain, the neck linker and a short coiled coil called the neck, which is necessary and sufficient for homodimerization (Berliner et al, 1995; supplementary information online). The second was a longer dimeric construct with the N-terminal 612 amino acids of kinesin (Kin612), which, in addition, contained the extended coiled coil 1 and two potentially flexible regions, called the swivel and the hinge (Grummt et al, 1998; Fig 1A). Finally, for control purposes, we also prepared a monomeric fragment containing the first 340 amino acids (Kin340), lacking both the neck region and the next coiled coil. The constructs contained either a carboxy-terminal monomeric green fluorescent protein (GFP) tag (Snapp et al, 2003; Zacharias et al, 2002) for single-molecule imaging (Fig 1A) or a C-terminal oligohistidine tag for surface immobilization in gliding assays (Fig 2B).
Figure 1.
Properties of single kinesins measured by single-molecule imaging. (A) Domain architecture of kinesin 1 constructs. (B) Scheme showing an immobilized microtubule on biotin-PEG functionalized glass, enabling single-molecule imaging by total internal reflection fluorescence microscopy. (C) Examples of space–time plots (kymographs) for each construct as indicated. Monomeric Kin340GFP is not processive. Scale bar, 2 μm (horizontal), 2 s (vertical). (D) Histogram of the mean velocity (left) and dwell time (right) with model fits for the long construct Kin612GFP in low ionic strength buffer (B12). (E) Motility data of Kin612GFP and Kin401GFP in low ionic strength buffer. Numbers in parentheses represent the 95% confidence interval. CC1, coiled coil 1; CC2, coiled coil 2; H, hinge; M, motor domain; N, neck; PEG, polyethylene glycol; S, swivel; T, tail.
Figure 2.
Mutual inhibition of collective microtubule transport by kinesin constructs. (A) Chemical structure of Ni-Tris-NTA-PEG coupled to glass (left) and schematic illustration of histidine (His)-tagged motors immobilized on a Ni-Tris-NTA-PEG surface (right). (B) Domain architecture of the two kinesin 1 constructs used. Symbols are as in Fig 1A. (C) Scheme of several immobilized kinesins transporting a microtubule. (D) Sequence of three filtered images showing the transport of fluorescently labelled microtubules by Kin612His immobilized on a Ni-Tris-NTA-PEG surface. The fourth image is a processed image, in which the microtubule at t=0 s is shown in white and the segments of alternating colour show the trajectories of microtubule movement, each segment representing the distance covered during 6 s. Scale bar, 10 μm. (E) Molecular surface densities of immobilized Kin612His (blue) and Kin401His (red) as calculated from the motor mass on the surface as determined by RIfS (inset), plotted as a function of the concentration used to incubate Ni-Tris-NTA-PEG surfaces for 10 min to allow functional immobilization. (F) Dependence of the average microtubule-gliding velocity v on the measured density of Kin612His (blue) and Kin401His (red) in medium ionic strength buffer (B80). (G) Dependence of the average microtubule-gliding velocity v on the measured density of Kin401His in medium (B80, dark red) and low (B12, light red) ionic strength buffer. Inset: variation of the microtubule transport velocity at a density of 11,000 Kin401His molecules per square micrometre in response to a sequential buffer change, as indicated, in the same experiment. Error bars of measured velocities are standard deviations. Ni-Tris-NTA-PEG, nickel-Tris-nitrilotriacetic acid polyethylene glycol; RIfS, reflectometric interference spectroscopy.
Using single-molecule imaging (Fig 1B), we first established that the dimeric constructs behaved indistinguishably with respect to their motile properties at the individual motor level. Kin612GFP and Kin401GFP showed processive motility (Fig 1C; supplementary Movie 1 online) with an identical average velocity of 0.67 μm s−1 and an average dwell time of 1.8 s in low ionic strength buffer (Methods; Fig 1D,E). By contrast, monomeric kinesin Kin340GFP did not show processive runs (supplementary Movie 1 online; Fig 1C), as expected (Vale et al, 1996).
