Abstract
The tubulin vinca domain is the target of widely different microtubule inhibitors that interfere with the binding of vinblastine. Although all these ligands inhibit the hydrolysis of GTP, they affect nucleotide exchange to variable extents. The structures of two vinca domain antimitotic peptides—phomopsin A and soblidotin (a dolastatin 10 analogue)—bound to tubulin in a complex with a stathmin-like domain show that their sites partly overlap with that of vinblastine and extend the definition of the vinca domain. The structural data, together with the biochemical results from the ligands we studied, highlight two main contributors in nucleotide exchange: the flexibility of the tubulin subunits' arrangement at their interfaces and the residues in the carboxy-terminal part of the β-tubulin H6–H7 loop. The structures also highlight common features of the mechanisms by which vinca domain ligands favour curved tubulin assemblies and destabilize microtubules.
Keywords: antimitotics, GTPase, microtubules, nucleotide exchange, structure
Introduction
Microtubules are cytoskeletal polymers of tubulin αβ heterodimers that are essential in several cellular functions such as cell division and morphogenesis. The dynamics of microtubules is described in terms of dynamic instability (Mitchison & Kirschner, 1984). This behaviour is a consequence of GTP hydrolysis in the β-subunit, which accompanies tubulin polymerization and generates nonlinearity in the assembly kinetics (Carlier et al, 1984), the α-tubulin nucleotide being non-hydrolysable and non-exchangeable. On microtubule disassembly, GDP tubulin is released. Exchange of GDP for GTP regenerates polymerizable GTP tubulin. In solution, dynamic instability leads to oscillatory polymerization kinetics (Carlier et al, 1987; Mandelkow et al, 1988), with a period that depends on the rate of GDP dissociation from tubulin. The rate of nucleotide exchange is therefore an important parameter in the regulation of microtubule dynamics.
Tubulin is the target of antimitotic small molecules. Among them, a group of compounds inhibits tubulin binding of the anticancer drug vinblastine; their sites define what has been termed the vinca domain (Hamel, 1992). These compounds have widely different chemical formulas and different effects on tubulin; in particular, although nucleotide exchange is partly inhibited by vinblastine, it is much more extensively prevented by other vinca domain ligands (Bai et al, 1990a). These observations have raised the possibility that vinca domain ligands bind to distinct locations (Hamel, 1992) and challenged their possible mechanism of action. To address these questions, we have used the only soluble αβ-tubulin complex shown so far to yield crystals that diffract to a sufficient resolution to identify bound ligands, namely a complex with the RB3 stathmin-like domain (RB3-SLD). We report the 4.1Å and 3.8 Å resolution X-ray structures of tubulin–colchicine-RB3-SLD [(Tc)2R] (Ravelli et al, 2004) in complex with two vinca domain antimitotic peptides, phomopsin A (Tonsing et al, 1984) and soblidotin (Natsume et al, 2007; a dolastatin 10 analogue also named TZT-1027 or auristatin PE; Fig 1A; supplementary Table 1 online). In addition to clarifying the relationship of the vinca domain ligand-binding sites and their mechanisms of microtubule inhibition, the structures elucidate the structural mechanism for GTP exchange on tubulin.
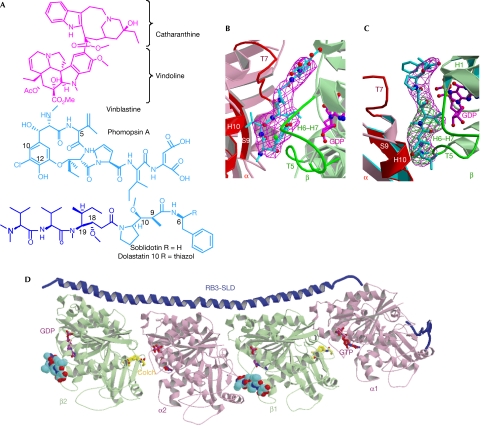
Figure 1.
