Abstract
In both human and mouse, the Igf2 gene, localized on chromosomes 11 and 7, respectively, is expressed from the paternally inherited chromosome in the majority of tissues. Insulin-like growth factor-II (IGF-II) plays an important role in embryonic growth, and aberrant IGF2 expression has been documented in several human pathologies, such as Beckwith–Wiedemann syndrome (BWS), and a wide variety of tumors. Human and mouse genetic data strongly implicate another gene, CDKN1C (p57kip2), located in the same imprinted gene cluster on human chromosome II, in BWS. p57KIP2 is a cyclin-dependent kinase inhibitor and is required for normal mouse embryonic development. Mutations in CDKN1C (p57kip2) have been identified in a small proportion of patients with BWS, and removal of the gene from mice by targeted mutagenesis produces a phenotype with elements in common with this overgrowth syndrome. Patients with BWS with biallelic expression of IGF2 or with a CDKN1C (p57kip2) mutation, as well as overlapping phenotypes observed in two types of mutant mice, the p57kip2 knockout and IGF-II-overexpressing mice, strongly suggest that the genes may act in a common pathway of growth control in situations where Igf2 expression is abnormal. Herein, we show that p57kip2 expression is reduced on IGF-II treatment of primary embryo fibroblasts in a dose-dependent manner. In addition, p57kip2 expression is down-regulated in mice with high serum levels of IGF-II. These data suggest that the effects of increased IGF-II in BWS may, in part, be mediated through a decrease in p57kip2 gene expression.
The Igf2 gene, predominantly expressed from the paternal allele in mice and humans, encodes a growth factor that plays an important role in embryonic growth (1–4). IGF-II is highly expressed during development, is produced by many tissues, and functions in an autocrine/paracrine manner. In addition, like many hormones, it is also secreted into the serum where it is sequestered almost completely by IGF binding proteins (5). IGF-II binds to two receptors with a high affinity. The type I IGF receptor (IGF1R) mediates most of the in vivo biological effects (3). The role of the other receptor, the mannose-6-P/IGF-II receptor (IGF2R), in IGF-II-mediated transmembrane signaling is unclear. Its main IGF-II-associated function is to bind this protein at the cell surface and internalize it, resulting in the lysosomal degradation of IGF-II. Hence, this receptor has been proposed to clear IGF-II from the circulation (6); in its absence, circulatory levels of IGF-II are increased (7, 8). In mice, the Igf2R gene, located on chromosome 17, is imprinted reciprocally to Igf2, being expressed from the maternally inherited allele (9).
Mice with paternal inheritance of a null Igf2 allele are viable but experience severe growth retardation (60% of normal birth weight; ref. 1). In contrast, mice that overexpress IGF-II are bigger than normal littermates. The phenotypes associated with excess IGF-II are variable and depend on genetic background and IGF-II level. For example, mice that overexpress IGF-II because of a loss of imprinting (the maternal allele is expressed) are viable and 10% bigger than normal (10). In addition, mice with excess IGF-II caused by the absence of Igf2R die late in gestation and show an overgrowth phenotype associated with several developmental abnormalities (7, 8). These mutant phenotypes are rescued in an Igf2 null background (7, 11, 12).
In humans, accumulating evidence suggests that excess IGF-II during fetal development is involved in the pathogenesis of Beckwith–Wiedemann syndrome (BWS), a growth disorder characterized by defects including prenatal and postnatal overgrowth, abdominal wall defects, and macroglossia (13). IGF-II is expressed at highest levels in the tissues that are most affected (14). In addition, IGF2 imprinting is lost in the majority of BWS cases, resulting in biallelic expression of IGF-II (14, 15). BWS is a genetically heterogeneous disorder. Most cases are sporadic, but approximately 15% are familial, and a small number of patients with BWS have cytogenetic abnormalities involving chromosome 11p15. Paternal uniparental disomy and paternal duplication of this region, as well as translocations involving maternal 11p15.5 (where IGF2 resides) have been shown for BWS (16, 17). Finally, mouse genetic models have reinforced the evidence for an involvement of IGF-II in BWS. Indeed, mice chimeric for cells containing extra copies of the Igf2 gene, inducing 2- to 3-fold overexpression of IGF-II, have some BWS features, including prenatal overgrowth, polyhydramnios, and fetal and neonatal lethality but not the omphalocele, renal medullary dysplasia, and adrenal cortical hyperplasia often seen in patients with BWS (18). This incomplete BWS pattern suggests that other genes are involved in this disease and/or that excess of IGF-II in this model is not sufficient to induce all of the BWS characteristics.
