Abstract
Cleavage of the amyloid precursor protein (APP) is a crucial event in Alzheimer disease pathogenesis that creates the amyloid-β peptide (Aβ) and liberates the carboxy-terminal APP intracellular domain (AICD) into the cytosol. The interaction of the APP C terminus with the adaptor protein Fe65 mediates APP trafficking and signalling, and is thought to regulate APP processing and Aβ generation. We determined the crystal structure of the AICD in complex with the C-terminal phosphotyrosine-binding (PTB) domain of Fe65. The unique interface involves the NPxY PTB-binding motif and two α helices. The amino-terminal helix of the AICD is capped by threonine T668, an Alzheimer disease-relevant phosphorylation site involved in Fe65-binding regulation. The structure together with mutational studies, isothermal titration calorimetry and nuclear magnetic resonance experiments sets the stage for understanding T668 phosphorylation-dependent complex regulation at a molecular level. A molecular switch model is proposed.
Keywords: Alzheimer disease, amyloid precursor protein, APP intracellular domain, Fe65, phosphotyrosine-binding domain
Introduction
Alzheimer disease is a neurodegenerative disorder and the main cause of senile dementia in the present world. Pathologically, the disease is characterized by the formation of senile plaques and neurofibrillary tangles in the brain, accompanied by substantial neuronal and synaptic loss in the neocortex, which is likely to represent the main reason for cognitive impairment in Alzheimer disease (Mattson, 2004). Strong biochemical and genetic evidence support the hypothesis that accumulation of the amyloid-β peptide (Aβ), the main constituent of senile plaques, is a crucial event in Alzheimer disease pathogenesis. Aβ formation results from sequential cleavage of its precursor protein (APP), an integral and ubiquitously expressed type I transmembrane protein (Wolfe & Guenette, 2007), by the β-site-cleaving enzyme 1 and the γ-secretase complex (Haass, 2004).
Important binding partners of APP interact with the intracellular APP carboxyl terminus, modulating transport and signalling events (Wolfe & Guenette, 2007). The C terminus of APP adopts only transient structures when not bound to a binding partner (Ramelot et al, 2000). It contains the highly conserved Y682ENPTY motif (underlined residue numbering corresponds to the neuronal APP spliceform APP695, UniPROT entry: P05067-4) where several adaptor proteins bind through their phosphotyrosine-binding (PTB) domains (Wolfe & Guenette, 2007). As a consequence of the γ-secretase cleavage (ɛ-cleavage), the APP intracellular domain (AICD; 49–50 residues) is cleaved off and liberated into the cytosol (Weidemann et al, 2002), and is believed to have a function in gene regulation (McLoughlin & Miller, 2008). The APP C terminus can alternatively be cleaved by caspases at residue D664 generating a strong neurotoxic peptide comprising the C-terminal 31 amino acids of APP (AICD-C31), which could be linked to increased synaptic loss and neuronal death in Alzheimer disease (Galvan et al, 2002).
The APP-interacting protein that has generated the most interest is Fe65, as knockout studies in worms and mice resulted in phenotypes markedly similar to those seen when APP genes were knocked out (Zambrano et al, 2002; Guenette et al, 2006). Fe65 is a brain-enriched adaptor protein that is important for brain development (Guenette et al, 2006), and contains one WW domain and two PTB domains. Recently, the high-resolution structures of the WW domain (Meiyappan et al, 2007) and the amino-terminal PTB domain (PTB1; Radzimanowski et al, 2008a) have been determined. The C-terminal PTB domain (PTB2) binds to the C terminus of APP (Russo et al, 2005). Binding of Fe65 to the APP C terminus is thought to influence APP processing and Aβ generation (McLoughlin & Miller, 2008). The APP C terminus contains eight putative phosphorylation sites, with seven of them being phosphorylated in Alzheimer disease-affected brains (Lee et al, 2003). The most important and brain-limited phosphorylation occurs at threonine T668 (Pastorino & Lu, 2006). Phosphorylation of T668 is linked to neurite extension, anterograde transport of vesicular cargo, nuclear signalling and regulation of Fe65 binding. By contrast, enhanced phosphorylation of APP is believed to be a pathological trait of Alzheimer disease, as it seems to correlate with an increased generation of Aβ.