The tubulin vinca domain. (A) Chemical formulas of vinblastine, phomopsin A and soblidotin. The amino-terminal tripeptide of soblidotin is highlighted in darker blue. (B) View of the vinca domain in (Tc)2R, with bound phomopsin A and its σa-weighted Fobs−Fcalc omit map; the map is contoured at 3.5σ and overlapped with phomopsin A. (C) View of the vinca domain in (Tc)2R, with bound soblidotin and its σa-weighted Fobs−Fcalc omit map contoured at 4σ (magenta). The σa-weighted Fobs−Fcalc omit map of crystals soaked with the soblidotin N-terminal peptide, contoured at 3.5σ (green), is overlapped with it. A stereoscopic version of this panel is provided as Supplementary Fig 6 online. (D) The structure of (Tc)2R with bound phomopsin A (cyan), represented as a space-filling model. Two sites are identified, one at the β1–α2 interface and the other one at the β2 end of the complex. The RB3-SLD (dark blue) and colchicine (yellow) domains are also represented. Colch, colchicine; RB3-SLD, RB3 stathmin-like domain.
Results And Discussion
The vinca domain and its ligand-binding modes
The ligands were identified by inspection of difference electron-density maps calculated using diffraction data from soaked (Tc)2R crystals (phomopsin A: the highest peak was 8.5σ, the first peak away from phomopsin A being 5.9σ; soblidotin: the highest peak was 8.6σ, the first peak at a different location being 5.1σ). The orientation of phomopsin A was unambiguous in its asymmetric electron density (Fig 1B), as opposed to that of soblidotin, which is a more symmetrically shaped molecule. Therefore, we independently determined the structure of the amino-terminal tripeptide of soblidotin bound to (Tc)2R. The localization of the amino acids in common between the parent molecule and its shorter part unambiguously defines the orientation of soblidotin and shows that its four N-terminal amino acids are ordered in the complex (Fig 1C). Although the details of their interactions differ, as their chemical structures are different, phomopsin A and soblidotin share a binding site at the interface of the two tubulins in (Tc)2R (Fig 1). They interact with residues in a common core of secondary structure elements: helix H10, strand S9 and loop T7 in the α2-subunit and loops T5 and H6–H7 in the β1-subunit (for the nomenclature of tubulin secondary structure elements, see Nogales et al, 1998, and for the labelling of tubulin subunits, see Fig 1D). In addition, soblidotin contacts helix H1 in the β1-subunit.
Several lines of evidence detailed below suggest that the binding of the ligands to tubulin is similar to their binding to (Tc)2R. In the case of phomopsin A, some evidence comes from structural data. Indeed, in addition to the site at the inter-dimer β1α2 interface, another phomopsin A molecule is bound to the β-subunit of the second heterodimer (Fig 1D; supplementary Fig 1 online). We find that the positions in the tubulin sequence of the residues contacted by the ligand at both sites are identical, whereas the site at the β2 end of (Tc)2R lacks an α-interface or any other extensive interface that might have resulted from crystal packing. The structure is also in good agreement with activity data on ustiloxins, which are phomopsin A analogues that have closely related 13-member rings (Li et al, 2008). For example, the substitution in ustiloxins of the hydroxyl at C12 of phomopsin A (Fig 1A; supplementary Fig 2 online) by a bulkier methoxyl group leads to markedly reduced activity. In the structure, this hydroxyl interacts tightly with loops T5 and H6–H7 of the β-subunit, potentially establishing an H bond with Pro β222 carbonyl. By contrast, substitution of the C5 isopropenyl substituent by bulkier or shorter hydrophobic groups is accompanied by a gradual decrease in activity. The interaction of this substituent with tubulin, in a previously identified hydrophobic pocket contributed by the α-tubulin S9 β-strand and H10 α-helix (Gigant et al, 2005), seems looser than that of the hydroxyl at C12. Interestingly, the C10 appendage of ustiloxins A and B (phomopsin A numbering, see Fig 1A), as compared with ustiloxins D and F (and with phomopsin A), leads to more active compounds (Koiso et al, 1998); this appendage might be accommodated in the vindoline subsite, which abuts on the phomopsin A macrocycle (see below).