More recent data suggest that the p57kip2 gene, a cyclin-dependent kinase inhibitor that is tightly linked to Igf2 and expressed from the maternally inherited allele, could also be involved in this disease (19, 20). Several patients with BWS carrying a mutant CDKN1C (p57kip2) have been identified, with an incidence estimated at 5–10% (21–23). In most patients, the status of IGF2 expression has not been determined, but in the few informative cases analyzed, IGF2 imprinting is maintained. In addition, mice carrying a defective maternal p57kip2 allele have been generated (24, 25). These mice show some BWS phenotypes, which include those lacking in the IGF-II models, suggesting an involvement of both of these gene products in the syndrome. However, this result does not explain the syndrome in patients with two paternal and one maternal copy of 11p15 who therefore have two paternally expressed Igf2 alleles and an intact maternal CDKN1C (p57kip2) allele. Nor does it explain the small number of reported patients with BWS with CDKN1C (p57kip2) mutations alone. One possible explanation is that a relationship exists between p57KIP2 and IGF-II (24, 26–30). This hypothesis has been reinforced by another mouse model in which the level of IGF-II is 7- to 11-fold higher than in the normal situation. Importantly, these mice have most of the BWS phenotypes, even those that seem to be specific to the p57kip2 knockout mice, such as omphalocele (30). Taken together, these data suggest that an increased level of IGF-II may directly or indirectly affect p57kip2.
Two approaches have been taken to address this hypothesis. First, p57kip2 expression was analyzed in vitro in the presence of increasing amounts of IGF-II. Second, we have used an in vivo model in which serum IGF-II is elevated because of the absence of IGF2R. In both situations, p57kip2 expression is affected by abnormally high levels of IGF-II protein. We suggest that IGF-II and p57KIP2 may have both independent and interactive functions in mammalian growth and development.
Materials and Methods
Mice.
(CBA × C57BL/6J)F1 female Thp mice heterozygous for the Thp deletion were crossed with BALB/c or F1 males. The results were the same on both genetic backgrounds. The Thp mutant is easily identified by its short-tailed phenotype (31, 32). However, we also characterized each Thp mutant by the absence of Igf2r expression (9). In the control normal-tailed littermate, Igf2r is expressed.
Cell Culture.
Normal embryos were obtained at embryonic day (e)14.5. The primary embryonic fibroblast (PEF) cultures were generated as described (33). Cells were plated in 100-mm dishes at 7 × 105 cells per plate and cultured in DMEM (GIBCO/BRL) containing 10% (vol/vol) heat-inactivated FCS. After 24 h, exponentially growing cells were washed with PBS and cultured in DMEM containing either 10% (vol/vol) (high serum conditions) or 0.5% (vol/vol) (low serum conditions) heat-inactivated FCS with increasing concentrations of IGF-II (Sigma). Cells were harvested for isolation of nucleic acids after 24 h.
Cell Growth in Culture.
PEF cells and NIH 3T3 fibroblasts (clone A31) were cultured in triplicate as described above. At the beginning of or before the termination of the experiment, a pulse of BrdUrd was added to each dish for 1 h. Cells that incorporated BrdUrd into their DNA were identified by using a BrdUrd labeling and detection kit (Roche Molecular Biochemicals) and counted in situ. The percentage of BrdUrd-positive cells was determined by scoring a total of 300 cells in triplicate and represents the ratio of the number of BrdUrd-positive cells to the total cell number multiplied by 100.
Embryo Growth Analysis.
For embryo staging, e0.5 was taken as noon on the day of vaginal plug appearance. For analysis of growth, embryos were dissected and patted dry with absorbent paper, and wet weights were measured. For determination of dry weights, preweighed embryos were placed into tubes and dried by incubation at 60°C for 48 h and 100°C for 24 h (34). Statistical analysis was performed on 15 embryos for each age by using Student's t test (P < 0.05).
Northern Blot Analysis.
Total embryonic RNA was prepared by the guanidinium thiocyanate/phenol chloroform procedure (35). The Gapdh and p57kip2 probes were generated by PCR. The gene-specific oligonucleotide primers used were, for Gapdh, 5′ primer, ACA GTC CAT GCC ATC ACT GCC ACTC, and 3′ primer, CCA GCC CCA GCA TCA AAG GTG G, and for p57kip2, 5′ primer, CTA GGC CCG ACT GAG AGC AA, and 3′ primer, TGA GGT CAG ATC TGA AGC CG.
The Igf2r probe, provided by D. Barlow (Netherlands Cancer Institute, Amsterdam), is a 1.8-kilobase cDNA corresponding to exons 25 to 36 of the Igf2R gene. The amount of p57kip2 mRNA has been estimated by quantitative Northern blot analysis with Gapdh as control. The relative intensities of autoradiographic signal have been quantified by densitometric scanning on a Chromoscan 3 (Joyce–Loebl) or on a Storm PhosphorImager (Molecular Dynamics).
Western Blot Analysis.