Here, we report the crystal structure of the complete neurotoxic part of the AICD in complex with the human Fe65-PTB2 domain at a resolution of 2.1 Å, and describe the mechanism of the Alzheimer disease-relevant, and phosphorylation-dependent complex regulation.
Results and Discussion
AICD/Fe65-PTB2 shows a unique binding interface
The AICD (ɛ-cleavage product) has been shown to be intrinsically disordered in solution (Ramelot et al, 2000). Nuclear magnetic resonance (NMR) experiments, however, indicate that parts of the peptide undergo a major structural change on addition of Fe65-PTB2 (supplementary Fig S1 online). The NMR signals of 15N-labelled AICD reveal that 15–20 residues remain unstructured leading to the identification of a minimal-construct, AICD-C32 (Fig 1B), which is amenable to structural studies. Addition of either 50-mer or 32-mer AICD peptides to 15N-labelled Fe65-PTB2 results in similar spectral changes, indicating that the molecular interface between Fe65-PTB2 and both AICD peptides is identical. Isothermal titration calorimetry (ITC) experiments confirmed that both peptides bind with similar affinities and with a Kd value of 0.2 μM (Table 1). By contrast, a short 11-mer AICD peptide (N680GYENPTYKFF) covering the classical PTB-binding site (Zhang et al, 1997; Yun et al, 2003) binds with an unusually high Kd value of about 100 μM, as derived from NMR experiments.
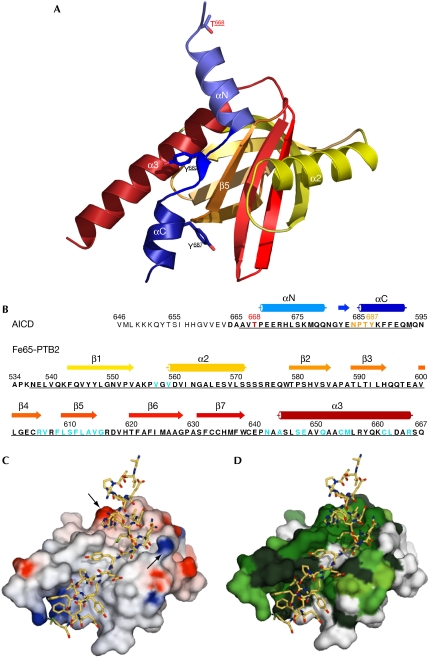
Figure 1.
The AICD/Fe65-PTB2 complex. (A) Overall structure of the complex in a view on the binding cleft with the AICD shown in blue and Fe65-PTB2 shown in yellow to red. The phosphorylation-amenable residues of the AICD are either on the side (T668 and Y687) or central (Y682) to the interface. (B) Numbered primary sequences and labelled secondary structures of the peptides in the same colour code used in (A). Residues of the cloned constructs (AICD-C32 and Fe65-PTB2) are shown in bold and underlined if included in the model. The crucial complex-regulating residue T668 is highlighted in red, the N684PTY consensus PTB-binding motif in orange and Fe65 residues involved in the interface in blue. (C) Electrostatic surface potential of Fe65-PTB2 in the same view as in (A), highlighting the general hydrophobicity of the interaction and showing the charge clamp for the helix dipole of AICD helix αN (indicated by arrows). (D) Surface of Fe65 showing the conservation (green) of helix α3 and of the binding sites for both AICD helices. The central part of the interface corresponding to strand β5 and the Y687 binding pocket is less conserved. AICD, amyloid precursor protein intracellular domain; PTB2, C-terminal phosphotyrosine-binding domain.
Table 1.