In the case of soblidotin, the structures correlate well with abundant biochemical data that have been collated to establish the tubulin-binding mode of dolastatin 10, which differs from soblidotin only by an additional thiazole ring substituent at position 6 (Fig 1A). Several segments of dolastatin 10 were prepared, the shortest to inhibit microtubule assembly being a tripeptide (Bai et al, 1990b), which we identified in soaked (Tc)2R crystals. Eighteen isomers of dolastatin 10 at five out of its nine chiral centres were also synthesized (Bai et al, 1990b); among them, reversal of configuration at positions 18 and 19 affects binding to tubulin. Both the methoxyl and s-butyl substituents at these positions in the well-ordered part of soblidotin in (Tc)2 R are in contact with tubulin, at residue Tyrβ224 (Fig 2A). By contrast, reversal of configuration at positions 6, 9 and 10 leads to derivatives that essentially behave as dolastatin 10 in the inhibition of microtubule assembly (Bai et al, 1990b); these three positions are adjacent to, or in the part of, the molecule that is not visible in the soblidotin electron density and are hence disordered. It is nevertheless possible that modifications in the disordered part of a molecule alter its affinity for a binding site; indeed, reversal of configuration at positions 9 and 10 of dolastatin 10 has been reported to alter its properties other than inhibition of microtubule assembly (Bai et al, 1993). Finally, dolastatin 10 photo labels the N-terminal part of the β-subunit, most likely by establishing a disulphide bond with residue Cys β12 through its thiazole ring (Bai et al, 2004). In the structure of its complex with (Tc)2R, the corresponding part of soblidotin is in contact with residue Gln β15 (distance: 3 Å) and reaches close to Cys β12 (Fig 2A).
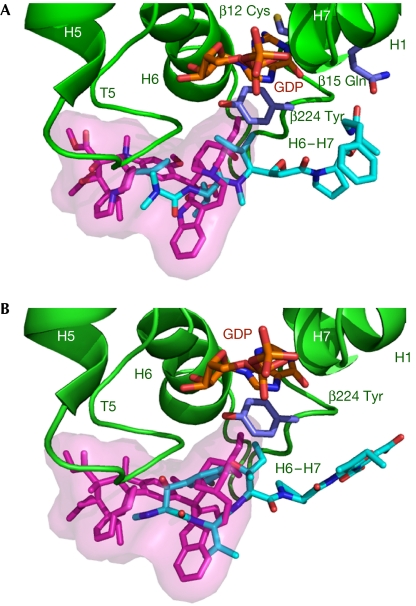
Figure 2.
The overlap of tubulin-bound soblidotin (A, cyan) and phomopsin A (B, cyan) with vinblastine (magenta); all are shown as sticks together with the β-tubulin structural elements with which they are in contact. A semitransparent vinblastine surface is drawn to aid visualization of the overlap.
Phomopsin A, as the soblidotin close analogue dolastatin 10, induces the formation of tubulin assemblies, rings and spirals. The diameter of these assemblies, around 40 nm (Tonsing et al, 1984; Bai et al, 1999), is similar to the 44 nm diameter of the projection of a helix built from the repetition of tubulin–RB3-SLD (T2R) complexes (Gigant et al, 2000) or to the 46±6 nm diameter of the vinblastine-induced spirals of tubulin in complex with Rhel; Rhel is an RB3-SLD construct lacking its N-terminal part and therefore not preventing its tubulin complexes from associating into curved protofilaments (Gigant et al, 2005). We also checked that the vinca domain ligands studied here induce similar curved assemblies with tubulin when in complex with Rhel (supplementary Fig 3 online). Therefore, taken together, the structural and biochemical data suggest that soblidotin and phomopsin A bind to tubulin in solution in a manner similar to that observed in (Tc)2R, and the structures we determined allow us to rationalize the interference of these drugs with tubulin properties.
