Summary
In this paper, we discuss recent advances in our knowledge of the dengue virus life cycle based on new structural data of the virus and its proteins. Specifically, we focus on the structure of the pre-membrane protein, prM and its role in virus assembly, the first full-length structure of a multi-domain dengue virus replication protein, NS3, and the recently solved structures of NS5 methyltransferase and polymerase domains. These structures provide a basis for describing function and predicting putative host interactions.
Introduction
Dengue fever (DF) and dengue hemorrhagic fever (DHF)/dengue shock syndrome (DSS) have become a global health burden in recent decades affecting more than 50 million people a year in southeast Asia, the western Pacific, Central and South America. Dengue virus (DENV), the causative agent of these diseases is continuing to spread and has emerged as a significant health threat throughout the world [1,2]. Currently, there is no vaccine or effective antiviral against the virus. However, significant effort has been put forth towards developing vaccines and antivirals by increasing our understanding of the viral life cycle, the interaction of the virus with the host, and identifying factors important for transmission of the virus. At the forefront of these studies are structural analyses of the virus and viral proteins that have provided valuable insight into the mechanism of DENV pathogenesis.
DENV is a member of the Flaviviridae family and is grouped within the flavivirus genus together with other pathogenic viruses including West Nile virus (WNV), Japanese encephalitis virus (JEV), tick-borne encephalitis virus (TBEV) and yellow fever virus (YFV) [3]. The viral genome consists of a positive-sense RNA of ~11kb. This RNA encodes 3 structural proteins (C, prM and E) that form the components of the virion, and 7 non-structural proteins (NS1, NS2A/B, NS3, NS4A/B, NS5) involved in viral RNA replication (Figure 1). The atomic structures for many of these viral proteins have been solved. In this review, we will highlight recent advances in structural studies on DENV and its proteins. We will discuss the relevance of these studies with regard to the functional mechanisms of the viral proteins, their interactions with host proteins, and their role in the viral life cycle.
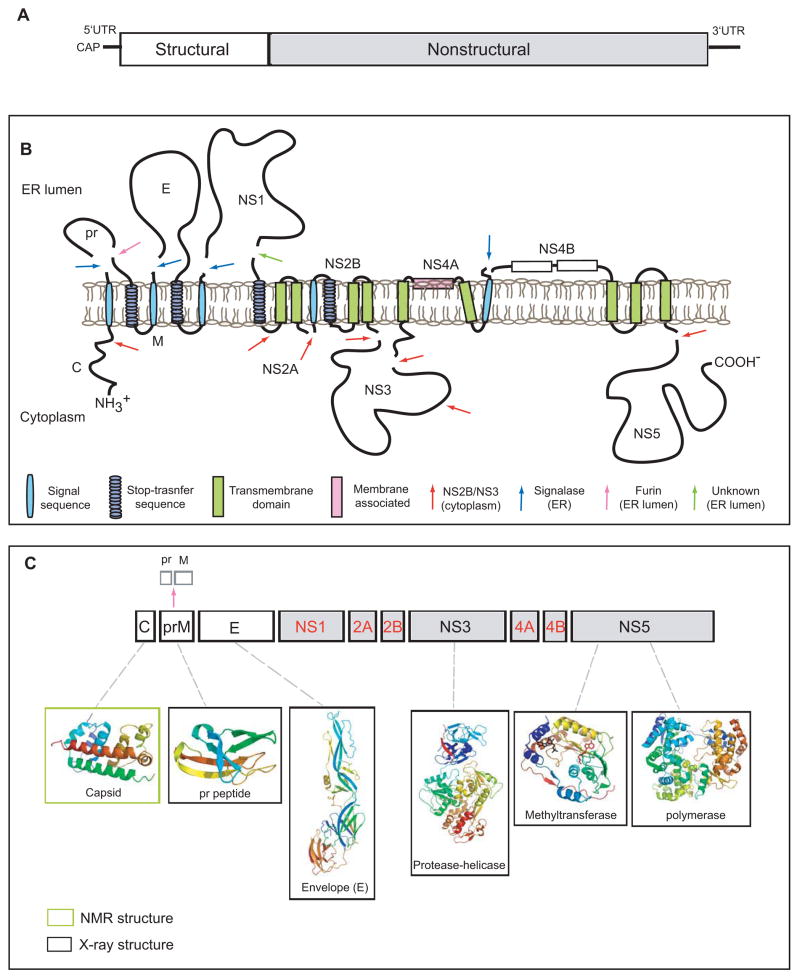
Figure 1. Schematic diagram of the dengue virus genome and polyprotein.