We then asked whether the two dimeric constructs that behave identically as single molecules show differences when working as an ensemble. We immobilized oligohistidine-tagged kinesins on nickel-Tris-nitrilotriacetic acid that was covalently linked to a glass surface through a high-density polyethylene glycol (PEG) layer (Lata & Piehler, 2005; Fig 2A–C). Owing to multivalent interaction with the oligohistidine tag, these multivalent chelator heads ensure stable (supplementary Fig 1A online) and specific (supplementary Fig 1B online) molecular complex formation (Lata et al, 2005). Compared with other methods of specific motor protein immobilization such as the attachment of biotinylated motors to physisorbed streptavidin (Berliner et al, 1994) or of otherwise tagged motors to physisorbed antibodies (Case et al, 1997), our new method for motor immobilization has several advantages. The PEG polymer brush prevents motor denaturation, even of short kinesin constructs (supplementary Fig 2A–C online), and provides mechanically well-defined (Oesterhelt et al, 1999) surface attachment. Furthermore, the absence of adaptor or blocking proteins facilitates the precise determination of the amount of immobilized motor protein by solid-phase detection methods.
We then performed microtubule-gliding experiments (Fig 2C,D) over a broad range of motor densities that spanned several orders of magnitude. In contrast to previous studies in which the density of surface-adsorbed motors was estimated, here we determined the density of immobilized motors experimentally by reflectometric interference spectroscopy (Hanel & Gauglitz, 2002; Fig 2E). Using the long construct Kin612His in medium ionic strength buffer, we confirmed previous observations by showing that the velocity of collective microtubule transport did not depend on the density of full-length kinesin-1 within the range of previously reported densities (up to around 1,000 motors μm−2; Fig 2F, data shown in blue; Howard et al, 1989; Hancock & Howard, 1998). We noted, however, a small decrease in transport velocity at very high densities. Interestingly, the minimal dimeric construct Kin401His showed a different behaviour. The velocity of gliding decreased with increasing motor densities (Fig 2F, data shown in red; supplementary Movie 2 online), although the microtubule transport velocities of both constructs were similar at low motor densities and corresponded to the velocity of individual motors in single-molecule experiments (Fig 1E). Therefore, the two kinesin constructs of differing lengths that behave identically as single molecules show marked differences in their transport efficiency when working as a mechanically coupled ensemble.
It is unlikely that the marked slowdown of microtubule transport by Kin401His at high motor densities is caused by steric interference, because we observed such a slowdown to a much lesser extent with the longer construct that was studied over a similar range of densities. We also did not see an indication for motor inactivation at high densities. Instead, all microtubules moved smoothly. Furthermore, very long microtubules (>25 μm) moved more slowly than very short microtubules (∼1 μm) at intermediate densities of Kin401His (data not shown).
These results indicate that the total number of motors interacting with a microtubule is responsible for the observed effect on the velocity of gliding. As it is known that the affinity of kinesin-1 for microtubules increases with decreasing ionic strength (Thorn et al, 2000; supplementary Table 1 online), we tested this prediction by performing gliding assays in low salt buffer over the same range of fixed motor densities. Interestingly, the decrease in ionic strength and thus the increase in motor affinity led to a more marked density-dependent decrease in the microtubule transport velocity for both constructs used (Figs 2G, 3C,D). These changes in transport velocity depending on ionic strength were reversible (Fig 2G, inset), arguing against an irreversible inactivation of motors at high surface densities. Taken together, these results support the idea that it is the average number of motors bound to a microtubule that determines its transport velocity. In conclusion, our results indicate that tight mechanical coupling between very short kinesin constructs causes mutual interference between stepping motors at high surface densities, probably as a consequence of uncoordinated stepping (Leduc et al, 2007).
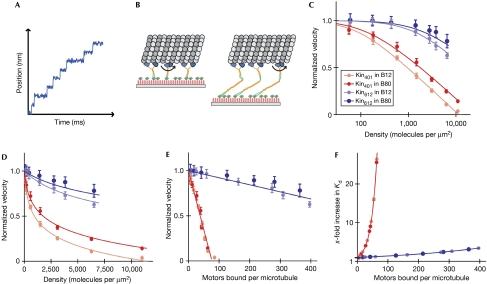
Figure 3.