The compounds we studied inhibit the binding of vinblastine to tubulin (Bai et al, 1990a; Natsume et al, 2000). Indeed, when the structures of the complexes are superimposed on that of (Tc)2R-vinblastine (Gigant et al, 2005), the ligands overlap significantly: the 13-member ring of phomopsin A overlaps with the catharanthine moiety of vinblastine, as do the first two amino acids of soblidotin (Fig 2). This defines the core of the vinca domain. The binding sites of the three ligands also differ in two respects. First, although vinblastine contacts α- and β-subunits to a similar extent, burying around 260 Å2 of each surface, the contact of phomopsin A and soblidotin with the β1-subunit is more extensive than that with α2 (350 Å2 compared with 200 Å2). Second, the non-overlapping parts of these ligands—vinblastine on the one hand, and soblidotin and phomopsin A on the other—extend in opposite directions (Fig 2). The vindoline part of vinblastine reaches towards the N-terminal end of the β-subunit T5 loop, whereas phomopsin A and soblidotin make extensive contacts with the H6–H7 loop (residues 216–224), mostly with its C-terminal end.
Inhibition of nucleotide exchange
Many vinca domain ligands have been shown to inhibit tubulin nucleotide exchange (Hamel, 1992). Remarkably, vinblastine interferes with nucleotide exchange to a much lesser extent than do phomopsin A or soblidotin (Bai et al, 1990a; Natsume et al, 2000; data not shown). Vinca domain ligands do not contact the β-tubulin nucleotide. In the exchangeable nucleotide site, Tyr β224, the C-terminal residue of the H6–H7 loop, is one of the residues sandwiching the guanine base (Fig 2). Vinblastine makes little contact with Tyr β224, as only one of its atoms is distant from it by less than 4 Å, whereas soblidotin and phomopsin A interact extensively with this residue—for example, eight atoms of soblidotin fulfil the above distance criterion. The interaction of soblidotin or phomopsin A with the conserved Tyr β224 buttresses it against the nucleotide (Fig 2), and this might be sufficient to account for inhibition by these compounds of nucleotide exchange in tubulin.
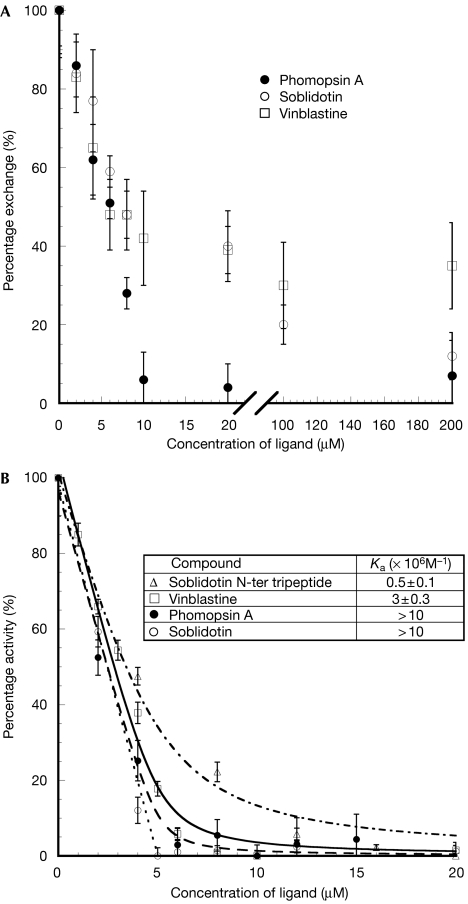
Nucleotide exchange is slowed down at least 10-fold in the (Tc)2R-like stathmin–tubulin complex (Amayed et al, 2000). The three vinca domain ligands we have studied further inhibit nucleotide exchange in (Tc)2R (Fig 3A). On account of the high affinity of these compounds for their site at the interface between the two tubulin molecules (see below, inhibition of GTPase), saturation of this site occurs when a stoichiometric amount of ligand is added to the complex at the 5 μM concentration of (Tc)2R used in these experiments. Inhibition of half of the exchange that is reached for a nearly stoichiometric amount of added ligand reflects this saturation. The simplest interpretation, which involves only local effects of the vinca ligands, is that at stoichiometry all three vinca domain ligands completely inhibit exchange of the β1 nucleotide in (Tc)2R. This contrasts with the levelling off at 25% of the inhibition of tubulin nucleotide exchange at a high concentration of vinblastine (Bai et al, 1990a). In the restricted flexibility environment of the vinca domain in (Tc)2R, where the two tubulin molecules are held together by the RB3 long C-terminal helix, the additional tubulin–tubulin interactions mediated by vinblastine together with the limited interference of this drug with the C-terminal part of the H6–H7 loop prevent nucleotide exchange. No further inhibition of nucleotide exchange is observed when more vinblastine is added, which might reflect its low affinity for the β2 moiety of the vinca domain or its lower propensity to inhibit exchange when bound to the tubulin β-subunit. By contrast, complete inhibition of (Tc)2R nucleotide exchange by phomopsin A is observed. Soblidotin is much less efficient, as inhibition by this compound is still not fully complete at a 40-fold excess over (Tc)2R concentration. This reflects a much lower affinity of soblidotin than phomopsin A for its β2-subunit half site and is consistent with the observation that, in soaked crystals, significant electron density at that site is observed only with the latter.