Total embryonic or cell protein extracts were lysed in 1% SDS/10 mM Tris⋅HCl, pH 6.8. The lysates were quantified by using the Bio-Rad Protein Assay (Bio-Rad). Samples containing 50 μg of protein were analyzed by SDS/PAGE followed by Western blot analysis. The membrane was blocked with 5% (wt/vol) nonfat dry milk in PBS buffer/0.05% (vol/vol) Tween 20/1.5% (vol/vol) rabbit serum for 1 h at room temperature. The membrane was incubated directly overnight at 4°C with an anti-mouse p57KIP2 goat polyclonal antibody (1:1,000; M-20 Santa Cruz Biotechnology). After washing, the membrane was incubated with horseradish peroxidase anti-goat IgG antibody (1:1,000; Sigma). The reaction was visualized by using an enhanced chemiluminescence kit (Amersham Pharmacia). To ascertain further the quantity of protein, the membrane was washed for 30 min at 50°C with the stripping buffer [100 mM β-mercaptoethanol/2% (wt/vol) SDS/62.5 mM Tris⋅HCl, pH 6.8] and incubated subsequently with the α-tubulin antibody (1;1,000; Sigma T5168). Blood serum samples were obtained after centrifugation of blood obtained from the umbilical vessels. Serum (2.5 μl) was mixed directly with loading buffer. Human IGF-II mouse monoclonal IgG (Amano Pharmaceutical, Nagoya, Japan) was used as primary IGF-II antibody. IGF-II Western blot analysis was performed as described by Ludwig et al. (7).
Results
IGF-II Affects the p57kip2 Expression in Vitro.
To evaluate the possible effect of IGF-II peptide on p57kip2 expression, we used an in vitro assay with PEF cell cultures derived from wild-type embryos (33). Growth of PEFs and expression of Igf2 in the presence of low or high concentrations of serum have been described (33, 36). PEF cultures were treated with different concentrations of IGF-II over 24 h.
Effect of IGF-II Peptide on Cell Proliferation in Vitro.
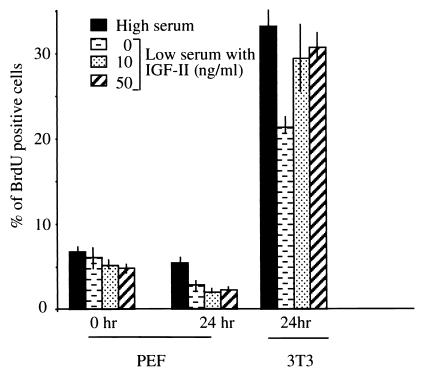
Because IGF-II and p57KIP2 have potentially antagonistic functions on cell proliferation (IGF-II is a growth promoter and p57KIP2 a negative regulator of growth), we measured BrdUrd incorporation in exponentially growing cells in high or low serum medium supplemented with a range of concentrations of IGF-II. At 1 h after the transfer into test medium, BrdUrd incorporation is similar between PEF cultures, irrespective of the concentration of serum present. In contrast, after 24 h of IGF-II treatment, BrdUrd incorporation during a 1-h pulse label is significantly lower in cells shifted to low serum conditions compared with cells in high serum conditions (Fig. 1). However, in low serum, BrdUrd incorporation is not significantly different in PEF cells cultivated in the presence or absence of different concentrations of IGF-II. In contrast, IGF-II is able to increase the fraction of 3T3 fibroblasts in S phase in low serum. This result is consistent with data from similar experiments that used the same NIH 3T3 cells conducted by Campisi and Pardee (37). Our findings suggest that, under these conditions, IGF-II treatment has no significant mitogenic effect on PEFs.
Figure 1.
Effect of IGF-II treatment on proliferation as measured by BrdUrd incorporation into DNA. Histogram representation of the ratio of the number of BrdUrd-positive cells to the total cell number multiplied by 100. Data were obtained in triplicate and are given for both PEF and NIH 3T3 cells (clone A31; ref. 37).
Down-Regulation of p57kip2 Expression in Cells Cultivated in the Presence of IGF-II.