A. The viral genome is a positive sense RNA of ~11kb in length. It is capped at the 5′ end but lacks a poly-(A) at its 3′-end. The structural proteins (open boxes) are encoded at the 5′ one third of genome followed by the non-structural proteins (grey boxes). B. Membrane topology of the polyprotein. The viral RNA is translated as a polyprotein and processed by cellular and viral proteases (denoted by arrows). The structural proteins include capsid (C), membrane protein (prM/M) and envelope (E). prM and E are released from the polyprotein by signalase cleavage in the ER, but remain anchored on the luminal side of the membrane. The C is also anchored in the ER membrane (on the cytoplasmic side) by a conserved hydrophobic signal sequence at its C-terminal end. This signal sequence is cleaved by the viral NS2B-NS3 protease. During virus maturation, prM is further cleaved by furin in the TGN into the pr peptide and M protein. The non-structural proteins are processed mainly by the viral protease NS2B-NS3 in the cytoplasm with the exception of NS1, which is released from NS2A by a yet unidentified protease in the lumen of the ER. NS2A/2B and NS4A/4B are anchored in the ER as transmembrane proteins. The topology of NS4A and NS4B are predicted through biochemical and cellular analyses [62,67]. C. Structural proteome of dengue virus. NMR and X-ray structures are shown for C, prM, E, NS3 (full-length) and the NS5 methyltransferase and polymerase domains (PDB identifiers: 1R6R, 3C5X, 1OKE, 2VBC, 2P1D, 2J7U, respectively)[4, 5–7, 12, 13, 18, 32, 36]. Structures are currently not available for the proteins denoted in red.
Virus assembly and maturation
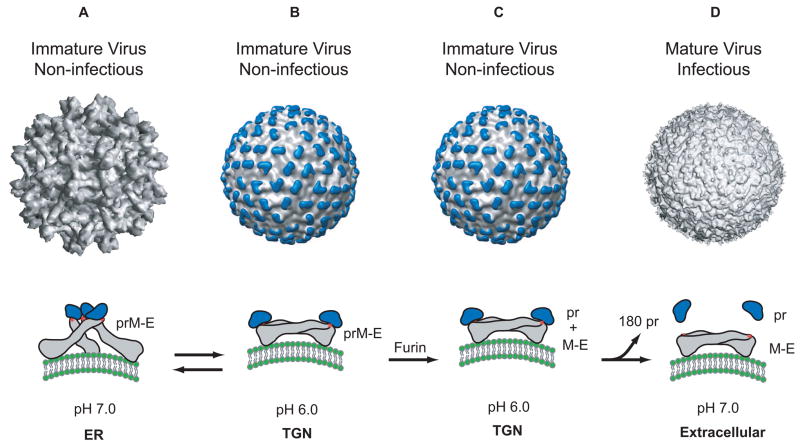
The structure of DENV was previously solved through a combination of cryo-electron microscopy and X-ray crystallography [4–13]. Recently, the structures of intermediates in the assembly process have also been obtained and these provide insight into the assembly and maturation process of DENV [13,14]. In supernatants of infected cells, the virus is found either as a mature or immature particle with a diameter of about 50 nm and 60 nm, respectively. Both particles consist of an outer glycoprotein shell and an internal host derived lipid bilayer. Within this bilayer is an RNA-protein core consisting of genome RNA and capsid proteins (C). The glycoprotein shell is well defined and consists of 180 copies each of an envelope (E) and membrane protein (prM/M). These two proteins have different conformations in the immature and mature DENV particles and therefore, confer unique structural features to both forms of particles. In the immature virion, prM and E form 90 heterodimers that extend as 60 trimeric spikes from the particle surface (figure 2A). In the mature virion, E is found as 90 homodimers that lie flat against the viral surface forming a ‘smooth’ protein shell (figure 2D). The ‘pr’ peptide is cleaved from prM during maturation and M remains in the mature particle as a transmembrane protein beneath the E protein shell. The structural transitions from immature (‘spiky’) to mature (‘smooth’) morphology (described in figure 2) occur while in transit through the Trans-Golgi Network (TGN) and are driven predominantly by conformational changes in the E protein [5,6,12]. These conformational changes in E are triggered by low pH (~5.8–6.0) and occur prior to the maturation cleavage of prM by a host encoded furin protease (figure 2B) [14]. It was demonstrated that these conformational changes were reversible (based on pH). This suggested that the immature particle could exist reversibly in either ‘spiky’ or ‘smooth’ forms depending on the pH of the cellular environment. This is a remarkable observation considering that such reversible conformational changes require the E protein to toggle between forming heterodimeric interactions with prM in spikes that projects vertically outwards from the particle to forming homodimeric structures that lie flat against the viral surface. The molecular reorganization required for these changes are extensive, the details of which still remain illusive. Following maturation however, the mature particle cannot revert back to its immature morphology. Once cleaved, the pr peptide remains associated with E (figure 2C) until the mature particle is released into the neutral pH of the extracellular environment. Therefore, it was proposed that the pr peptide functioned as a cap-like structure that protected the fusion peptide on E from undergoing premature fusion prior to virus release [14].