Figure 3.
Effects of vinca domain ligand binding on (Tc)2R nucleotides. (A) Inhibition of nucleotide exchange. The variations of the normalized percentage of (Tc)2R nucleotide exchanged as a function of phomopsin A (filled circles), soblidotin (open circles) and vinblastine (open squares) concentrations are presented. Each data point is the average of three measurements, performed as described in Methods. (B) Inhibition of GTPase activity. The variation of the specific GTPase activity of (Tc)2R is presented as a function of phomopsin A concentration (filled circles, dashed), soblidotin (open circles, dotted) and its amino-terminal tripeptide (open triangles, alternating dash–dot). The lines represent fits of the data, calculated as described by Wang et al (2007), from which the vinblastine data (open squares, continuous line) are taken. Insert: equilibrium binding constants Ka of these ligands, deduced from the fit of the inhibition curves, as described (Wang et al, 2007). For phomopsin A and soblidotin, at the concentration of (Tc)2R at which data were measured (5 μM), the variation of the GTPase activity is linear over most of the range explored, which allows only a lower limit of Ka to be determined.
GTPase inhibition and affinity for the vinca domain
Tubulin hydrolyses GTP during microtubule assembly. In addition, non-microtubular tubulin has a GTPase activity that is enhanced by colchicine (David-Pfeuty et al, 1979) and by stathmin-like domains (Wang et al, 2007). The hydrolysis of GTP by tubulin is due to intermolecular tubulin–tubulin interactions: α-subunit residues of one molecule enhancing the activity of the β-subunit of another one (Nogales et al, 1998; Wang et al, 2007). In solution, this activity is due to the formation of assemblies (Wang et al, 2007), probably curved, similar to those that have been identified by electron microscopy (Saltarelli & Pantaloni, 1982). Phomopsin A and soblidotin inhibit this activity (supplementary Fig 4 online) as well as GTP hydrolysis by (Tc)2R, with an efficiency similar to that of vinblastine (Fig 3B). In (Tc)2R, these two ligands act as a wedge at the β1–α2 interface and restrict its flexibility, similarly to vinblastine (Wang et al, 2007). This in turn prevents α-subunit catalytic residues from being properly positioned to enhance hydrolysis of the β-tubulin nucleotide and probably explains the inhibitory effects of these ligands. The inhibition of (Tc)2R GTPase activity, as it specifically measures the interference of ligands with assembled α- and β-subunits of two different heterodimers (Wang et al, 2007), provides a convenient way to measure binding to the vinca domain. Rate variation is perfectly accounted for by the binding of one inhibitor to (Tc)2R with an affinity greater than 107 M−1 (Fig 3B). The stoichiometry is consistent with the observation that (Tc)2R comprises a single catalytic site at the interface of the two tubulin heterodimers (Wang et al, 2007). In the case of phomopsin A, this means that the site at the inter-dimer interface is of high affinity, whereas the site that is contributed only by β-tubulin residues has significantly lower affinity. Finally, it is interesting to note that the soblidotin N-terminal tripeptide binds to (Tc)2R at least 20 times less well than the whole molecule (Fig 3B), whereas the overlap of soblidotin with vinblastine is limited to its N-terminal dipeptide; this suggests that significant binding affinity is gained by extending vinca domain ligands so that they interact with tubulin beyond the core of this domain.