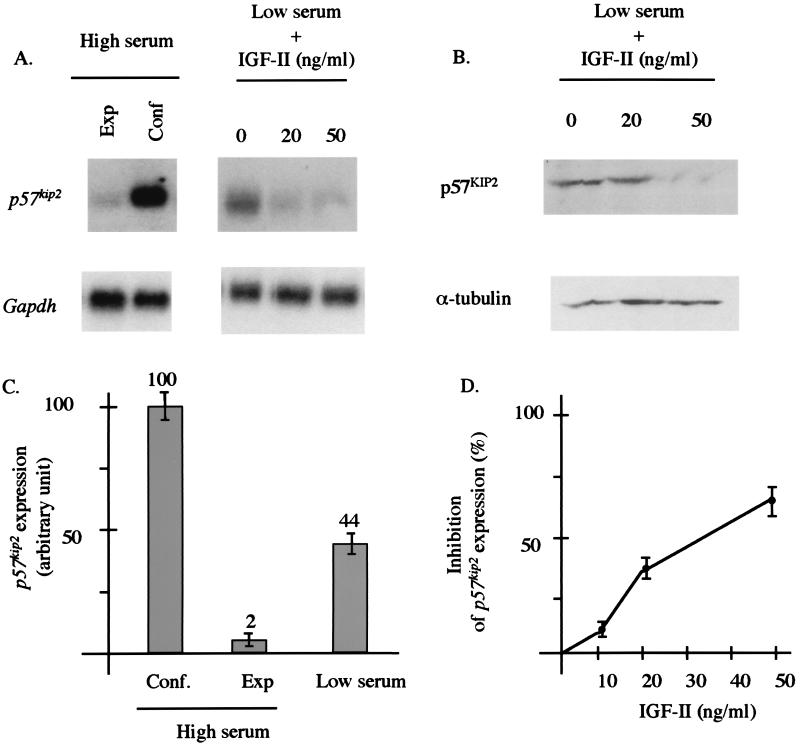
Expression of p57kip2 in control and IGF-II-treated cells is shown in Fig. 2. As expected for a cell-cycle inhibitor (38), actively dividing cells cultivated in the presence of serum have a very low p57kip2 expression. In contrast, after a 24-h serum deprivation or 2 days after reaching confluence, these cell cultures show growth arrest accompanied by a 22- and 50-fold increase in p57kip2 mRNA expression, respectively (Fig. 2 A and C). Interestingly, cells cultivated in low serum in the presence of different concentrations of IGF-II show a decrease in p57kip2 mRNA expression compared with untreated cells. Indeed, the p57kip2 expression from cells cultivated with 10, 20, or 50 ng/ml of IGF-II peptide decreased 10%, 37%, and 63%, respectively (Fig. 2 A and D). To determine whether the down-regulation of p57kip2 mRNA transcription observed in vitro results in a decrease in p57KIP2 protein levels, we performed Western analysis on cell extracts from IGF-II-treated or untreated PEFs. The reduction in p57kip2 transcription in cultured cells on IGF-II treatment is also associated with a reduced protein level (Fig. 2B). The reduction is inversely proportional to the IGF-II concentration. This down-regulation is not due to a relative increase in the number of actively dividing cells (see above) but represents a direct or indirect effect of IGF-II via an IGF-II related pathway.
Figure 2.
Expression of p57kip2 in cultured cells with or without IGF-II. (A) Poly(A)+ RNA was extracted from confluent (Conf) or exponentially growing (Exp) cells cultivated in the presence of 10% (vol/vol) FCS (high serum) or from cells that had been serum starved for 24 h (low serum) in the presence of different concentrations of IGF-II. mRNA (0.5 μg) was analyzed by Northern analysis with a p57kip2 probe. (B) Western blot analyses with p57KIP2 antibody. The blot containing protein extracts from PEFs treated with 0, 10, or 50 ng of IGF-II was probed with anti-p57KIP2 and with anti-α-tubulin as a loading control. The data shown are representative of three independent experiments. (C) p57kip2 mRNA expression in cells cultivated with 10% (vol/vol) (high serum) or 0.5% (vol/vol) (low serum) FCS. Data represent the p57kip2 signal normalized with the Gapdh control signals. (D) The percentage of inhibition of p57kip2 expression corresponds to the ratio of the p57kip2 signal normalized with the Gapdh control signals between the IGF-II-treated and untreated cells.
Down-Regulation of p57kip2 mRNA in Embryos with Increased IGF-II Levels.
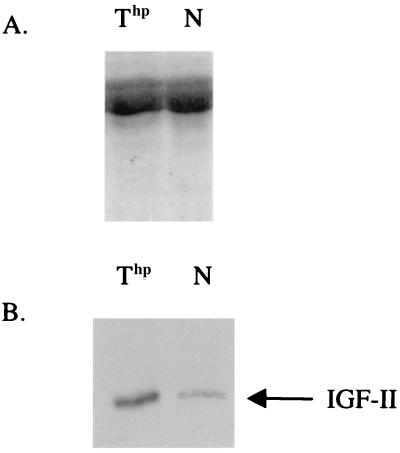
To determine whether this observed effect could occur in vivo, we used the Thp mutant (31, 32). This mutant carries the Tme deletion on chromosome 17 encompassing the Igf2R gene. Maternally inherited Thp heterozygous embryos on this genetic background die in utero at about e16. Paternal inheritance of the deletion results in viable embryos, because the paternal Igf2R allele is normally repressed (9). In the Thp mutant, IGF-II serum levels were measured and shown to be increased 3- to 4-fold (Fig. 3); a level comparable to that observed in Igf2R knockout mice (7, 8). The phenotype of Thp embryos overlaps that described for mice lacking the Igf2R gene, with both mutations resulting in a complex maternally inherited phenotype, including disproportionate overgrowth and embryonic lethality (7, 8, 31). The lethality of both mutants can be rescued in a Igf2−/− background, indicating that lethality results from the accumulation of toxic levels of IGF-II (7, 11, 12).
Figure 3.