Figure 2.
Structure of the dengue virion and conformations of the E protein. A. The cryo-EM reconstruction of the immature virion at neutral pH [12]. In this structure, the E protein exists as a heterodimer with prM, and these heterodimers form 60 trimeric spikes that extend away from the surface of the virus. This arrangement of E gives the virus a ‘spiky’ morphology and represents the initial particle that buds into the ER. The conformation of the E protein (grey) within a spike is shown below the virion. The ‘pr’ peptide is shown in blue protecting the fusion peptide on E (shown as a red star). B. The cryo-EM reconstruction of the immature virion at low pH [14]. During its transit through the secretory pathway, the virus encounters low pH in the TGN. Under these conditions, the prM-E heterodimers dissociate from their trimeric spike-like organization and form 90 dimers that lie flat against the viral surface. This orientation of prM-E proteins gives the virion a ‘smooth’ morphology. Upon raising the pH (reverse reaction), this ‘smooth’ particle can revert back to its ‘spiky’ morphology. C. While in the TGN, the prM protein is cleaved into its ‘pr’ peptide and M protein by the host endoprotease, furin. The cleaved ‘pr’ peptide maintains its position as a ‘cap’ on E and the E proteins remain as 90 homodimers lying parallel to the virion surface. M, not shown in this figure lies embedded in the viral membrane beneath the E protein shell. D. The cryo-EM reconstruction of the mature virion [10]. Following furin cleavage, the mature virion is secreted into the extracellular milleu and the pr peptide is released from mature particle.
Recently, the atomic structures of prM were solved at pH 5.5 (2.2Å) and 7.0 (2.6Å) [13]. Both structures were similar indicating that pH did not affect the tertiary structure of prM. The construct consisted of a prM-E fusion protein where the prM protein was fused to the ectodomain of E. The transmembrane domain of prM was replaced by a linker. The furin cleavage site was also mutated. The resulting structure of prM has a unique fold and consists of 7 antiparallel β-strands, stabilized with three S-S bonds. The glycosylation modification at asparagine 69 was also observed. The structure of E in this construct was similar to its pre-fusion dimeric conformation [12]. As predicted previously, the pr peptide was positioned as a 'cap' on E protecting the hydrophobic fusion loop [13]. These recent studies on the immature virions and proteins have provided new details of DENV assembly and maturation. They have also opened new avenues that can be explored toward understanding the molecular mechanism behind E protein dynamics and the role of the immature particles in the virus life cycle.
The E protein also provides the first point of contact between the virus and the host cell. Several cellular proteins and carbohydrate molecules that function as attachment factors mediating viral entry have been identified, and these molecules have been shown to interact with the E protein [15–21]. These attachment factors assist in concentrating the virus on the cell surface increasing its access to specific cellular receptor/s. Structural insight into the interaction of E with one of these attachment factors, dendritic-cell-specific ICAM3-grabbing non-integrin (DC-SIGN) [22] has been obtained. Unfortunately, a specific cellular receptor for dengue has not yet been identified. It is possible that currently identified attachment molecules could function as specific cellular receptors for dengue. However, it would be necessary to demonstrate that they mediate clathrin-mediated endocyotsis of the particle and this has not been shown for any of these putative receptors to date.
Viral RNA replication
Replication of the viral genome primarily occurs in the cytoplasm of infected cells. Initially, the incoming viral RNA is translated into a polyprotein, which is then directed to the endoplasmic reticulum (ER). Signal sequences within the polyprotein translocate NS1 and the ectodomains of prM and E into the lumen of the ER while the C, NS3 and NS5 proteins are localized to the cytoplasm. NS2A/B and NS4A/B remain predominantly as transmembrane proteins. Processing of this polyprotein is a fundamental process that must occur before viral RNA replication can proceed (figure 1). This task is carried out by host signalases that reside in the lumen of the ER and the viral NS3 protein and its co-factor, NS2B that reside in the cytoplasm [23–25]. Structural insights into the viral replication proteins have been limited to NS2B/NS3 and NS5. However, a number of ultrastructural studies have also been done that demonstrate substantial re-arrangements of internal membranes permitting facile virus RNA synthesis and assembly [26]. The NS4A protein has been implicated in this alteration of intracellular host membranes but the mechanism by which this occurs is unknown [27].