Global fit of the kinetic model for mutual interference of mechanically coupled motors to the experimental data. (A) Schematic illustration of the stepping behaviour of an individual motor. (B) Schematic illustration of the spatial dimensions of immobilized Kin401His (left) and Kin612His (right) transporting a microtubule. Polyethylene glycol (PEG; red), coiled coil segments of kinesin (yellow), motor domains (blue) and the microtubule (grey) are drawn roughly to scale. (C) Global least-squares fit of the kinetic model of mutual interference (lines) to four sets of experimental data (circles) as indicated. The normalized microtubule transport velocity is plotted as a function of the motor density shown on a logarithmic scale. Error bars of the velocities represent standard deviations. (D) Global fit and data as in (C), shown on a linear density scale. (E) Normalized microtubule transport velocities and global fit (same data as in (C,D)) replotted as a function of the number of motors bound per microtubule as predicted by the kinetic model. (F) Increase in the dissociation constant as predicted by the model as a function of the number of motors bound per microtubule. Colour code in (C–F) as in Fig 2F,G.
The observed collective effect of negative interference can be described quantitatively by a kinetic model based on the known biophysical properties of kinesin-1 at the individual motor level. The model is based on the observation that the duration of each individual step of a processive kinesin is about several tens of microseconds (Carter & Cross, 2005), which is three orders of magnitude shorter than the average waiting time of tens of milliseconds between individual steps (Fig 3A). This separation of timescales leads to the expectation that in the absence of coordination (Leduc et al, 2007), each motor, while making a step, works against the other motors that are at that moment statically bound to the microtubule. The central assumption of the model is therefore a resulting counterforce that is proportional to the number of attached motors and that can vary between the two kinesin constructs used (Fig 3B). An additional essential feature of the model is that the probability of stepping decreases with increasing load (Svoboda & Block, 1994; Coppin et al, 1997; Carter & Cross, 2005). For simplicity, we assume a linear force–velocity relationship. Furthermore, we assume that the rate of dissociation of kinesin from the microtubule increases exponentially with applied load (Coppin et al, 1997; for details, see Methods and the supplementary information online).
This model quantitatively describes the phenomenon of negative interference of mechanically coupled motors over a broad range of experimental conditions, as shown by a global fit to our experimental velocity–density curves for both kinesin constructs in either low or medium ionic strength buffer (Fig 3C (logarithmic scale), 3D (linear scale)). The principal result of this analysis is that the minimal dimeric kinesin construct lacking the first extended coiled coil produces a counterforce of about 0.09 pN per microtubule-bound motor, whereas the longer construct produces a counterforce about 20 times smaller. Furthermore, we obtain a detachment force of 1.8 pN from the fit, which is assumed to be identical here for all constructs and buffer conditions. This value is in the same range as that found in single-molecule optical trapping experiments (Coppin et al, 1997; Schnitzer et al, 2000). The model can be used to predict the number of motors bound to the microtubule at various surface densities (supplementary information online). A plot of the experimentally determined transport velocities compared with the predicted number of attached motors shows a linear relationship (Fig 3E). This is a consequence of the proportionality between the total counterforce and the number of attached motors, and of the assumed linear force–velocity relationship. This analysis shows that the two kinesin constructs of differing lengths have completely different sensitivities to the number of microtubule-attached motors. In the case of the minimal dimeric kinesin, as few as 10 attached motors leads to a significant slowdown of microtubule transport, whereas more than 100 longer kinesins can attach to the same microtubule without strong mutual interference. Furthermore, one can calculate the increase in the dissociation constant as a function of the number of attached motors (supplementary information online), again showing characteristic differences between the two constructs (Fig 3F).
