Abstract
The calcineurin/NFAT (nuclear factor of activated T-cells) signalling pathway is essential for many aspects of vertebrate development and is the target of the widely used immunosuppressive drugs FK506 and cyclosporine A. The basis for the therapeutic specificity of these drugs has remained unclear, as calcineurin is expressed ubiquitously. By inactivating calcineurin during haematopoietic development, we found that although this signalling pathway has an important, non-redundant role in the regulation of lymphocyte developmental checkpoints, it is not essential for the development of blood myeloid lineages. These studies have shown that the specificity of calcineurin inhibitors arises from the selective use of calcineurin at distinct developmental stages. The requirement for calcineurin/NFAT in the development of the adaptive but not of the innate immune system is consistent with the idea that the evolutionary appearance of this pathway was involved in the emergence of vertebrates.
Keywords: haematopoiesis, calcineurin, NFAT, signalling
Introduction
The calcineurin/NFAT (nuclear factor of activated T-cells) signalling pathway, originally discovered in lymphocytes, is important in many aspects of vertebrate development, including axonal guidance, vasculogenesis and cardiac morphogenesis. Ligation of many receptors leads to a rise in intracellular calcium concentration and to the activation of calcineurin phosphatase by Ca2+-bound calmodulin. Dephosphorylation of the cytoplasmic subunits (NFATc) of NFAT transcription complexes by calcineurin reveals the nuclear localization sequence and results in their import into the nucleus, where they bind to DNA and regulate transcription cooperatively with other transcription factors (Crabtree & Olson, 2002). The important role of calcineurin/NFAT signalling in the regulation of immune function is shown by the fact that this pathway is the common target of two of the most effective immunosuppressive drugs, FK506 and cyclosporine A (Flanagan et al, 1991). The molecular basis of specificity of these drugs in blocking immune responses is not clear; however, two theories have been advanced. Specificity might arise from the selective use of the ubiquitous pathway for specific developmental steps. Alternatively, the main gain-of-function mechanism of action of both cyclosporine A and FK506 would render cells with lower concentrations of calcineurin paradoxically more sensitive to the drugs. We investigated the role of this pathway in the development of the haematopoietic system by deleting the regulatory subunit calcineurin B1 (Cnb1) specifically in bone marrow-derived stem cells. In adult mice all blood cells derive from haematopoietic stem cells (Iwasaki & Akashi, 2007); therefore, deletion of a genomic locus in these cells results in deletion in all haematopoietic lineages, thereby allowing identification of the steps requiring a specific gene.
Results And Discussion
Expression of Cnb1 and Cnb2 in haematopoietic lineages
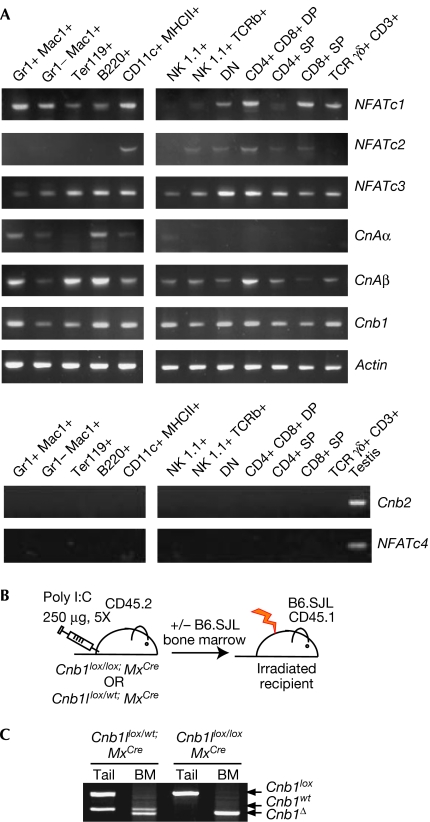
The calcineurin complex is composed of a catalytic subunit, calcineurin A (CNA), and a regulatory subunit, calcineurin B (CNB). Three genes encode catalytic subunits (CnAα, CnAβ and CnAγ), and two genes encode regulatory subunits (Cnb1 and Cnb2; Rusnak & Mertz, 2000). Although CnAα, CnAβ and Cnb1 are expressed ubiquitously, expression of CnAγ and Cnb2 is restricted to the germ line. Deletion of Cnb1, therefore, results in a lack of calcineurin enzymatic activity in non-germline cell types. Expression of NFATc, CnA and CnB genes was verified by using reverse transcription–PCR (RT–PCR) in several blood lineages (Fig 1A). Although Cnb1 was expressed in all cell types analysed, expression of Cnb2 was detected, as expected, only in testis. This indicates that genetic inactivation of Cnb1 would eliminate all calcineurin enzymatic activity in bone marrow-derived blood lineages.