NS3 protease-helicase
NS3 is a multifunctional protein of 618 amino acids that functions both as a chymotrypsin like serine protease as well as an RNA helicase and RTPase/NTPase. The protease domain is N-terminal in NS3 (residues 1–180) and cleaves the viral polyprotein at several sites as depicted in figure 1. The enzyme consists of 6 β-strands that form two β-barrels with the catalytic triad (His-51, Asp-75 and Ser-135) sandwiched between them. Activity of the protease is critically dependent upon the presence of its co-factor, NS2B which is conserved among the flaviviruses [24,28,29]. Recently, the structure of the full-length NS3 of DENV-4 was solved to 3.15Å resolution (figure 3) [30]. This structure suggests that NS3 is an extended molecule with the protease domain spatially oriented on top of subdomains I and II of the helicase. The NS3 protein included residues 49–66 from the NS2B which were linked to the N-terminus of full-length NS3 by a Gly-Ser linker. However, its protease domain was inactive but retains the same active site conformation and overall fold as an active form of the DENV-2 protease domain that included an extended region of NS2B (residues 40–80) [31]. Analysis of these two structures as well as the structure of the substrate-bound form of the WNV protease domain [32] indicated that residues 67–80 of NS2B are critically important for protease activity of NS3. This is due to NS2B wrapping around the protease domain as a 'belt-like' structure and forming an integral part of the protease active site. While the central region of NS2B (residues 67–80) interacts with the protease, flanking hydrophobic regions of NS2B are predicted to anchor the NS2B-NS3 complex in the ER membrane [33]. This geometry places the protease active site close to predicted transmembrane domains that must be cleaved within the polyprotein. The consensus cleavage site of NS3 requires a dibasic (Arg/Lys)-Arg motif at the P2 and P1 positions respectively and a small amino acid (Gly) at the P1′ position. The substrate specificity as well as the cis/trans activity of many of the flavivirus proteases have also recently been characterized [34,35]. These studies provided evidence of a required cis-cleavage of the NS3 helicase domain at a site hidden from exposure to the protease domain. This indicated that the NS3 is structurally dynamic and may indeed require an extended conformation (as observed in this recent structure) for its function. Such a conformation of NS3 could explain the ability of the NS2B-NS3 protease to function on sites not readily accessible to it. These same studies have also suggested that the cis-trans activity of the protease may play an important role in controlling the dynamics of viral protein translation versus RNA replication by controlling the availability of viral proteins.
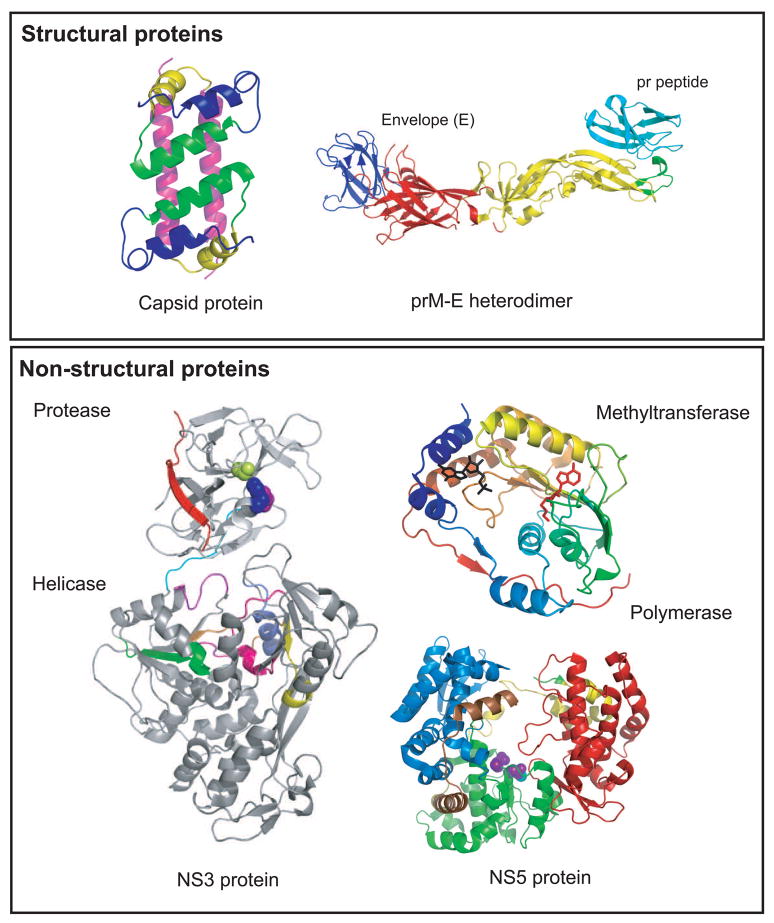
Figure 3.