High levels of IGF-II in Thp mutant serum. Serum (2.5 μl) from mutant (Thp) and normal (N) embryos at e15.5 was analyzed. (A) Coomassie blue staining of SDS/polyacrylamide gel containing serum from mutant and normal embryos. (B) Immunoblot analysis of IGF-II carried out as described (7). Signals were scanned by densitometry. Ratios between mutant (Thp) and normal (N) were 3.0, 3.4, and 3.1 for three independent experiments.
Because IGF-II overexpression is always associated with a somatic overgrowth phenotype, we measured total embryonic wet and dry weights versus developmental age. The wet weight of Thp mutant embryos is at least 20% greater than that of control littermates from e12.5 onward. This increase reaches a maximum at e13.5 and is maintained thereafter (Table 1). These results are consistent with previous measurements (34). To determine how much of the increase is due to excess fluid, dry weights were also measured. At e12.5, there was a comparable and significant increase in dry weight, suggesting that the increase in wet weight is not primarily due to fluid. However, at e13.5 onward, increase in water content does contribute significantly to the weight increase, although there is still somatic overgrowth in Thp embryos compared with the normal littermates. This result is consistent with the obvious edema observed in these mutants (34).
Table 1.
Growth measurements of mutant (Thp) and normal embryos
| Measurement | Thp (no.) | Normal | Thp/normal ratio, % |
|---|---|---|---|
| Wet weight, mg | |||
| e12.5 | 104 ± 6 (13) | 86 ± 6 (11) | 120∗ |
| e13.5 | 184 ± 31 (8) | 132 ± 15 (8) | 140∗ |
| e14.5 | 345 ± 41 (17) | 252 ± 30 (16) | 137∗ |
| e15.5 | 556 ± 30 (8) | 402 ± 26 (8) | 138∗ |
| Dry weight, mg | |||
| e12.5 | 9 ± 1 (13) | 8 ± 1 (11) | 118∗ |
| e13.5 | 18 ± 2 (8) | 14 ± 2 (8) | 133∗ |
| e14.5 | 34 ± 5 (17) | 27 ± 5 (16) | 124∗ |
| e15.5 | 57 ± 6 (8) | 45 ± 3 (8) | 125∗ |
| Water content† | |||
| e12.5 | 9.9 | 9.8 | 101 |
| e13.5 | 9.4 | 8.4 | 112∗ |
| e14.5 | 9.1 | 8.3 | 109∗ |
| e15.5 | 8.7 | 7.8 | 111∗ |
Statistically significant difference between Thp and normal embryo (Student's t test, P < 0.05). Mean values ± SEM were calculated from measurements of the number of embryos (shown in parentheses).
† Water content per unit dry weight = (wet weight − dry weight)/dry weight.
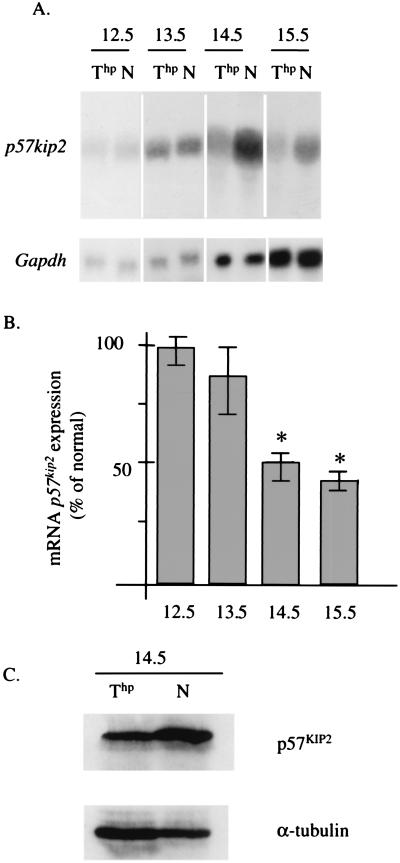
To investigate whether there is an inverse relationship between p57KIP2 and IGF-II levels in vivo, we compared the p57kip2 expression between the mutant and normal embryos from e12.5 to e15.5 (Fig. 4 A and B). At e12.5 and e13.5, no statistically significant difference in expression was observed between the mutant and normal embryos. However, at e14.5 and e15.5, mutant embryos show an average reduction in expression of 50% compared with the normal embryos derived from the same litter. The reduced expression is also associated with reduced p57KIP2 protein (Fig. 4C). Hence, reduction in p57kip2 mRNA and protein occurs after the cessation of increased somatic growth shown in Table 1.
Figure 4.