Isothermal titration calorimetry data for AICD/Fe65-PTB2 complexes
| Ligand | Kd (μM) | ΔH (kcal/mol) | ΔS (cal/K) | Relative affinity |
|---|---|---|---|---|
| AICD-C50 | 0.21±0.08 | −2.50 × 104±400 | −55.9±2.2 | 1 |
| AICD-C32 | 0.22±0.02 | −2.42 × 104±3400 | −53.5±11.7 | 1 |
| AICD-C32 T668A | 0.33±0.01 | −2.08 × 104±200 | −42.6±0.8 | 0.65 |
| AICD-C32 T668E | 0.34±0.02 | −2.06 × 104±800 | −42.0±2.7 | 0.63 |
| AICD-C32 pT668 | 1.56±0.18 | −1.17 × 104±300 | −14 | 0.14 |
| AICD 11-mer (NMR) | 103±25 | — | — | 0.002 |
| AICD, amyloid precursor protein intracellular domain; NMR, nuclear magnetic resonance; PTB2, C-terminal phosphotyrosine-binding domain. | ||||
The AICD-C32/Fe65-PTB2 complex was crystallized as described (Radzimanowski et al, 2008b) and the structure was solved using the Molecular Replacement method. Refinement statistics are given in Table 2. As part of the pleckstrin homology domain superfold, Fe65-PTB2 (residues A534 to Q667) shows the canonical PTB domain fold that consists of seven antiparallel β strands forming two orthogonal β sheets and a C-terminal α helix (α3; Fig 1A,B). Fe65-PTB2 contains another α helix (α2) inserted between strands β1 and β2, which is also observed in the structures of the X11 (Zhang et al, 1997) and mammalian disabled (mDab) PTB domains (Yun et al, 2003; supplementary Figs S2 and S3 online). Fe65-PTB2 in the complex structure is similar to the structure of free Fe65-like Fe65L2 (PDB code 2dyq; 44% identity in the PTB domains; root mean square deviation of 1.24 Å for 122 of 124 Cα-atoms; supplementary Figs S2 and S3 online), indicating that the domain interacts as a rigid body.
Table 2.
Refinement statistics
| Resolution (Å) | 33–2.1 |
| Number of reflections | 30,579 |
| Rwork/Rfree | 20.2/23.8 |
| Number of atoms | |
| Protein | 2,415 |
| Water molecules | 236 |
| B-factors (Å2) | |
| Overall | 41.3 |
| Protein | 40.8 |
| Fe65-PTB2 | 39.1 |
| AICD-C32 | 47.5 |
| Water | 47.1 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.321 |
| Ramachandran plot quality (%) | |
| Most favoured | 96.3 |
| Additionally allowed | 3.7 |
| AICD, amyloid precursor protein intracellular domain; PTB2, C-terminal phosphotyrosine-binding domain. | |
The peptide-binding site (approximately 1000 Å2) shows an extended cleft formed by strand β5, the C-terminal helix α3 and the preceding loop, and residues from the β1-α2 loop and the N terminus of the α2 helix (Fig 1C,D and supplementary Fig S4 online). The binding cleft, and especially helix α3, is mainly hydrophobic and highly conserved in the protein family. A total of 28 amino acids (residues A666 to M693) of the AICD peptide fold on binding to the Fe65-PTB2 domain in an extended conformation built up by a β strand orientated antiparallel to strand β5 and α helices at the N- and C-termini (αN and αC) (Fig 1A,B). The structured C-terminal half of the AICD, therefore, is involved in a unique protein–protein interaction, which is three times larger than known peptide/PTB domain complexes. Independently of our work, an NMR structure has been solved of an AICD-C32/Fe65L1-PTB2 structure from mouse (identities with human proteins: AICD-C32 100%, Fe65-PTB2 54.4%) by Yokoyama and co-workers (Li et al, 2008). Although the structures are very similar overall (root mean square deviation of 1.3 Å for 158 Cα-atoms), in the NMR structure, the C terminus of Fe65L1-PTB2 is truncated (helix α3 is three turns shorter), AICD helix αC is not formed and the NPTY motif is described as a type I β-turn. The reduced interface might explain the decrease in binding affinity as determined by ITC (Kd=0.79 μM).
AICD recognition by Fe65-PTB2
The AICD/Fe65-PTB2 interaction is divided into three parts according to the AICD secondary structure. The conserved N684PTY motif locates to the N terminus of AICD helix αC. Tyrosine Y687 is positioned into the (phospho-)tyrosine binding pocket and, besides the Van-der-Waals interactions, the tyrosine is not coordinated further (Fig 2A). The amphipathic αC helix of the AICD (T686YKFFEQM) is a new feature for peptides interacting with PTB domains, and is stabilized mainly by hydrophobic interactions with helix α3 (Fig 2A). As in other AICD peptide/PTB domain structures the peptides are lacking C-terminal residues (Zhang et al, 1997; Yun et al, 2003), it is not known if helix αC folds in all complexes.