Structures of dengue virus proteins. Top panel (structural proteins): The NMR structure of the capsid protein dimer (1R6R) is shown on the left with helix 1, blue; helix 2, green, helix 3, yellow and helix 4, pink [4]. The X-ray structure of the prM-E protein is shown on the right [13]. The three domains of the E protein are shown: domain I, red; domain II, yellow; domain III, blue. prM is shown in cyan forming a cap protecting the fusion peptide (green) on E. Bottom panel (non-structural proteins): the X-ray structure of the full-length NS3 protein (2VBC) is shown on the left [18]. The protease domain (residues 1–168) is N-terminally located in the molecule. The catalytic triad is shown in blue (His 51), purple (Asp 75) and limon (Ser 135). NS2B is shown in red. The linker between the protease and helicase domains (residues 169–179) is shown in cyan. The helicase domain forms the C-terminal two-thirds of the molecule (residues 180–618). The active site motifs are colored: motif I (196–202), purple; motif II (284–287), magenta; motif III (312–324), orange; motif IV (359–367), yellow; motif V (407–420), pink; motif VI (452–463), slate blue. NS5 is shown on the right. The methytransferase (2P1D) is shown colored in rainbow from N-terminal to the C-terminal end [32]. The CAP structure is shown in black and the by-product of the methylation reaction, S-adenosyl-homocysteine (SAH) is shown in red. The polymerase of DENV-3 (2J7U) is shown with the fingers domain (273–315; 416–496; 543–600) in blue, palm domain (497–542; 601–705) in green, and thumb domain (706–900) in red. The nuclear localization signal (NLS) is shown in yellow (b NLS, 320–368) and brown (a/b NLS, 369–405).
The helicase domain of NS3 (residues 180–618) has seven structural motifs reminiscent of superfamily 2 helicases [36]. It has three subdomains with significant sequence identity and structural similarity to other flavivirus helicases [30,37–39]. Subdomains I and II are also structurally similar to the corresponding domains in hepatitis C virus suggesting a common functional mechanism [40,41], however, the fold of subdomain III is unique to the flaviviruses, and may be a site for protein binding. Both subdomains I (residues 181–326) and II (residues 327–481) are composed of a central six-stranded parallel β-sheet, which is flanked by 4 α-helices. Subdomain III (residues 482–618) has 4 approximately parallel α-helices surrounded by three shorter α-helices and two solvent-exposed antiparallel β-strands. In the recent structure of full-length NS3 [30], the helicase domain adopts a similar structure to the isolated DENV helicase [37], with the exception that several regions previously disordered in the isolated domain structure are now visible. However, the helicase activity of the full-length NS3 protein is ~30-fold higher than the isolated domain [37] indicating that the protease domain may influence the enzymatic activities of the helicase. The protease and helicase are linked by an interdomain linker that shares little conservation between the flaviviruses, but plays an important role in the association between the two domains in NS3 (the buried surface area between the domains is ~1380Å2 and ~380Å2 with and without the linker, respectively). The helicase domain of NS3 is also implicated in interacting with the polymerase, NS5 [42] and in WNV implicated in virulence [43]. Recently, a residue in the NS3 helicase domain (W349) was shown to be involved in virus assembly suggesting that NS3 plays an additional role in the life cycle following viral RNA replication [44]. The NS3 protein has also been implicated in inducing apoptosis in infected cells [45]. Overall, the full-length structure of NS3 represents a significant milestone in flavivirus biology as crystallization of full-length viral proteins has long been a challenge.
NS5 methyltransferase-polymerase
The largest (900 residues, 104kDa) and the most conserved protein in DENV is NS5 (67% sequence identity among DENV serotypes 1–4). It is also a bifunctional enzyme with a methyltransferase domain (MTase; residues 1–296) at its N-terminal end and a RNA-dependent RNA polymerase (RdRp; residues 320–900) at its C-terminal end. The structure of the MTase domain was previously solved for DENV [46] and recently solved for WNV [47]. Both structures have an S-adenosyl-methionine-dependent MTase core structure that folds into a α/β/β sandwich cradled between N-and C-terminal subdomains (figure 3). Overall, the two MTase structures are very similar and this domain has recently been shown to sequentially catalyze both guanine N-7 and ribose 2′-O-methylation [46,48]. The primary difference between the two structures is observed in the SAM-binding site and the cap-binding site, with the former having a more open conformation in DENV versus WNV, and the latter being more open in WNV [46,47]. It is suggested that the differences in the SAM-binding site may reflect two distinct states of the enzyme, with the closed conformation (in WNV) representing tight SAM-binding, and the open state representing the release of the by-product of the methylation reaction, S-adenosyl-homocysteine (SAH). Both enzymes have a highly positively charged surface at the GTP and SAM binding sites to accommodate capped RNA substrates.