These results indicate that the two different dimeric constructs show a distinct coupling stiffness. Such a construct-specific coupling stiffness can be detected in our experimental setup, because the nitrilotriacetic acid polyethylene glycol (NTA-PEG) linkers provide only marginal flexibility. Theoretical models as well as experimental data (Oesterhelt et al, 1999) predict that the PEG chains used here can be extended a maximum of 2 nm by the highest forces generated by the motors in our experiment (supplementary information online). This extension is considerably smaller than the 8 nm step size of kinesin. Therefore, the motor domains of the minimal dimeric kinesin can be considered as tightly coupled to the surface. Large counterforces at high surface densities lead to a marked slowdown of the short construct. After extended time periods, these forces even resulted in the disruption of the microtubules (supplementary Fig 3, Movie 3 and information online). By contrast, the longer dimeric construct, with its additional coiled coil of about 20 nm that is probably linked flexibly to the neck, provides an additional ‘passive lever arm' accommodating movements of the kinesin motor domains that are considerably larger than the step size (Fig 3B). Therefore, the hindering load exerted by longer kinesins is smaller than that exerted by shorter kinesins. Consequently, only at very low ionic strength, which further increases the number of bound motors due to an increase in affinity, is a slowdown of microtubule transport observed for the longer kinesin construct at very high motor densities.
In the past, the gliding velocity of a processive motor was considered to be independent of the density of motors (Howard, 1997). Our experiments show that this behaviour is restricted to the case of loose mechanical coupling between the immobilized motors. Apparently, loose mechanical coupling was ensured when experiments were previously performed either with full-length kinesin or with kinesin fragments that were surface-immobilized through adaptor proteins that probably provided additional flexibility and/or leverage.
In conclusion, we observed that minimal dimeric motor constructs inhibit each other in a number-dependent manner when collectively transporting a microtubule. Our results indicate that loose mechanical coupling resulting from elongated and/or potentially unstructured regions ensures efficient cargo transport by teams of motors. This might be one reason for the existence of the commonly found long stretches of coiled coil interrupted by short potentially unstructured stretches in kinesin motor proteins. In cells, motors are often highly concentrated locally, especially in microtubule bundles, such as in antiparallel microtubule overlaps in the mid-zone of late mitotic spindles or at the kinetochore–microtubule bundle interface. Loose mechanical coupling might, therefore, be necessary to help in reducing the problems caused by the stochastic stepping mechanism of processive motors when working in large interconnected ensembles in the meshwork of the microtubule cytoskeleton.
Methods
Protein biochemistry. Proteins were purified as described in the supplementary information online.
Buffers. Medium ionic strength buffer (B80) was 80 mM K-PIPES, 2 mM MgCl2, 1 mM EGTA, 2 mM MgATP, 2 mM mercaptoethanol, pH 6.8. Low ionic strength buffer (B12) contained 12 mM instead of 80 mM K-PIPES.
Preparation of multivalent NTA-PEG functionalized surfaces. Tris-NTA-PEG functionalized glass coverslips or reflectometric interference spectroscopy transducer slides were prepared as described earlier (Lata & Piehler, 2005), dried and stored at −20°C. On the day of the experiment, coverslips for microscopy were rewetted and loaded with Ni2+ ions by washing with the following series of solutions: (i) 100 mM HCl incubated for 2.5 min, (ii) wash buffer (20 mM HEPES, pH 7.5, 150 mM NaCl) incubated for 15 min, (iii) 20 mM NiCl2 in wash buffer incubated for 5 min and (iv) 200 mM imidazole in wash buffer incubated for 5 min. The Ni-loaded slides were equilibrated again with wash buffer and kept in a humid container.
Motor immobilization and time-lapse fluorescence microscopy. A flow chamber was built from a multivalent NTA-PEG functionalized coverslip and a poly-L-lysine (PLL)-PEG passivated counter glass (Bieling et al, 2007), separated by two strips of double sticky tape (Tesa; Hamburg, Germany). The flow chamber was equilibrated with 50 μl B80 while positioned on an ice-cold metal block. Then 20 μl of B80 containing 1 nM–2.5 μM histidine-tagged kinesins was flowed into the chamber. Unbound motors were washed out with 30 μl B80 after 10 min. For motor concentrations lower than 200 nM, residual Ni-NTA-binding sites were saturated by incubating the flow chamber after motor immobilization with 10 μM enhanced GFP-His10 in B80 for 5 min to prevent electrostatic repulsion of microtubules from the otherwise charged surface. The chamber was allowed to warm up to room temperature and five chamber volumes of microtubule mix (paclitaxel-stabilized microtubules either in B80 or B12) containing 50–200 nM polymerized tubulin, an oxygen scavenger system (20 mM glucose, 20 μg ml−1 glucose oxidase and 10 μg ml−1 catalase) and 10 μM paclitaxel (Sigma-Aldrich; Taufkirchen, Germany) were flowed through the chamber. Microtubules were visualized by shuttered digital time-lapse fluorescence microscopy with frame rates of 0.33 or 2 s−1 (Axiovert 200 microscope (Zeiss; Jehn, Germany) with a × 63/1.4 Plan Apochromat objective (Zeiss), CoolSnap HQ camera (Photometrix; Tucson, AZ, USA) or a IX-71 total internal reflection fluorescence (TIRF) microscope (Olympus) with a × 60/1.45 TIRFM objective (Olympus; Hamburg, Germany) and a Hamamatsu C8484 camera). The temperature was maintained at 25±1°C.