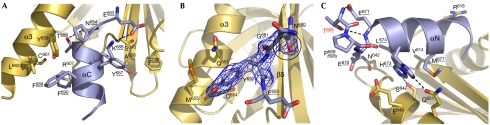
Figure 2.
The AICD/Fe65-PTB2 interface. (A) The interaction of AICD helix αC including the N684PTY sequence with Fe65-PTB2. (B) The central interface including the antiparallel β sheet in trans formed by the AICD Y682E sequence and Fe65-PTB2 strand β5. The hydrophobic binding cavity for Y682 created by helix α3 and the hinge glycine G681 (with encircled peptide bond) linking helix αN and the β-strand of the AICD are highlighted. The final 2mFo-DFc map at a 1.5 σ level is given for G681 and Y682. (C) The complex-regulating interaction of the T668PEE capping box and helix αN of the AICD with Fe65-PTB2. T668 points inside the helix, P669 in trans configuration, and the glutamates are well separated and stabilize the whole arrangement. H673 is shown in its alternative interactions, and L674 and M677 are accommodated in hydrophobic cavities separated by V614. Residues E648 and R616 clamping the helix dipole as shown in Figure 1C are given. AICD, amyloid precursor protein intracellular domain; PTB2, C-terminal phosphotyrosine-binding domain.
Hydrogen bonding of the short β-strand forming part of the AICD (Y682E) occurs exclusively to the backbone of the underlying Fe65 β5 strand, explaining the lower conservation on the Fe65 side (Fig 1D). Tyrosine Y682, which can be phosphorylated in vivo and that is important for the binding of Shc (Src homology 2 domain containing) and Grb2 (Growth-factor receptor bound 2) proteins (Tarr et al, 2002; Russo et al, 2005), lies in a hydrophobic binding pocket created by the α3 helix (Fig 2B). Binding is similar to that seen in the complex with mDab (Yun et al, 2003), but in a different rotamer conformation compared with the complex with the X11-PTB domain (Zhang et al, 1997; supplementary Fig S5 online). The X11 binding mode is excluded in the AICD/Fe65-PTB2 complex, as the space is occupied by the αN helix of the AICD. Lau et al (2000) showed by glutathione S-transferase (GST)-fusion protein binding assays that Fe65 can compete with the binding of X11 to APP, but that the reverse does not occur. Interestingly, short APP peptides corresponding to our 11-mer AICD (Kd value to Fe65-PTB2 of 103 μM), bind to X11 with affinities in the low micromolar range (10-mer: 4.56 μM; 14-mer: 0.32 μM), as determined by Surface Plasmon Resonance (Zhang et al, 1997). Further studies are necessary to understand the various binding properties, and the competition of Fe65 and X11 for APP.
The β-strand-forming part of the AICD is flexibly connected to helix αN through the conserved glycine G681 (Fig 2B), which is essential for the interaction (Cao & Sudhof, 2004). On a structural basis, this constraint clearly originates from sterical reasons, as there is no space for a side chain, and from main chain flexibility, as G681 is found in the otherwise disallowed region of the Ramachandran plot (35°/129°). Interestingly, in the other known complex structures with shorter AICD peptides, X11-PTB (Zhang et al, 1997) and mDab-PTB (Yun et al, 2003), glycine is peptide-flipped and various chain conformations indicate a hinge function (supplementary Fig S5 online).
The most intriguing feature in the AICD structure is helix αN (P669EERHLSKMQQ), which crosses perpendicularly over the N terminus of the α3 helix (Fig 1 and 2C). The helix is fixed through the hydrophobic interactions of L674 and M677, which reach into two adjacent hydrophobic cavities on the Fe65-PTB2 surface and through several hydrogen bonds along the helix. In addition, the helix dipole arising from the aligned peptide bonds in the helix is clamped in between the charges of Fe65 residues R616 and E648 (Fig 1C and 2C). As previously observed by NMR studies, residues T668PEE form a capping box at the N terminus of the helix (Ramelot et al, 2000). The side chain of T668 is hydrogen-bonded to the main chain of the residue ‘i+3' (E671), and P669 is in trans configuration. E671 is tied back by hydrogen bonding to the main-chain nitrogen of T668. The charges of the two consecutive glutamates E670–671 are therefore well separated and the residues stabilize either the interaction with Fe65-PTB2 or the αN helix cap by hydrogen bonding.