The crystal structures for both DENV [49] and WNV [50]polymerases have been solved. The polymerase domain of NS5 assumes a structure similar to other RdRp molecules, and is composed of a canonical right hand conformation with palm, fingers, thumb subdomains (reviewed in [51]). It also shares a common catalytic mechanism for the incorporation of nucleotides utilizing two metal ions coordinated by structurally conserved aspartic acid residues (also known as the GDD motif). The RdRps differ from DNA-dependent RNA polymerases by the existence of the 'finger tips' that connect the fingers and thumb subdomains to create a fully encircled active site. The DENV RdRp displays a more flexible fingers subdomain and therefore through a rotation of ~8° away from the thumb subdomain, forms a more ‘open’ conformation compared to WNV [49,50]. Interestingly, the flavivirus RdRps have a nuclear localization signal between residues 320–405. This NLS region in NS5 was previously thought to be a flexible interdomain linker, but is now known to be a well-defined structural component of the RdRp [49,50,52,53]. Specifically, residues 320–368 are strictly conserved among the flaviviruses and bind β-importin. These residues are also implicated in interacting with NS3 [42,52]. In DENV infections, the NS5 protein is primarily localized within the nucleus. However, not all flavivirus RdRps localize to the nucleus. The rationalization for a viral RdRp localizing to the nucleus when its actual enzymatic functions in the virus life cycle are required in the cytoplasm is currently unknown but is actively being investigated [54]. However, these observations do suggest that apart from its enzymatic functions, NS5 may also engage in virus-host interactions and actively interact with the host environment.
Other viral non-structural proteins
Unfortunately, there is no structural information available for viral proteins NS1, NS2A and NS4A/4B. NS1 is a 45kDa glycoprotein that is translocated into the lumen of the ER and secreted from the cell [55–57]. It is implicated in functions within the viral RNA replication complex [58–60] as well as in viral defense through inhibition of complement activation [61]. Although the protein forms stable oligomers (dimers and hexamers) in solution, structural analyses of this protein and has been quite challenging [62,63]. This is also true for the hydrophobic proteins NS2A and NS4A/4B. NS2A is a ~22kDa protein that is implicated to form part of the replication complex [64]. As a result of an internal NS2B-NS3 cleavage, two forms are observed, NS2A and NS2Aα Both forms are important for virus production [65]. NS4A (16kDa) and NS4B (27kDa) are integral membrane proteins. NS4A is proposed to induce membrane alterations important for virus replication [27,66]. NS4B is implicated in assisting viral RNA replication through its direct interaction with NS3 [67]. It is also suggested to block IFN α/β-induced signal transduction [68,69]. Structural analayses of these three proteins have been unsuccessful since they all possess multiple transmembrane hydrophobic segments. However, the membrane topology of NS4A and NS4B has been predicted through biochemical analyses (figure 1B) [27,70].
Concluding remarks
Advances have been recently made towards obtaining structural insight into the function of many DENV proteins. Although this has been useful, the multi-functional nature of these proteins requires additional information to understand how these proteins interact to form active replication and assembly complexes. Parallel work on identifying viral and host protein interactions will be essential in developing a complete molecular description of the viral life cycle.
Acknowledgments
We thank Anita Robinson and Jennifer Yoder for assistance. We acknowledge support from the Public Health Service Program Project Grant (AI055672) from the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest within the annual period of review have been highlighted as:
•of special interest
•• of outstanding interest
- 1.Morens DM, Fauci AS. Dengue and hemorrhagic fever: a potential threat to public health in the United States. Jama. 2008;299:214–216. doi: 10.1001/jama.2007.31-a. [DOI] [PubMed] [Google Scholar]
- 2.Gould EA, Solomon T. Pathogenic flaviviruses. Lancet. 2008;371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn RJ. The Flaviviruses. In: Acheson NH, editor. Fundamentals of Molecular Virology. 2004. in press. [Google Scholar]
- 4.Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proc Natl Acad Sci U S A. 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the Dengue virus envelope protein after fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 7.Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. Journal of Virology. 2005;79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the Flavivirus life cycle. Nat Rev Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn RJ. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Corver J, Chipman PR, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG. Structures of immature flavivirus particles. J EMBO. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Lok S-M, Yu I-M, Zhang Y, Kuhn R, Chen J, Rossmann MG. Structure of the flavivirus precursor membrane-envelope protein complex and its implication for maturation. Science. 2008 doi: 10.1126/science.1153263. submitted. [DOI] [PubMed] [Google Scholar]