Inhibition of microtubule assembly
Phomopsin A and soblidotin, as vinblastine, inhibit microtubule assembly (Bai et al, 1990a; Natsume et al, 2000). Our data suggest two explanations for this effect, common to the three ligands. First, as shown by their inhibition of GTPase, they all bind tightly to (Tc)2R at the interface of two tubulin molecules in a helical assembly (Gigant et al, 2000). They also induce the formation of spirals (vinblastine; Gigant et al, 2005) or rings of tubulin (phomopsin A or dolastatin 10; Tonsing et al, 1984; Bai et al, 1999). Therefore, they stabilize such curved assemblies. Second, when they bind to tubulin, the ligands studied here prevent it from assembling in straight protofilaments, as this would lead to steric clashes with these drugs (supplementary Fig 5 online). Such protofilaments are required to establish the lateral interactions that assist microtubule formation (Nogales et al, 1999). Therefore, preventing tubulin from forming straight structures and stabilizing curved tubulin assemblies both contribute to phomopsin A, soblidotin and vinblastine impeding microtubule assembly.
In summary, the structures of tubulin complexes with phomopsin A and soblidotin extend the part of the vinca domain that is structurally defined, clarify the mechanism for the exchange of tubulin nucleotide and elucidate the ways by which vinca ligands prevent microtubule assembly.
Methods
Crystallization and structure determination. Crystals of (Tc)2R were obtained as described earlier (Gigant et al, 2000; Dorléans et al, 2007) and then soaked by the vinca domain ligands studied here. For soblidotin and its N-terminal tripeptide, we used a 24-h soaking time in a 2 mM solution of the vinca domain molecule. For phomopsin A, crystals were soaked in a 0.7 mM phomopsin A solution in the crystallization buffer supplemented with 15% dimethylsulphoxide to enhance the solubility of this ligand. The crystals were flash-cooled in liquid nitrogen and diffraction data sets were collected at the European Synchrotron Radiation Facility (Grenoble, France) on beamline ID14eh4. For structure determination, the structure of (Tc)2R was used as a starting model and further refined as described by Gigant et al (2005) with REFMAC using the TLS option (Winn et al, 2001). Details are given in the supplementary information online. The coordinates and structure factors have been deposited with the Protein Data Bank with accession codes 3du7 (phomopsin A) and 3e22 (soblidotin).
Inhibition of nucleotide exchange. The exchange of GDP for GTP at the β-tubulin nucleotide site was estimated by measuring the displacement of ‘cold' GDP by [α-32P]GTP following a procedure adapted from Bai et al (1990a). (Tc)2R (5 μM) was incubated for 5 min on ice with the candidate inhibitor at varying concentrations in P-buffer (80 mM PIPES-K, pH 6.8, 1 mM MgCl2 and 0.5 mM EGTA). [α-32P]GTP (50 μM) was added and the reaction mixture was incubated for another 15 min. (Tc)2R was separated from unbound nucleotides by rapid gel filtration on a Micro Bio-Spin P6 column (Bio-Rad; Marnes-La-Coquette, France) previously equilibrated with the same buffer (Penefsky, 1977). There was no release of tubulin-bound nucleotide during centrifugation (Caplow & Zeeberg, 1980) and we checked that tubulin was totally eluted; the result was corrected for eluted free nucleotide. The radioactivity of the tubulin-containing eluted fraction was counted and compared with the radioactivity of a known concentration of [α-32P]GTP. The exchange measured with (Tc)2R alone, that is without vinca site ligand, was normalized to 100%. This corresponds to about 0.45 GDP/GTP exchange per tubulin. The exchange measured with tubulin without inhibitor and without RB3-SLD is around 0.7 [α-32P]GTP per tubulin.