The TPEE helix cap regulates complex dissociation
Phosphorylation of T668 was shown to induce cis-isomerization of proline P669, resulting in a destabilization of the helix cap T668PEE (Ramelot & Nicholson, 2001). The importance of isomerization is emphasized by the interaction of the phosphorylated APP C terminus with prolyl isomerase 1 (Pin1), which accelerates isomerization 1,000-fold and directly influences APP processing and Aβ production (Pastorino et al, 2006). To test the importance of residue T668 for Fe65-PTB2 binding and APP C terminus conformation, we solved the AICD/Fe65-PTB2 structures of the T668A and T668E point mutants, which lack a polar side chain or have been thought to mimic a phospho-threonine, respectively (supplementary Table S1 online). The two mutations in the full-length APP have been found previously to impair Fe65 binding both in vitro and in vivo (Ando et al, 2001). In both structures, the helical cap and helix αN are destabilized, as judged from a relative increase of the temperature factors, although P669 seems to remain in trans configuration (hardly visible), and helix αN is still intact and bound to Fe65-PTB2 (data not shown). However, the side chain of E671 loses its hydrogen bond to the main chain of the mutated T668 in both cases and is rotated towards E670, which is still hydrogen-bonded to Fe65-PTB2. ITC measurements with either the wild-type or mutant proteins showed only a slight weakening of the interaction, with Kd values increasing from 0.22±0.02 μM (same for AICD-C50) to 0.34±0.02 μM (Table 1). However, the T668-phosphorylated AICD-C32 construct revealed a more than sevenfold increase of the Kd value to 1.56±0.18 μM, an effect that indicates much larger structural rearrangements and that, under in vivo conditions, will notably change the complex equilibrium. The gain of entropy for both mutants, and even more for phosphorylated AICD-C32 with respect to wild-type protein, is indicative of reduced folding on complex formation. NMR spectral changes observed in Fe65-PTB2 on titration with either the unmodified or the phosphorylated peptide show the largest differences for residues at the N-terminal end of helix α3 in Fe65-PTB2, which corresponds to the region that is in contact with the T668PEE capping box (supplementary Fig S6 online).
Integrating our results with previous data, we propose a ‘molecular switch' model for complex regulation (Fig 3A,B). In the unphosphorylated state, the APP C terminus binds to Fe65-PTB2 with high affinity. The T668PEE capping box stabilizes helix αN (Ramelot et al, 2000) and the APP C terminus/Fe65-PTB2 interaction. Our structures of the T668E and T668A mutants reveal the destruction of the helix capping box resulting in a destabilization of helix αN. On T668 phosphorylation, the helix forms with lower propensity (Ando et al, 2001) and P669 partitions from an all-trans to a 9% cis conformer distribution (Ramelot & Nicholson, 2001). Although the backbone dihedrals of the phosphorylated form in trans conformation have been shown to be similar to the unphosphorylated protein (Ramelot & Nicholson, 2001) and therefore helix αN should be retained, we found a sevenfold reduced binding to Fe65-PTB2, probably owing to steric and electrostatic repulsion of the accumulated negatively charged functional groups. In addition, according to a comparison of our data with the NMR structure of the cis peptide (Ramelot & Nicholson, 2001), we conclude that this conformation binds to Fe65 with much lower affinity. Although the impact of this molecular switch on complex regulation is evident, the consequences of its deregulation on APP processing and Aβ generation, and therefore Alzheimer disease pathogenesis, still need to be validated.
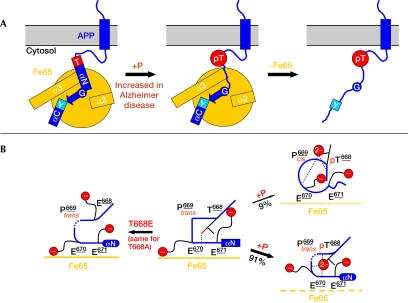
Figure 3.