- 14.Yu I-M, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn R, Rossmann MG, Chen J. Structure of immature dengue virus at low pH primes proteolytic maturation. Science. 2008 doi: 10.1126/science.1153264. submitted. [DOI] [PubMed] [Google Scholar]
- 15.Navarro-Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, Virelizier JL, Arenzana-Seisdedos F, Despres P. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 2003;4:723–728. doi: 10.1038/sj.embor.embor866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL, et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. 2003;197:823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren J, Ding T, Zhang W, Song J, Ma W. Does Japanese encephalitis virus share the same cellular receptor with other mosquito-borne flaviviruses on the C6/36 mosquito cells? Virol J. 2007;4:83. doi: 10.1186/1743-422X-4-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnan MN, Sukumaran B, Pal U, Agaisse H, Murray JL, Hodge TW, Fikrig E. Rab 5 is required for the cellular entry of dengue and West Nile viruses. J Virol. 2007;81:4881–4885. doi: 10.1128/JVI.02210-06. Epub 2007 Feb 4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jindadamrongwech S, Thepparit C, Smith DR. Identification of GRP 78 (BiP) as a liver cell expressed receptor element for dengue virus serotype 2. Arch Virol. 2004;149:915–927. doi: 10.1007/s00705-003-0263-x. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Chiou SS, Chen WJ. Differential binding efficiency between the envelope protein of Japanese encephalitis virus variants and heparan sulfate on the cell surface. J Med Virol. 2004;72:618–624. doi: 10.1002/jmv.20025. [DOI] [PubMed] [Google Scholar]
- 21.Miller JL, Dewet BJ, Martinez-Pomares L, Radcliffe CM, Dwek RA, Rudd PM, Gordon S. The Mannose Receptor Mediates Dengue Virus Infection of Macrophages. PLoS Pathog. 2008;4:e17. doi: 10.1371/journal.ppat.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson A, Kuhn RJ, Rossmann MG. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell. 2006;124:485–493. doi: 10.1016/j.cell.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 23.Falgout B, Markoff L. Evidence that flavivirus NS1-NS2A cleavage is mediated by a membrane-bound host protease in the endoplasmic reticulum. J Virol. 1995;69:7232–7243. doi: 10.1128/jvi.69.11.7232-7243.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falgout B, Pethel M, Zhang Y-M, Lai C-J. Both nonstructural proteins NS2B anbd NS3 are required for the proteolytic processing of Dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chambers TJ, McCourt DW, Rice CM. Production of yellow fever virus proteins in infected cells: identification of discrete polyprotein species and analysis of cleavage kinetics using region-specific polyclonal antisera. Virology. 1990;177:159–174. doi: 10.1016/0042-6822(90)90470-c. [DOI] [PubMed] [Google Scholar]
- 26.Mackenzie JM, Jones MK, Westaway EG. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. Journal of Virology. 1999;73:9555–9567. doi: 10.1128/jvi.73.11.9555-9567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem. 2007;282:8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 28.Chambers TJ, Nestorowicz A, Amberg SM, Rice CM. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993;67:6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu CF, Wang SH, Sun CM, Hu ST, Syu WJ. Activation of dengue protease autocleavage at the NS2B-NS3 junction by recombinant NS3 and GST-NS2B fusion proteins. J Virol Methods. 2003;114:45–54. doi: 10.1016/j.jviromet.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Luo D, Xu T, Hunke C, Gruber G, Vasudevan SG, Lescar J. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 32.Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16:795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clum S, Ebner KE, Padmanbhan R. Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J Biol Chem. 1997;272:30715–30723. doi: 10.1074/jbc.272.49.30715. [DOI] [PubMed] [Google Scholar]
- 34.Bera AK, Kuhn RJ, Smith JL. Functional characterization of cis and trans protease activity of the flavivirus NS2B/NS3 protease. J Biol Chem. 2007;282:12883–12892. doi: 10.1074/jbc.M611318200. [DOI] [PubMed] [Google Scholar]
- 35.Shiryaev SA, Ratnikov BI, Aleshin AE, Kozlov IA, Nelson NA, Lebl M, Smith JW, Liddington RC, Strongin AY. Switching the substrate specificity of the two-component NS2B-NS3 flavivirus proteinase by structure-based mutagenesis. J Virol. 2007;81:4501–4509. doi: 10.1128/JVI.02719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorbalenya AE, Koonin EV. Helicases: Amino acid sequencecomparisons and structure-function relationships. Current Opinions in Structural Biology. 1993;34:419–429. [Google Scholar]