Single-molecule imaging. In flow chambers with biotin-PEG instead of Tris-NTA-PEG functionalized glass (Bieling et al, 2007), biotinylated, Alexa568-labelled and pactitaxel-stabilized microtubules were allowed to adhere to the surface through NeutrAvidin (Invitrogen; Karlsruhe, Germany) for 5 min and the remaining free microtubules were washed out. Kinesin constructs were diluted to a final concentration of 10–20 pM in either B12 or B80 containing the oxygen scavenger system and added to the chamber. Imaging was performed on a custom TIRF microscope (Bieling et al, 2007) at a frame rate of 10 s−1.
Data analysis. Microtubule-gliding velocities were determined as described in the supplementary information online. A model for collective microtubule transport (see below) was fitted to the plots of the measured velocities as a function of the motor density using Matlab. Single-molecule movements were quantified by automated tracking analysis implemented in a commercial software package (Kalaimoscope; TransInsight, Dresden, Germany). Histograms were generated by pooling around 1,000 tracks, and were model-fitted using the curve fitting tool in Matlab (MathWorks; Natick, MA, USA).
Kinetic model for collective transport by mechanically coupled motors. The model is based on three main assumptions: (i) a linear counterforce–velocity relationship for stepping motors; (ii) proportionality between the number of microtubule-attached motors and the counterforce, and (iii) an exponential increase in the dissociation rate with force. The combination of these assumptions results in the following expression that was used for fitting the model to the measured relationship between the motor density D and the (loaded) velocity v (see the supplementary information online):
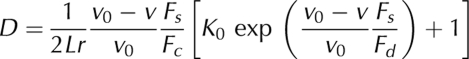 |
with the unloaded velocity v0, the stall force Fs, the counterforce Fc, the detachment force Fd, the unloaded dissociation constant K0, the microtubule length L and the motor reach r. Fs, Fd, L and r are independent of the length of the kinesin construct and of the buffer used. Fc is assumed to vary with the length of the kinesin construct, whereas K0 and v0 vary with ionic strength. The values of Fc, Fd and K0 result from the global fit to the four velocity–density relationships in Fig 3. The values of v0, Fs, L and r are fixed (see the supplementary information online).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information 1
Supplementary Information 2
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Acknowledgments
We thank Gerhardt Spatz-Kümbel for the synthesis of multivalent chelator heads, Mathias Utz for technical assistance, Kota Miura, Francois Pouthas and Marianne Uteng for PLL-PEG synthesis, Jan Vos for helping to initiate this project, Uros Krzic for help with data analysis, Jennifer Lippincott-Schwartz for the monomeric GFP clone, Francesco Pampaloni for discussions, Analytik Jena for the BIAffinity system and for technical support, the Deutsche Forschungsgemeinschaft and the German Federal Ministry of Education and Research (BMBF) for financial support. I.A.T. was supported by the European Commission (STREP ‘Active Biomics') and the Swiss National Science Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Berliner E, Mahtani HK, Karki S, Chu LF, Cronan JE Jr, Gelles J (1994) Microtubule movement by a biotinylated kinesin bound to streptavidin-coated surface. J Biol Chem 269: 8610–8615 [PubMed] [Google Scholar]
- Berliner E, Young EC, Anderson K, Mahtani HK, Gelles J (1995) Failure of a single-headed kinesin to track parallel to microtubule protofilaments. Nature 373: 718–721 [DOI] [PubMed] [Google Scholar]
- Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, Surrey T (2007) Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450: 1100–1105 [DOI] [PubMed] [Google Scholar]
- Carter NJ, Cross RA (2005) Mechanics of the kinesin step. Nature 435: 308–312 [DOI] [PubMed] [Google Scholar]
- Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD (1997) The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell 90: 959–966 [DOI] [PubMed] [Google Scholar]