Figure 1.
Inducible deletion of Cnb1 in bone marrow-derived lineages. (A) Expression of components of the calcineurin/NFAT pathway was analysed by RT–PCR on sorted cell populations. (B) Cnb1 conditional knockout Mx-1Cre mice were treated with poly I:C to induce the expression of Cre; whole or sorted bone marrow fractions from Cnb1-deficient and control donors were transplanted into irradiated CD45.1+ B6.SJL congenic recipients alone or in conjunction with CD45.1+ helper B6.SJL bone marrow. (C) PCR of genomic DNA from tail or Cnb1-deficient and control bone marrow after treatment with poly I:C. BM, bone marrow; NFAT, nuclear factor activating T-cells; poly I:C, poly inosilic acid; RT–PCR, reverse transcription PCR.
Inducible deletion of Cnb1 in bone marrow cells
Cnb1 conditional knockout mice (Cnb1flox/flox; Neilson et al, 2004) were crossed with mice expressing the Cre recombinase gene under the control of the interferon-inducible Mx-1 promoter (Kuhn et al, 1995). This promoter drives deletion in all haematopoietic cells and is induced by the administration of viral mimics such as poly inosilic acid (poly I:C). After treatment, whole or sorted bone marrow fractions from CD45.2+ control or CNB1-deficient donors were isolated and transplanted into congenic CD45.1+ recipients (B6.SJL; Fig 1B). The mismatched surface markers CD45.1 and CD45.2 were used to monitor the efficiency of reconstitution.
Whole bone marrow from mice treated with poly I:C was analysed to assess deletion of the Cnb1 genomic locus. The Cnb1 locus was completely deleted in CNB1-deficient bone marrow (Fig 1C). Control and experimental mice treated with poly I:C showed no sign of toxicity and survived for more than 6 months after treatment (data not shown).
CNB1 is not required for myeloid development
Traditionally, blood cells are categorized into lymphoid and myeloid lineages. The lymphoid lineage comprises T, B and natural killer cells. The myeloid lineage includes granulocytes, monocytes, erythrocytes and megakaryocytes. Dendritic cells can derive from both pathways (Iwasaki & Akashi, 2007). To analyse the role of calcineurin in the development of myeloid lineages, CD45.1+ lethally irradiated B6.SJL mice were injected with CD45.2+ control or CNB1-deficient bone marrow. All transplanted chimaeras survived with no sign of disease and were analysed 4 months after transplantation. All principal myeloid blood lineages were effectively reconstituted by control and CNB1-deficient bone marrow (Fig 2A–C).
Figure 2.
Development of myeloid lineages from CNB1-deficient bone marrow. (A) Percentage of indicated cellular subsets in bone marrow and spleen of mice transplanted with control or CNB1-deficient bone marrow. Mice were analysed by using flow cytometry 4 months after transplantation. (B) Absolute numbers of CD45.2+ cells of the indicated phenotype in bone marrow of transplanted recipients. (C) Absolute numbers of CD45.2+ cells of the indicated phenotype in the spleen of transplanted recipients. (D) Genomic PCR of the Cnb1 locus in tissues isolated from transplanted mice. (E) Immunoblot analysis of CNB1 and CNB2 protein expression in tissues isolated from transplanted mice. (F) Relative ratio of CD45.2+ to CD45.1+ cells in mice transplanted with CD45.2+ control or CNB1-deficient bone marrow mixed at a 1:1 ratio with B6.SJL bone marrow. Each symbol represents an individual animal. Bars represent mean values. The grey line indicates the predicted 1:1 ratio (logarithm of 1:1 equals 0). BM, bone marrow; Sp, spleen; Thy, thymus.
When tissues obtained from mice reconstituted with CNB1-deficient bone marrow were analysed, we observed complete deletion of the Cnb1 genetic locus and a lack of protein expression in bone marrow and spleen (Fig 2D,E). This indicated that cells that escaped deletion were not responsible for the observed reconstitution.