‘Molecular switch' model of APP/Fe65 complex regulation. (A) Scheme for complex regulation by the phosphorylation of T668. The two-helix capping sequences are labelled by their phosphorylation-amenable residues (T for T668PEE and Y for N684PTY). The hinge glycine G681 is indicated by an encircled letter G. The destruction of the helix cap T by T668 phosphorylation (pT) is shown by the transition from a square to a circle. The destabilization of helix αN is shown by the transition of the box to a simple line. (B) Simplified sketch of the structural rearrangements in the T668PEE helix capping box. The middle panel corresponds to the crystal structure in the unphosphorylated state (edges correspond to the stable cap). The structure of the T668E mutant (left, same for T668A) shows a destabilization of the helix cap (as indicated by round corners and dashed lines). Phosphorylation of T668 (right panels) leads to a partitioning in trans and cis conformers of P669 (Ramelot & Nicholson, 2001) and results in a decrease in Fe65-PTB2 binding affinity. In the trans conformation, Fe65-PTB2 binding is reduced owing to electrostatic repulsion (indicated by dashed lines for Fe65). In the cis conformation the helix cap is destroyed (circle and simple line), resulting in a low binding affinity (dotted lines for Fe65). APP, amyloid precursor protein.
Methods
Structural determination. Expression, purification and crystallization was performed as described previously (Radzimanowski et al, 2008b). The structure of Fe65-PTB2 (UniPROT entry of human Fe65: O00213) in complex with the AICD (UniPROT entry of human APP: P05067-4) was determined by Molecular Replacement using the program PHASER (Read, 2001) and the mDab1-PTB structure (1oqn) as a search model. The model was built using the program Coot (Emsley & Cowtan, 2004) and refinement was carried out with REFMAC5 (Murshudov et al, 1997). The quality of the model was checked using WHAT-IF (Vriend, 1990). Structural figures were generated with PYMOL, GRASP (Nicholls et al, 1991) and CONSURF (Landau et al, 2005). Crystallization of the mutants was performed as described for the wild-type complex.
Isothermal titration calorimetry. Isothermal titration calorimetry experiments were carried out in a buffer containing 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulphonic acid, pH 7.5, and 150 mM NaCl. Binding experiments were performed using a VP-ITC microcalorimeter (MicroCal, Northampton, MA, USA). In a typical experiment (carried out in triplicate), the cell was filled with 20 μM Fe65-PTB2 and 200 μM of the peptide was used as the ligand (titrant) in the syringe. Data processing was performed with Origin 7.0 software.
NMR experiments. NMR experiments are described in the supplementary information online.
Accession codes. Coordinates and structure factors have been deposited in the Protein Data Bank (accession codes 3DXC, 3DXD and 3DXE).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We especially thank P. Soba for his contributions in the initial phase of the project. We acknowledge access to beamlines at the European Synchrotron Radiation Facility (ESRF) in Grenoble (France) and the excellent support by the beamline scientists. We are grateful to S. Kins and S. Ravaud for scientific contribution. This work was financially supported by the Deutsche Forschungsgemeinschaft (DFG) grants WI2649/1-1 and WI2649/1-2 to K.W.
Footnotes
The authors declare to have a patent pending (no. 08 012 257.5) at the European Patent Office (Munich/Germany).
References
- Ando K, Iijima KI, Elliott JI, Kirino Y, Suzuki T (2001) Phosphorylation-dependent regulation of the interaction of amyloid precursor protein with Fe65 affects the production of beta-amyloid. J Biol Chem 276: 40353–40361 [DOI] [PubMed] [Google Scholar]
- Cao X, Sudhof TC (2004) Dissection of amyloid-beta precursor protein-dependent transcriptional transactivation. J Biol Chem 279: 24601–24611 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Galvan V, Chen S, Lu D, Logvinova A, Goldsmith P, Koo EH, Bredesen DE (2002) Caspase cleavage of members of the amyloid precursor family of proteins. J Neurochem 82: 283–294 [DOI] [PubMed] [Google Scholar]