GTPase activity measurements. The effect of the vinca domain ligands on the GTPase activity of (Tc)2R was evaluated by quantifying the release of free inorganic phosphate from [γ-32P]GTP during a 24-min time course as described for vinblastine (Wang et al, 2007). The procedure is detailed in the supplementary information available online.
Electron microscopy. The (Tc)2Rhel complex (5 μM) was incubated with 25 μM vinblastine, soblidotin or phomopsin A for 30 min at 25°C. Sample aliquots (5 μl) were applied to a glow-discharged carbon-coated 400-mesh per inch copper grid, allowed to adsorb for 30 s, washed twice with water, and negatively stained for 20 s with 1% (w/v) uranyl acetate. Specimens were examined in a Fei Technai 12 BioTwin TEM operated at 80 kV. Micrographs were recorded with an Eagle 4k CCD camera at a nominal × 68,000 magnification.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr T. Natsume and Dr M. Kobayashi (ASKA Pharmaceutical Co., Ltd) for providing us with soblidotin and its N-terminal tripeptide, as well as for their comments on the paper, and Professor C. Wang (Tongji University, Shanghai) for comments on the paper. This study was supported by the Centre National de la Recherche Scientifique, La Ligue Contre Le Cancer (équipe labellisée 2006) and l'Agence Nationale de la Recherche (grant ANR-05-BLAN-0292).
Footnotes
The authors declare that they have no conflict of interest.
References
- Amayed P, Carlier MF, Pantaloni D (2000) Stathmin slows down guanosine diphosphate dissociation from tubulin in a phosphorylation-controlled fashion. Biochemistry 39: 12295–12302 [DOI] [PubMed] [Google Scholar]
- Bai RL, Pettit GR, Hamel E (1990a) Binding of dolastatin 10 to tubulin at a distinct site for peptide antimitotic agents near the exchangeable nucleotide and vinca alkaloid sites. J Biol Chem 265: 17141–17149 [PubMed] [Google Scholar]
- Bai RL, Pettit GR, Hamel E (1990b) Structure–activity studies with chiral isomers and with segments of the antimitotic marine peptide dolastatin 10. Biochem Pharmacol 40: 1859–1864 [DOI] [PubMed] [Google Scholar]
- Bai R, Roach MC, Jayaram SK, Barkoczy J, Pettit GR, Luduena RF, Hamel E (1993) Differential effects of active isomers, segments, and analogs of dolastatin 10 on ligand interactions with tubulin. Correlation with cytotoxicity. Biochem Pharmacol 45: 1503–1515 [DOI] [PubMed] [Google Scholar]
- Bai R, Durso NA, Sackett DL, Hamel E (1999) Interactions of the sponge-derived antimitotic tripeptide hemiasterlin with tubulin: comparison with dolastatin 10 and cryptophycin 1. Biochemistry 38: 14302–14310 [DOI] [PubMed] [Google Scholar]
- Bai R, Covell DG, Taylor GF, Kepler JA, Copeland TD, Nguyen NY, Pettit GR, Hamel E (2004) Direct photoaffinity labeling by dolastatin 10 of the amino-terminal peptide of beta-tubulin containing cysteine 12. J Biol Chem 279: 30731–30740 [DOI] [PubMed] [Google Scholar]
- Caplow M, Zeeberg B (1980) Stoichiometry for guanine nucleotide binding to tubulin under polymerizing and nonpolymerizing conditions. Arch Biochem Biophys 203: 404–411 [DOI] [PubMed] [Google Scholar]
- Carlier MF, Hill TL, Chen Y (1984) Interference of GTP hydrolysis in the mechanism of microtubule assembly: an experimental study. Proc Natl Acad Sci USA 81: 771–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Melki R, Pantaloni D, Hill TL, Chen Y (1987) Synchronous oscillations in microtubule polymerization. Proc Natl Acad Sci USA 84: 5257–5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Pfeuty T, Simon C, Pantaloni D (1979) Effect of antimitotic drugs on tubulin GTPase activity and self-assembly. J Biol Chem 254: 11696–11702 [PubMed] [Google Scholar]