We also examined the ability of CNB1-deficient bone marrow to reconstitute myeloid lineages in a competitive setting by transplanting CD45.2+ control or CNB1-deficient bone marrow in a ratio of 1:1 with CD45.1+ B6.SJL whole bone marrow into irradiated B6.SJL mice. Chimaeras were analysed 5–6 months after transplantation. CNB1-deficient bone marrow competed efficiently with wild-type bone marrow to generate granulocytes, megakaryocytes, early erythroblasts and dendritic cells (Fig 2F).
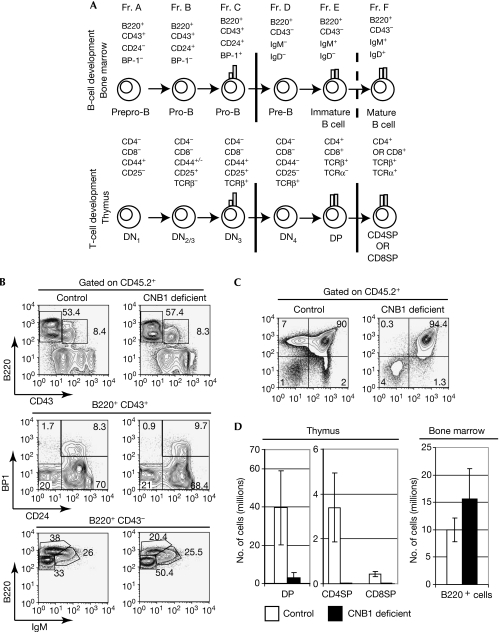
CNB1 is required for B- and T-cell development
Common lymphoid progenitors can give rise to both T cells, in the thymus, and B cells, in the bone marrow (Iwasaki & Akashi, 2007). B-cell precursors at all stages of development can be found in the adult mouse bone marrow and categorized into several fractions designated A to F according to the expression of cell surface markers (Hardy & Hayakawa, 2001). Two principal checkpoints exist in B-cell development (Fig 3A). The first corresponds to the rearrangement and selection of the immunoglobulin heavy chain by pairing with the surrogate light chain to form the pre-B-cell receptor (BCR; transition from Hardy's fraction C to D). The second corresponds to rearrangement and pairing of the immunoglobulin light chain with the immunoglobulin heavy chain to form the BCR (transition from Hardy's fraction E to F; Hayakawa & Hardy, 2000).
Figure 3.
CNB1-deficient B- and T-cell development in a non-competitive setting. (A) Diagram illustrating stages and surface marker in B- and T-cell development. Bars indicate principal developmental checkpoints. (B) CD45.1+ B6.SJL lethally irradiated mice were injected with CD45.2+ control or CNB1-deficient bone marrow and then analysed 4 months after transplantation by flow cytometry. Figure shows the percentage of developing B-cell subsets in the bone marrow of transplanted recipients. (C) Percentage of developing T-cell subsets in the thymus of transplanted recipients. (D) Absolute numbers of CD45.2+ developing T and B cells in transplanted mice. DN, double negative; DP, double positive.
The development of immature T cells, or thymocytes, closely parallels that of B cells (Melchers, 2005; Fig 3A). For thymocytes, the first developmental checkpoint corresponds to rearrangement of the T-cell receptor β-chain (TCRβ) and pairing with the pre-Tα to form the pre-TCR. Similar to the pre-BCR, the pre-TCR delivers survival/differentiation signals that allow the transition to the next developmental stage (transition from the double negative 3/4 stage to the double positive stage; Bhandoola et al, 2007). The second checkpoint corresponds to rearrangement and expression of the TCRα chain to form the αβTCR. At this stage, thymocytes are positively and negatively selected by peptide-major histocompatibility complex molecules, a process unique to developing T cells (Starr et al, 2003).
First, we examined the role of calcineurin/NFAT signalling in the development of B and T cells in a non-competitive transplantation experiment, by transplanting whole CD45.2+ control or CNB1-deficient bone marrow in CD45.1+ lethally irradiated B6.SJL mice. As reported previously (Neilson et al, 2004), the deletion of Cnb1 resulted in a block of positive selection, and impaired transition between the double-negative and double-positive stages of development of thymocytes, but it had no major effect on B-cell development in the bone marrow (Fig 3B–D).