- Guenette S, Chang Y, Hiesberger T, Richardson JA, Eckman CB, Eckman EA, Hammer RE, Herz J (2006) Essential roles for the FE65 amyloid precursor protein-interacting proteins in brain development. EMBO J 25: 420–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C (2004) Take five—BACE and the gamma-secretase quartet conduct Alzheimer's amyloid beta-peptide generation. EMBO J 23: 483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N (2005) ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 33: W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau KF, McLoughlin DM, Standen CL, Irving NG, Miller CC (2000) Fe65 and X11beta co-localize with and compete for binding to the amyloid precursor protein. Neuroreport 11: 3607–3610 [DOI] [PubMed] [Google Scholar]
- Lee MS, Kao SC, Lemere CA, Xia W, Tseng HC, Zhou Y, Neve R, Ahlijanian MK, Tsai LH (2003) APP processing is regulated by cytoplasmic phosphorylation. J Cell Biol 163: 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H et al. (2008) Structure of the C-terminal PID domain of Fe65L1 complexed with the cytoplasmic tail of APP reveals a novel peptide binding mode. J Biol Chem, doi:10.1074/jbc.M803892200 [DOI] [PubMed] [Google Scholar]
- Mattson MP (2004) Pathways towards and away from Alzheimer's disease. Nature 430: 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin DM, Miller CC (2008) The FE65 proteins and Alzheimer's disease. J Neurosci Res 86: 744–754 [DOI] [PubMed] [Google Scholar]
- Meiyappan M, Birrane G, Ladias JA (2007) Structural basis for polyproline recognition by the FE65 WW domain. J Mol Biol 372: 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Pastorino L, Lu KP (2006) Pathogenic mechanisms in Alzheimer's disease. Eur J Pharmacol 545: 29–38 [DOI] [PubMed] [Google Scholar]
- Pastorino L et al. (2006) The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature 440: 528–534 [DOI] [PubMed] [Google Scholar]
- Radzimanowski R, Ravaud S, Schlesinger S, Koch J, Beyreuther K, Sinning I, Wild K (2008a) Crystal structure of the human Fe65-PTB1 domain. J Biol Chem 283: 23113–23120 [DOI] [PubMed] [Google Scholar]
- Radzimanowski R, Beyreuther K, Sinning I, Wild K (2008b) Overproduction, purification, crystallization and preliminary X-ray analysis of human Fe65-PTB2 in complex with the amyloid precursor protein intracellular domain. Acta Crystallogr F 64: 409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelot TA, Nicholson LK (2001) Phosphorylation-induced structural changes in the amyloid precursor protein cytoplasmic tail detected by NMR. J Mol Biol 307: 871–884 [DOI] [PubMed] [Google Scholar]
- Ramelot TA, Gentile LN, Nicholson LK (2000) Transient structure of the amyloid precursor protein cytoplasmic tail indicates preordering of structure for binding to cytosolic factors. Biochemistry 39: 2714–2725 [DOI] [PubMed] [Google Scholar]
- Read RJ (2001) Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D 57: 1373–1382 [DOI] [PubMed] [Google Scholar]
- Russo C, Venezia V, Repetto E, Nizzari M, Violani E, Carlo P, Schettini G (2005) The amyloid precursor protein and its network of interacting proteins: physiological and pathological implications. Brain Res Brain Res Rev 48: 257–264 [DOI] [PubMed] [Google Scholar]
- Tarr PE, Roncarati R, Pelicci G, Pelicci PG, D'Adamio L (2002) Tyrosine phosphorylation of the beta-amyloid precursor protein cytoplasmic tail promotes interaction with Shc. J Biol Chem 277: 16798–16804 [DOI] [PubMed] [Google Scholar]
- Vriend G (1990) WHAT IF: a molecular modeling and drug design program. J Mol Graph 8: 52–56, 29 [DOI] [PubMed] [Google Scholar]
- Weidemann A, Eggert S, Reinhard FB, Vogel M, Paliga K, Baier G, Masters CL, Beyreuther K, Evin G (2002) A novel epsilon-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with Notch processing. Biochemistry 41: 2825–2835 [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Guenette SY (2007) APP at a glance. J Cell Sci 120: 3157–3161 [DOI] [PubMed] [Google Scholar]
- Yun M, Keshvara L, Park CG, Zhang YM, Dickerson JB, Zheng J, Rock CO, Curran T, Park HW (2003) Crystal structures of the Dab homology domains of mouse disabled 1 and 2. J Biol Chem 278: 36572–36581 [DOI] [PubMed] [Google Scholar]
- Zambrano N, Bimonte M, Arbucci S, Gianni D, Russo T, Bazzicalupo P (2002) feh-1 and apl-1, the Caenorhabditis elegans orthologues of mammalian Fe65 and beta-amyloid precursor protein genes, are involved in the same pathway that controls nematode pharyngeal pumping. J Cell Sci 115: 1411–1422 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lee CH, Mandiyan V, Borg JP, Margolis B, Schlessinger J, Kuriyan J (1997) Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain. EMBO J 16: 6141–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information