- Dorléans A, Knossow M, Gigant B (2007) Studying drug–tubulin interactions by X-ray crystallography. In Microtubule Protocols, Zhou J (ed) Vol 137, pp 235–244. Totowa, NJ, USA: Humana [DOI] [PubMed] [Google Scholar]
- Gigant B, Curmi PA, Martin-Barbey C, Charbaut E, Lachkar S, Lebeau L, Siavoshian S, Sobel A, Knossow M (2000) The 4 Å X-ray structure of a tubulin:stathmin-like domain complex. Cell 102: 809–816 [DOI] [PubMed] [Google Scholar]
- Gigant B, Wang C, Ravelli RBG, Roussi F, Steinmetz MO, Curmi PA, Sobel A, Knossow M (2005) Structural basis for the regulation of tubulin by vinblastine. Nature 435: 519–522 [DOI] [PubMed] [Google Scholar]
- Hamel E (1992) Natural products which interact with tubulin in the vinca domain: maytansine, rhizoxin, phomopsin A, dolastatins 10 and 15 and halichondrin B. Pharmacol Ther 55: 31–51 [DOI] [PubMed] [Google Scholar]
- Koiso Y, Morisaki N, Yamashita Y, Mitsui Y, Shirai R, Hashimoto Y, Iwasaki S (1998) Isolation and structure of an antimitotic cyclic peptide, ustiloxin F: chemical interrelation with a homologous peptide, ustiloxin B. J Antibiot (Tokyo) 51: 418–422 [DOI] [PubMed] [Google Scholar]
- Li P, Evans CD, Wu Y, Cao B, Hamel E, Joullie MM (2008) Evolution of the total syntheses of ustiloxin natural products and their analogues. J Am Chem Soc 130: 2351–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow EM, Lange G, Jagla A, Spann U, Mandelkow E (1988) Dynamics of the microtubule oscillator: role of nucleotides and tubulin–MAP interactions. EMBO J 7: 357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW (1984) Dynamic instability of microtubule growth. Nature 312: 237–242 [DOI] [PubMed] [Google Scholar]
- Natsume T, Watanabe J, Tamaoki S, Fujio N, Miyasaka K, Kobayashi M (2000) Characterization of the interaction of TZT-1027, a potent antitumor agent, with tubulin. Jpn J Cancer Res 91: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T, Watanabe J, Ogawa K, Yasumura K, Kobayashi M (2007) Tumor-specific antivascular effect of TZT-1027 (Soblidotin) elucidated by magnetic resonance imaging and confocal laser scanning microscopy. Cancer Sci 98: 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Downing KH, Amos LA, Löwe J (1998) Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol 5: 451–458 [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH (1999) High-resolution model of the microtubule. Cell 96: 79–88 [DOI] [PubMed] [Google Scholar]
- Penefsky HS (1977) Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem 252: 2891–2899 [PubMed] [Google Scholar]
- Ravelli RBG, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M (2004) Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 428: 198–202 [DOI] [PubMed] [Google Scholar]
- Saltarelli D, Pantaloni D (1982) Polymerization of the tubulin–colchicine complex and guanosine 5′-triphosphate hydrolysis. Biochemistry 21: 2996–3006 [DOI] [PubMed] [Google Scholar]
- Tonsing EM, Steyn PS, Osborn M, Weber K (1984) Phomopsin A, the causative agent of lupinosis, interacts with microtubules in vivo and in vitro. Eur J Cell Biol 35: 156–164 [Google Scholar]
- Wang C, Cormier A, Gigant B, Knossow M (2007) Insight into the GTPase activity of tubulin from complexes with stathmin-like domains. Biochemistry 46: 10595–10602 [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57: 122–133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information