Down-regulation of the expression of p57kip2 gene in Thp embryos. (A) Northern blot analysis of total RNA isolated from normal (N) and mutant embryos (Thp) at different stages of gestation hybridized with p57kip2. Gapdh expression is used as a control. (B) Graphic representation shows the average of the p57kip2 activity for five embryos normalized to Gapdh expression, compared with p57kip2 expression of normal embryos from the same litter. Error bars represent the standard errors. Statistically significant differences are indicated by an asterisk (Student's t test; *, P < 0.05). (C) Western blot analyses with p57KIP2 antibody. The blot containing mutant (Thp) and normal (N) embryo protein extracts at e14.5 was probed with anti-p57KIP2 and with anti-α-tubulin as a loading control.
Discussion
The p57kip2 null and the overexpressing IGF-II mice, in conjunction with dosage changes in each of these genes resulting in the same human syndrome (BWS), have suggested a functional interaction between these two genes in abnormal situations (24, 26–30). It has been demonstrated that IGF-II gene expression is not affected in embryos carrying a maternally inherited defect in p57kip2, indicating that the p57KIP2 gene product does not affect the Igf2 gene expression (39). Recently, Caspary and collaborators (40) showed, by using a double-knockout mouse model overexpressing the Igf2 gene and carrying a null mutation in p57kip2 gene, that IGF-II and p57KIP2 may act in an antagonistic manner to control cell proliferation and development in a subset of the tissues affected in patients with BWS. Our work supports this conclusion and shows an additional level of interaction between these two imprinted genes where an increase in IGF-II protein is associated with changes in the transcriptional activity of p57kip2, resulting in a decrease in the level of p57KIP2 protein. Hence, in abnormal situations where IGF-II is elevated, p57kip2 levels are reduced. It is interesting to note that a similar effect is not observed in the placenta (data not shown), where genetic evidence suggests that IGF-II acts through a currently unidentified receptor (41). There is no evidence indicating whether the effect of elevated IGF-II on p57kip2 is direct or indirect. This regulation might be mediated through the Igf1R or InsR and/or via other factors, such as IGF binding proteins, as well.
The down-regulation observed in vivo is correlated with the increased level of IGF-II in the serum induced by the absence of the type 2 IGF receptor. Even if we cannot exclude the involvement of other genes, our in vitro assay indicates strongly that excess IGF-II affects p57kip2 expression. This effect depends on the level of IGF-II, which, in mice, is regulated in a number of ways. In our murine model, the decrease in p57kip2 expression is stage specific. This temporal relationship may be because the down-regulation requires a threshold level of IGF-II and/or because tissue(s) affected by IGF-II overexpression may express p57kip2 later during development. In vitro, the p57kip2 down-regulation is inversely proportional to the IGF-II concentration. This dosage effect may explain why p57kip2 reduction has not been observed in other investigations where IGF-II is perturbed. In these animals models, p57kip2 expression either has not been analyzed at these stages or may have been obscured by the presence of normal cells in chimeric animals (18).
By using the in vitro model, we made several observations. First, like p27KIP1, another member of the Cip/Kip cyclin-dependent kinase inhibitors, p57kip2 is induced by growth factor deprivation and contact inhibition, albeit by using different mechanisms—p27KIP1 is regulated posttranscriptionally under these conditions (42). Second, like IGF-I, IGF-II is not sufficient on its own to allow DNA synthesis in embryonic murine cells cultivated in low serum conditions. Nonetheless, cells in the presence of exogenous IGF-II express less p57KIP2 protein. This finding suggests that although p57kip2 does control cell proliferation in vitro (19), down-regulation of p57kip2 is not sufficient to induce DNA synthesis in PEFs in low serum with or without IGF-II.
BWS is likely to involve several imprinted genes located on chromosome 11p15, because some patients show paternal uniparental disomy and others show balanced germ-line chromosomal rearrangements involving the maternal chromosome. The IGF2, KVLQT1, CDKN1C, and H19 genes are all members of the cluster of imprinted genes located on chromosome 11p15, and lesions have been identified for all of them in different patients with BWS (14, 15, 21, 22, 43–46). The results, presented herein might explain how two different genetic lesions, gain of function of IGF2 and mutation in the CDKN1C gene, result in the same disease. This suggestion is reinforced by the fact that the phenotypes associated with p57kip2 mutation such as cleft palate and omphalocele are neither enhanced by overexpression of Igf2 nor rescued by its reduction, suggesting that IGF-II acts upstream of p57kip2 at least for these tissues. However, inconsistencies still exist if all of the mouse models, including this one, are considered together. Some BWS characteristics such as renal dysplasia or adrenal cytomegaly in mice carrying a p57kip2 null allele are not observed in other mice with increased IGF-II (30). These observations suggest either that p57kip2 levels are not sufficiently suppressed to cause these defects in IGF-II mutants or that some defects in BWS may be occurring through a p57kip2-dependent and IGF-II-independent pathway. In addition, mice carrying a defect in the active p57kip2 allele do not have all of the phenotypes of mutants overexpressing IGF-II, such as overgrowth (24, 25), suggesting the existence of IGF-II-dependent pathways independent of p57kip2. Nevertheless, none of the mouse models fully recapitulate the human BWS phenotype. Indeed, some patients with BWS have a defective p57kip2 gene without detectable changes in the epigenetic states of IGF2 (23), and they have a nearly full spectrum of BWS phenotypes including the overgrowth phenotype, which is absent in p57kip2 knockout mice. This discordance can be the result of functional differences between human and mouse, be due to differences in genetic background, or reflect the fact that this group of patients with BWS has mutations in other critical genes involved in BWS. Finally, highly variable phenotypes have been associated with BWS, and, with the exception of correlations between somatic isodisomy and hemihypertrophy and between exomphalos and CDKINC mutation (47, 48), no obvious correlation between phenotype and genotype has been established. It has been suggested that at least some tissues are highly sensitive to ratios of IGF-II and p57KIP2 (40). The relationship between Igf2 and p57kip2 demonstrated in the current study supports this suggestion.