- 37.Wu J, Bera K, Kuhn RJ, Smith JL. Structure of the flavivirus helicase: implications for catalytic activity, protein interactions, and proteolytic processing. J Virol. 2005;79:10268–10277. doi: 10.1128/JVI.79.16.10268-10277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita T, Unno H, Mori Y, Tani H, Moriishi K, Takamizawa A, Agoh M, Tsukihara T, Matsuura Y. Crystal structure of the catalytic domain of Japanese encephalitis virus NS3 helicase/nucleoside triphosphatase at a resolution of 1.8 A. Virology. 2008;373:426–436. doi: 10.1016/j.virol.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 39.Xu T, Sampath A, Chao A, Wen D, Nanao M, Chene P, Vasudevan SG, Lescar J. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J Virol. 2005;79:10278–10288. doi: 10.1128/JVI.79.16.10278-10288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao N, Hesson T, Cable M, Hong Z, Kwong AD, Le HV, Weber PC. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 41.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 42.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 43.Brault AC, Huang CY, Langevin SA, Kinney RM, Bowen RA, Ramey WN, Panella NA, Holmes EC, Powers AM, Miller BR. A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat Genet. 2007;39:1162–1166. doi: 10.1038/ng2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patkar CG, Kuhn RJ. Yellow Fever Virus NS3 Plays an Essential Role in Virus Assembly Independent of Its Known Enzymatic Functions. J Virol. 2008;82:3342–3352. doi: 10.1128/JVI.02447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramanathan MP, Chambers JA, Pankhong P, Chattergoon M, Attatippaholkun W, Dang K, Shah N, Weiner DB. Host cell killing by the West Nile Virus NS2B-NS3 proteolytic complex: NS3 alone is sufficient to recruit caspase-8-based apoptotic pathway. Virology. 2006;345:56–72. doi: 10.1016/j.virol.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 46.Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. Embo J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol. 2007;81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malet H, Egloff MP, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, et al. Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J Biol Chem. 2007;282:10678–10689. doi: 10.1074/jbc.M607273200. [DOI] [PubMed] [Google Scholar]
- 51.Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol. 2006;16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Johansson M, Brookes AJ, Jans DA, Vasudevan SG. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-b and the viral helicase, NS3. J Gen Virol. 2001;82:735–745. doi: 10.1099/0022-1317-82-4-735. [DOI] [PubMed] [Google Scholar]
- 53.Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, Vasudevan SG. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin b1 and importin a/b-recognised nuclear localisation signals (NLSs) J Biol Chem. 2002;277:36399–36407. doi: 10.1074/jbc.M204977200. [DOI] [PubMed] [Google Scholar]
- 54.Pryor MJ, Rawlinson SM, Butcher RE, Barton CL, Waterhouse TA, Vasudevan SG, Bardin PG, Wright PJ, Jans DA, Davidson AD. Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection. Traffic. 2007;8:795–807. doi: 10.1111/j.1600-0854.2007.00579.x. Epub 2007 May 2030. [DOI] [PubMed] [Google Scholar]
- 55.Mason PW. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology. 1989;169:354–364. doi: 10.1016/0042-6822(89)90161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlesinger JJ, Brandriss MW, Putnak JR, Walsh EE. Cell surface expression of yellow fever virus non-structural glycoprotein NS1: consequences of interaction with antibody. J Gen Virol. 1990;71(Pt 3):593–599. doi: 10.1099/0022-1317-71-3-593. [DOI] [PubMed] [Google Scholar]
- 57.Noisakran S, Dechtawewat T, Rinkaewkan P, Puttikhunt C, Kanjanahaluethai A, Kasinrerk W, Sittisombut N, Malasit P. Characterization of dengue virus NS1 stably expressed in 293T cell lines. J Virol Methods. 2007;142:67–80. doi: 10.1016/j.jviromet.2007.01.008. Epub 2007 Feb 2028. [DOI] [PubMed] [Google Scholar]
- 58.Westaway EG, Khromykh AA, Mackenzie JM. Nascent flavivirus RNA colocalized in situ with double-stranded RNA in stable replication complexes. Virology. 1999;258:108–117. doi: 10.1006/viro.1999.9683. [DOI] [PubMed] [Google Scholar]
- 59.Lindenbach BD, Rice CM. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindenbach BD, Rice CM. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999;73:4611–4621. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chung KM, Nybakken GE, Thompson BS, Engle MJ, Marri A, Fremont DH, Diamond MS. Antibodies against West Nile Virus nonstructural protein NS1 prevent lethal infection through Fc gamma receptor-dependent and -independent mechanisms. J Virol. 2006;80:1340–1351. doi: 10.1128/JVI.80.3.1340-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flamand M, Megret F, Mathieu M, Lepault J, Rey FA, Deubel V. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J Virol. 1999;73:6104–6110. doi: 10.1128/jvi.73.7.6104-6110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winkler G, Maxwell SE, Ruemmler C, Stollar V. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology. 1989;171:302–305. doi: 10.1016/0042-6822(89)90544-8. [DOI] [PubMed] [Google Scholar]
- 64.Chambers TJ, McCourt DW, Rice CM. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology. 1989;169:100–109. doi: 10.1016/0042-6822(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 65.Kummerer BM, Rice CM. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J Virol. 2002;76:4773–4784. doi: 10.1128/JVI.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol. 2006;80:4623–4632. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Umareddy I, Chao A, Sampath A, Gu F, Vasudevan SG. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J Gen Virol. 2006;87:2605–2614. doi: 10.1099/vir.0.81844-0. [DOI] [PubMed] [Google Scholar]
- 68.Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005;79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller S, Sparacio S, Bartenschlager R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J Biol Chem. 2006;281:8854–8863. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]