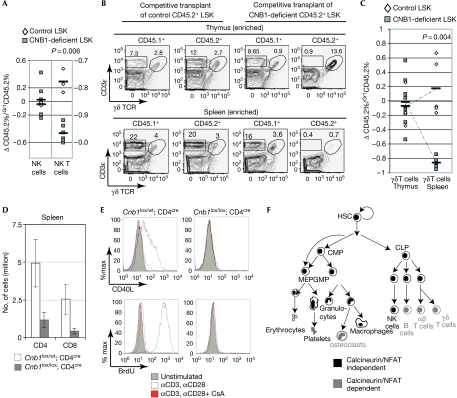
To minimize the possible confounding effect of long-lived B-cell precursors transplanted with unsorted whole bone marrow, B- and T-cell development was also analysed by transplanting sorted CD45.2+ control and CNB1-deficient progenitors (LSK, Lineage−/lowSca1highcKithigh) mixed with CD45.1+ helper bone marrow. Chimaerism was analysed 4–6 months after transplantation. As the amount of chimaerism varied between animals, the data were normalized to the percentage of chimaerism in the myeloid compartment. In this setting, a clear impairment in the progression past the pre-BCR (Fig 4A,B) and pre-TCR (Fig 4C,D) checkpoints was observed in B- and T-cell precursors that developed in the absence of calcineurin activity. Consistent with defective signal transduction downstream from the αβ TCR, the development of natural killer T cells, but not natural killer cells, was also impaired in the absence of calcineurin (Fig 5A).
Figure 4.
CNB1-deficient B- and T-cell development in a competitive setting. (A) Flow cytometry analysis of early B-cell development in a mouse reconstituted with CD45.2+ control or CNB1-deficient LSK progenitors and helper CD45.1+ bone marrow. The profile of early B-cell developmental markers is shown after gating on CD45.1+- and CD45.2+-positive cells. (B) Variation in CD45.2 chimaerism during B-cell development according to Hardy's nomenclature. Each symbol represents an individual mouse and the bars represents the average; P refers to two-tailed t-test. (C) Flow cytometry analysis of T-cell development in the thymus of a mouse reconstituted with CD45.2+ control or CNB1-deficient LSK progenitors and helper CD45.1+ bone marrow. The profile of T-cell developmental markers is shown after gating on CD45.1+- and CD45.2+-positive thymocytes. (D) Variation in CD45.2 chimaerism during T-cell development in the thymus. Each symbol represents an individual mouse and the bars represents the average; P refers to two-tailed t-test. DN, double negative; DP, double positive; LSK, Lineage−/lowSca1highcKithigh.
Figure 5.
CNB1-deficient natural killer- natural killer T- and γδ T-cell development in a competitive setting. (A) CD45.2 chimaerism in NK cells in the spleen and in NK T cells in the thymus normalized to myeloid reconstitution. Each symbol represents an individual mouse and the bars represents the average; P refers to two-tailed t-test. (B) Flow cytometry analysis of γδ T cell in the thymus and spleen of a mouse reconstituted with CD45.2+ control or CNB1-deficient LSK progenitors and helper CD45.1+ bone marrow. Before analysis, γδ T cells were enriched with anti-PE magnetic beads. The percentage of γδ T cells after gating on CD45.1+- and CD45.2+-positive thymocytes is shown. (C) CD45.2 chimaerism in γδ T cells in the thymus and spleen normalized to myeloid compartment. Each symbol represents an individual mouse and the bars represents the average; P refers to two-tailed t-test. (D) Absolute numbers of αβ T cells in the spleen in mice of the indicated phenotype (n=6, mean±s.d.). (E) Peripheral αβ T cells from mice of the indicated genotype were stimulated in vitro with anti-CD3 and anti-CD28 in the presence or absence of cyclosporine A, and then analysed by flow cytometry for the upregulation of CD40L and BrdU incorporation. (F) Summary of calcineurin-dependent and -independent haematopoietic lineages. BrdU, 5-bromodeoxyuridine; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; GMP, granulocyte-macrophage progenitor; HSC, haematopoietic stem cell; LSK, Lineage−/lowSca1highcKithigh; MEP, megakaryocyte progenitor (adapted from Iwasaki & Akashi, 2007); NK, natural killer; PE, phycoerythrin; TCR, T-cell receptor.