- Coppin CM, Pierce DW, Hsu L, Vale RD (1997) The load dependence of kinesin's mechanical cycle. Proc Natl Acad Sci USA 94: 8539–8544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt M, Woehlke G, Henningsen U, Fuchs S, Schleicher M, Schliwa M (1998) Importance of a flexible hinge near the motor domain in kinesin-driven motility. EMBO J 17: 5536–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock WO, Howard J (1998) Processivity of the motor protein kinesin requires two heads. J Cell Biol 140: 1395–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanel C, Gauglitz G (2002) Comparison of reflectometric interference spectroscopy with other instruments for label-free optical detection. Anal Bioanal Chem 372: 91–100 [DOI] [PubMed] [Google Scholar]
- Howard J (1997) Molecular motors: structural adaptations to cellular functions. Nature 389: 561–567 [DOI] [PubMed] [Google Scholar]
- Howard J, Hudspeth AJ, Vale RD (1989) Movement of microtubules by single kinesin molecules. Nature 342: 154–158 [DOI] [PubMed] [Google Scholar]
- Hunt AJ, Howard J (1993) Kinesin swivels to permit microtubule movement in any direction. Proc Natl Acad Sci USA 90: 11653–11657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerssemakers J, Howard J, Hess H, Diez S (2006) The distance that kinesin-1 holds its cargo from the microtubule surface measured by fluorescence interference contrast microscopy. Proc Natl Acad Sci USA 103: 15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Piehler J (2005) Stable and functional immobilization of histidine-tagged proteins via multivalent chelator headgroups on a molecular poly(ethylene glycol) brush. Anal Chem 77: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Lata S, Reichel A, Brock R, Tampe R, Piehler J (2005) High-affinity adaptors for switchable recognition of histidine-tagged proteins. J Am Chem Soc 127: 10205–10215 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ et al. (2004) A standardized kinesin nomenclature. J Cell Biol 167: 19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc C, Ruhnow F, Howard J, Diez S (2007) From the cover: detection of fractional steps in cargo movement by the collective operation of kinesin-1 motors. Proc Natl Acad Sci USA 104: 10847–10852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE (1999) Myosin-V is a processive actin-based motor. Nature 400: 590–593 [DOI] [PubMed] [Google Scholar]
- Mori T, Vale RD, Tomishige M (2007) How kinesin waits between steps. Nature 450: 750–754 [DOI] [PubMed] [Google Scholar]
- Oesterhelt F, Rief M, Gaub HE (1999) Single molecule force spectroscopy by AFM indicates helical structure of poly(ethylene-glycol) in water. New J Phys 1: 6.1–6.11 [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD (2006) Single-molecule analysis of dynein processivity and stepping behavior. Cell 126: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer MJ, Visscher K, Block SM (2000) Force production by single kinesin motors. Nat Cell Biol 2: 718–723 [DOI] [PubMed] [Google Scholar]
- Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, Lippincott-Schwartz J (2003) Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol 163: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Block SM (1994) Force and velocity measured for single kinesin molecules. Cell 77: 773–784 [DOI] [PubMed] [Google Scholar]
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM (1993) Direct observation of kinesin stepping by optical trapping interferometry. Nature 365: 721–727 [DOI] [PubMed] [Google Scholar]
- Thorn KS, Ubersax JA, Vale RD (2000) Engineering the processive run length of the kinesin motor. J Cell Biol 151: 1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T (1996) Direct observation of single kinesin molecules moving along microtubules. Nature 380: 451–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296: 913–916 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information 1
Supplementary Information 2
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3