Acknowledgments
We thank Dr. D. Barlow for the Igf2R probe, Dr. A. Efstratiadis for advice on the IGF-II antibody, and Prof. C. F. Graham. This work was supported by the Isaac Newton Trust (Cambridge, U.K.) and a European Molecular Biology Organization Fellowship awarded to V.G.
Abbreviations
- BWS
Beckwith–Wiedemann syndrome
- PEF
primary embryonic fibroblast
- en
embryonic day n
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080409297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080409297
References
- 1.DeChiara T M, Robertson E T, Efstratiadis A. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson-Smith A C, Cattanach B M, Barton S C, Beechey C V, Surani M A. Nature (London) 1991;351:667–670. doi: 10.1038/351667a0. [DOI] [PubMed] [Google Scholar]
- 3.Giannoukakis N, Deal C, Paquette J, Goodyer C G, Polychronakos C. Nat Genet. 1993;4:98–101. doi: 10.1038/ng0593-98. [DOI] [PubMed] [Google Scholar]
- 4.Efstratiadis A. Int J Dev Biol. 1998;42:955–976. [PubMed] [Google Scholar]
- 5.Murphy L J. J Mol Endocrinol. 1998;21:97–107. doi: 10.1677/jme.0.0210097. [DOI] [PubMed] [Google Scholar]
- 6.Kornfeld S. Annu Rev Biochem. 1992;61:307–330. doi: 10.1146/annurev.bi.61.070192.001515. [DOI] [PubMed] [Google Scholar]
- 7.Ludwig T, Eggenschwiler J, Fisher P, D'Ercole A J, Davenport M L, Efstratiadis A. Dev Biol. 1996;177:517–535. doi: 10.1006/dbio.1996.0182. [DOI] [PubMed] [Google Scholar]
- 8.Lau M M H, Stewart C E H, Liu Z, Bhatt H, Rotwein P, Stewart C L. Genes Dev. 1994;8:2953–2963. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- 9.Barlow D P, Stoger R, Herrmann B G, Saito K, Schweifer N. Nature (London) 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 10.Leighton P A, Ingram R S, Eggenschwiler J, Efstratiadis A, Tilghman S M. Nature (London) 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z Q, Fung M R, Barlow D P, Wagner E F. Nature (London) 1994;372:464–467. doi: 10.1038/372464a0. [DOI] [PubMed] [Google Scholar]
- 12.Filson A J, Louvi A, Efstratiadis A, Robertson E J. Development (Cambridge, UK) 1993;118:731–736. doi: 10.1242/dev.118.3.731. [DOI] [PubMed] [Google Scholar]
- 13.Beckwith J B. Birth Defects. 1969;131:293–294. [Google Scholar]
- 14.Weksberg R, Shen D R, Fei L Y, Song Q L, Squire J. Nat Genet. 1993;5:143–150. doi: 10.1038/ng1093-143. [DOI] [PubMed] [Google Scholar]
- 15.Joyce J A, Lam W K, Catchpoole D J, Jenks P, Reik W, Maher E R, Schofield P N. Hum Mol Genet. 1997;6:1543–1548. doi: 10.1093/hmg/6.9.1543. [DOI] [PubMed] [Google Scholar]
- 16.Elliot M, Maher E R. J Med Genet. 1994;31:560–564. doi: 10.1136/jmg.31.7.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ping A J, Reeve A E, Law D J, Young M R, Boehnke M, Feinberg A. Am J Hum Genet. 1989;44:711–719. [PMC free article] [PubMed] [Google Scholar]
- 18.Sun F L, Dean W L, Kelsey G, Allen N D, Reik W. Nature (London) 1997;389:809–815. doi: 10.1038/39797. [DOI] [PubMed] [Google Scholar]
- 19.Matsuoka S, Edwards M C, Bai C, Parker S, Zhang P, Baldini A, Harper J W, Elledge S J. Genes Dev. 1995;9:650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 20.Hatada I, Mukai T. Nat Genet. 1995;11:204–206. doi: 10.1038/ng1095-204. [DOI] [PubMed] [Google Scholar]
- 21.Hatada I, Ohashi H, Fukushima Y, Kaneko Y, Inoue M, Komoto Y, Okada A, Ohishi S, Nabetani A, Morisaki H, et al. Nat Genet. 1996;14:171–173. doi: 10.1038/ng1096-171. [DOI] [PubMed] [Google Scholar]