We also examined the development of γδ T lymphocytes, a subset of lymphocytes the lineage of which diverges from that of the αβ T-cell lineage at the double-negative 3 stage (Hayday, 2000). CNB1-deficient γδ T cells developed normally in the thymus, but were absent in the spleen of transplanted animals, suggesting that calcineurin might be required for γδ T-cell survival and/or proliferation in the periphery (Fig 5B,C). This is analogous to what we and others have found when examining the effects of inactivation of calcineurin on peripheral αβ T cells (Fig 5D,E; supplementary Fig 1 online; Manicassamy et al, 2008). The delayed deletion of the Cnb1 locus by means of a Cre recombinase driven by the CD4 promoter (Cnb1lox/lox;CD4cre) only mildly affects thymic selection (supplementary Fig 1 online). However, Cnb1lox/lox;CD4cre animals show severely reduced numbers of peripheral αβ T cells. Notably, the effects of calcineurin deletion in peripheral αβ T-cell activation and proliferation phenocopy the effects of calcineurin inhibition by treatment with cyclosporine A (Fig 5E; supplementary Fig 1 online).
Taken together, these data show that calcineurin is not essential for the development of myeloid lineages, whereas it is selectively required for the development of B and T cells. The development of both B and T cells is impaired at the checkpoint that is regulated by functionally related ‘pre-receptors' (Melchers, 2005), the pre-BCR and pre-TCR, respectively. In addition, the deletion of calcineurin blocks positive selection of thymocytes. It has been proposed that positive selection provides the same ‘proofreading' function that is performed by the surrogate light chain during B-cell development (Melchers, 2005). It is interesting to observe that both processes depend on calcineurin/NFAT signalling (Fig 3A, solid bars indicate developmental steps that require calcineurin). The observation that calcineurin is not required for the generation of γδ T cells in the thymus is consistent with the recent finding that ligand-dependent signalling by the γδ TCR is dispensable for the development of these cells (Jensen et al, 2008).
The calcineurin inhibitors FK506 and cyclosporine A are among the most effective of immunosuppressants and their introduction in the 1980s revolutionized transplant therapy. Although these drugs are relatively selective for the immune system, they show mechanism-based toxicity in the kidney and cardiovascular systems (Dumont, 2000); the basis of their selective action on lymphocytes remains unknown. One theory is that, as both drugs bind to intracellular receptors producing a composite surface that blocks activity of calcineurin, high levels of calcineurin might make cells paradoxically less sensitive to the drugs; indeed this seems to be the case with neurons (Graef et al, 2003). By contrast, the biologic and therapeutic selectively might simply be a matter of use of the ubiquitous pathway only by specific receptors. Our studies show that the latter is the case for the haematopoietic system, as we find only minor variations in the levels of calcineurin in haematopoietic lineages (Fig 1A).
In summary, calcineurin/NFAT signalling seems to have an important non-redundant role in the regulation of developmental checkpoints of the lymphocyte, but it is not required for the development of blood myeloid lineages. It is possible, however, that the function of these cells might be compromised when the calcineurin/NFAT pathway is inhibited.
Interestingly, the calcineurin/NFAT pathway is required for the development of osteoclasts, which represent a vertebrate-specific myeloid-derived lineage (reviewed in Takayanagi, 2007). These findings are consistent with the idea that the emergence of the calcineurin/NFAT pathway might have provided a new signalling pathway for the development of vertebrate-specific organs such as the combinatorial immune system and skeletal system (Wu et al, 2007).
Methods
Mice. Cnb1 conditional knockout mice (Neilson et al, 2004) and Mx-cre mice (Kuhn et al, 1995) have been characterized. For the induction of the Mx-cre recombinase in vivo, mice were treated with five i.p. injections of 250 μg of poly-inosilic acid (poly I:C; Invivogen, San Diego, CA, USA) every other day for 10 days. Two days after the last injection, the mice were killed and bone marrow cells were prepared using standard protocols. No consistent differences were observed among mice transplanted with bone marrow obtained from mice of the following genotypes: Cnb1+/+-MxCre; Cnb1flox/+-MxCre; Cnb1Δ/+-MxCre; Cnb1flox/flox or Cnb1flox/Δ; therefore, they are collectively referred to as ‘control mice'. In addition, no differences were observed between mice transplanted with bone marrow from Cnb1flox/flox-MxCre and Cnb1flox/Δ-MxCre, and these mice were used interchangeably for the experiments described in this study. Bone marrow derived from these mice is referred to as ‘CNB1-deficient' bone marrow. Mice that express the cre recombinase under the CD4 promoter (CD4cre; Lee et al, 2001) were purchased from Taconic (Hudson, NY, USA) and crossed with Cnb1 conditional knockout mice.