- 22.Lee M P, DeBaun M, Randhawa G, Reichard B A, Elledge S J, Feinberg A P. Am J Hum Genet. 1997;61:304–309. doi: 10.1086/514858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam W W K, Hatada I, Ohishi S, Mukai T, Joyce J A, Cole T R P, Donnai D, Reik W, Schofield P N, Maher E R. J Med Genet. 1999;36:518–523. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang P, Liegeois N J, Wong C, Finegold M, Hou H, Thompson J C, Silverman A, Harper J W, DePinho R A, Elledge S J. Nature (London) 1997;387:151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 25.Yan Y, Frisen J, Lee M H, Massague J, Barbacid M. Genes Dev. 1997;11:973–983. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- 26.Ferguson-Smith A C. Nat Genet. 1996;14:171–173. doi: 10.1038/ng1096-119. [DOI] [PubMed] [Google Scholar]
- 27.Swanger W J, Roberts J M. BioEssays. 1997;19:839–842. doi: 10.1002/bies.950191002. [DOI] [PubMed] [Google Scholar]
- 28.Reik W, Maher E R. Trends Genet. 1997;13:330–335. doi: 10.1016/s0168-9525(97)01200-6. [DOI] [PubMed] [Google Scholar]
- 29.Hastie N. Nature (London) 1997;389:785–787. doi: 10.1038/39732. [DOI] [PubMed] [Google Scholar]
- 30.Eggenschwiler J, Ludwig T, Fisher P, Leighton P A, Tilghman S M, Efstratiadis A. Genes Dev. 1997;11:3128–3142. doi: 10.1101/gad.11.23.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson D R. Genet Res. 1974;24:207–213. doi: 10.1017/s0016672300015226. [DOI] [PubMed] [Google Scholar]
- 32.Johnson D R. Genetics. 1974;76:795–805. doi: 10.1093/genetics/76.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eversole-Cire P, Ferguson-Smith A C, Sasaki H, Brown K D, Cattanach B M, Gonzales F A, Surani M A, Jones P A. Mol Cell Biol. 1993;13:4928–4938. doi: 10.1128/mcb.13.8.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babiarz B S, Donovan M J, Hathaway H J. Teratology. 1988;37:353–365. doi: 10.1002/tera.1420370409. [DOI] [PubMed] [Google Scholar]
- 35.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 36.Eversole-Cire P, Ferguson-Smith A C, Surani M A, Jones P A. Cell Growth Differ. 1995;6:337–345. [PubMed] [Google Scholar]
- 37.Campisi J, Pardee A B. Mol Cell Biol. 1984;4:1807–1814. doi: 10.1128/mcb.4.9.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama K I, Nakayama K. BioEssays. 1998;20:1020–1029. doi: 10.1002/(SICI)1521-1878(199812)20:12<1020::AID-BIES8>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 39.Caspary T, Cleary M A, Baker C C, Guan X J, Tilghman S M. Mol Cell Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caspary T, Cleary M A, Perlman E J, Zhang P, Elledge S J, Tilghman S M. Genes Dev. 1999;13:3115–3124. doi: 10.1101/gad.13.23.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker J, Lu J P, Robertson E J, Efstratiadis A. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- 42.Polyak K, Lee M H, Erdjument-Bromage H, Koff A, Roberts J M, Tempst P, Massague J. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 43.Lee M P, Hu R J, Johnson L A, Feinberg A P. Nat Genet. 1997;15:181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]
- 44.Reik W, Brown K W, Schneid H, Le Bouc Y, Bickmore W, Maher E R. Hum Mol Genet. 1995;4:2379–2385. doi: 10.1093/hmg/4.12.2379. [DOI] [PubMed] [Google Scholar]
- 45.Smilinich N J, Day C D, Fitzpatrick G V, Caldwell G M, Lossie A C, Cooper P R, Smallwood A C, Joyce J A, Schofield P N, Reik W, et al. Proc Natl Acad Sci USA. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee M P, DeBaun M R, Mitsuya K, Galonek H L, Brandenburg S, Oshimura M, Feinberg A P. Proc Natl Acad Sci USA. 1999;96:5203–5208. doi: 10.1073/pnas.96.9.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slatter R E, Elliott M, Welham K, Carrera M, Schofield P N, Barton D E, Maher E R. J Med Genet. 1994;31:749–753. doi: 10.1136/jmg.31.10.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam W, Hatada I, Ohishi S, Mukai T, Joyce J, Cole T, Donnai D, Reik W, Schofield P, Maher E. J Med Genet. 1999;36:518–523. [PMC free article] [PubMed] [Google Scholar]