Genomic PCR. Genomic PCR was performed on DNA extracted from various tissues using standard protocols. The primers used for Cnb1 genomic PCR have been described previously (Neilson et al, 2004).
Bone marrow transplantation. CD45.1+ B6.SJL mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Bone marrow chimaeras were prepared using standard protocols. Lethally irradiated (1250 rad) CD45.1+ B6.SJL mice were reconstituted with 5–10 million bone marrow cells from sex-matched CNB1-deficient and littermate control mice, alone or in a 1:1 mixture with CD45.1+ bone marrow from B6.SJL mice, as indicated. For the analysis of lymphoid development, the Lineage (Gr1, CD11b, CD3, B220, Ter119)− Sca1highcKithigh (LSK) fraction of CNB1-deficient and control bone marrow was sorted on BD FACSAria® and transplanted together with helper CD45.1+ bone marrow from B6.SJL mice (∼10,000 LSKs per mouse; 2:1 LSK equivalents).
Immunoblotting of total cell lysate. CNB1-deficient and control bone marrow cells and splenocytes were obtained by standard procedures. Cells were lysed in lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 1% Triton X-100 and 1 mM dithiothreitol containing a mixture of protease inhibitors). SDS–polyacrylamide gel electrophoresis and immunoblotting were carried out by standard procedures. The following antibodies were used for immunoblotting: anti-CNB1 (Upstate, Billerica, MA, USA), anti-Actin (Sigma, St Louis, MO, USA).
Flow cytometry and peripheral T-cell activation. Single cell suspensions from bone marrow, thymus and spleen were prepared and stained using standard protocols for flow cytometry. Antibody conjugates were purchased from eBioscience. Cellular subsets were identified as follows: granulocytes, Gr1+CD11b+; monocytes, Gr1−CD11b+; dendritic cells, MHC-II+ CD11c+; erythrocyte progenitors, Ter119+; megakaryocytes, CD41+; bone marrow B-cell subsets: fraction A, B220+CD43+ CD24−BP-1−; fraction B, B220+CD43+ CD24+BP-1−; fraction C, B220+CD43+ CD24+BP-1+; fraction D, B220+CD43− IgM−IgD−; fraction E, B220+CD43− IgM+IgD−; fraction F, B220+CD43− IgM+IgD+; T-cell subsets: double negative 1, Lineage−CD8−CD117+CD44+CD25−; double negative 2, Lineage−CD8−CD44+CD25+; double negative 3, Lineage−CD8−CD44−CD25+; double negative 4, Lineage−CD8−CD44−CD25−; double positive, CD4+CD8+; CD4SP, CD4+CD8−TCRβhigh; CD8SP, CD4−CD8+TCRβhigh; natural killer T cells, NK1.1+ TCRβ+; natural killer cells, NK1.1+ TCRβ−; γδ T cells; γδ TCR+ CD3ɛ+. Samples were acquired on BD LRS®, and/or BD FACSAria®, and analysed using FlowJo® software.
CNB1-deficient and control thymocytes and splenocytes were enriched for analysis of γδ T cells with phycoerythrin (PE)-conjugated γδ TCR antibody (eBioscience, San Diego, CA, USA) followed by anti-PE magnetic beads (Miltenyi Biotech, Auburn, CA, USA) according to the manufacturer's instructions.
For analysis of peripheral T-cell activation, cells were stimulated with plate-bound anti-CD3 (10 μg ml−1) and anti-CD28 (50 μg ml−1) for 48 h in the presence or absence of cyclosporine A (100 nM). Cells were pulsed with 5-bromodeoxyuridine (BrdU; 10 mM) for the last 24 h of stimulation. BrdU staining was performed with a BrdU BD Pharmingen kit according to the manufacturer's instructions.
Analysis of chimaerism in mice transplanted with LSK progenitors. CD45.1+ B6.SJL lethally irradiated mice were injected with CD45.2+ control or CNB1-deficient sorted LSK progenitors together with helper CD45.1+ bone marrow from B6.SJL mice. Mice were analysed 4–6 months after transplantation. The following formula was used to indicate CD45.2 chimaerism normalized to chimaerism in the myeloid compartment: (%CD45.2 in indicated population−%CD45.2 in Gr1+ granulocytes)/%CD45.2 in Gr1+ granulocytes.
RNA isolation and RT–PCR. Total RNA was collected from sorted populations with Trizol®, followed by purification and on-column DNase digestion with RNeasy mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer's recommendations. Complementary DNA was generated with SuperScript® II RT (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's recommendations. Primers used for amplification of cDNA are described in supplementary Table 1 online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Tom Serwold and the Weisman laboratory, and Kirk Jensen and the Yueh-hsiu Chen laboratory for providing help and reagents. E.M.G. and M.M.W. were supported by Stanford Graduate Fellowships. M.M.W. was additionally supported by a Howard Hughes Medical Institute pre-doctoral fellowship. L.H. was supported by an A*STAR Scholarship. This study was supported by grants from the National Institute of Health (to G.R.C.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bhandoola A, von Boehmer H, Petrie HT, Zuniga-Pflucker JC (2007) Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity 26: 678–689 [DOI] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN (2002) NFAT signaling: choreographing the social lives of cells. Cell 109(Suppl): S67–S79 [DOI] [PubMed] [Google Scholar]
- Dumont FJ (2000) FK506, an immunosuppressant targeting calcineurin function. Curr Med Chem 7: 731–748 [DOI] [PubMed] [Google Scholar]
- Flanagan WM, Corthesy B, Bram RJ, Crabtree GR (1991) Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature 352: 803–807 [DOI] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR (2003) Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell 113: 657–670 [DOI] [PubMed] [Google Scholar]
- Hardy RR, Hayakawa K (2001) B cell development pathways. Annu Rev Immunol 19: 595–621 [DOI] [PubMed] [Google Scholar]
- Hayakawa K, Hardy RR (2000) Development and function of B-1 cells. Curr Opin Immunol 12: 346–353 [DOI] [PubMed] [Google Scholar]
- Hayday AC (2000) γδ cells: a right time and a right place for a conserved third way of protection. Annu Rev Immunol 18: 975–1026 [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Akashi K (2007) Hematopoietic developmental pathways: on cellular basis. Oncogene 26: 6687–6696 [DOI] [PubMed] [Google Scholar]
- Jensen KD et al. (2008) Thymic selection determines γδ T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon γ. Immunity 29: 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K (1995) Inducible gene targeting in mice. Science 269: 1427–1429 [DOI] [PubMed] [Google Scholar]
- Lee PP et al. (2001) A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15: 763–774 [DOI] [PubMed] [Google Scholar]
- Manicassamy S, Gupta S, Huang Z, Molkentin JD, Shang W, Sun Z (2008) Requirement of calcineurin A β for the survival of naive T cells. J Immunol 180: 106–112 [DOI] [PubMed] [Google Scholar]
- Melchers F (2005) The pre-B-cell receptor: selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nat Rev Immunol 5: 578–584 [DOI] [PubMed] [Google Scholar]
- Neilson JR, Winslow MM, Hur EM, Crabtree GR (2004) Calcineurin B1 is essential for positive but not negative selection during thymocyte development. Immunity 20: 255–266 [DOI] [PubMed] [Google Scholar]
- Rusnak F, Mertz P (2000) Calcineurin: form and function. Physiol Rev 80: 1483–1521 [DOI] [PubMed] [Google Scholar]
- Starr TK, Jameson SC, Hogquist KA (2003) Positive and negative selection of T cells. Annu Rev Immunol 21: 139–176 [DOI] [PubMed] [Google Scholar]
- Takayanagi H (2007) The role of NFAT in osteoclast formation. Ann NY Acad Sci 1116: 227–237 [DOI] [PubMed] [Google Scholar]
- Wu H, Peisley A, Graef IA, Crabtree GR (2007) NFAT signaling and the invention of vertebrates. Trends Cell Biol 17: 251–260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information