Summary
DExD/H-box proteins are ubiquitously involved in RNA-mediated processes and use ATP to accelerate RNA conformational changes. However, their mechanisms of action, and what determines which RNA species are targeted, are not well understood. Here we show that the DExD/H-box protein CYT-19, a general RNA chaperone, mediates ATP-dependent unfolding of both the native and a long-lived misfolded conformation of a group I catalytic RNA with efficiencies that depend on the stabilities of the RNA species but not on specific structural features. CYT-19 then allows the RNA to re-fold, changing the distribution from equilibrium to kinetic control. Because misfolding is favored kinetically, conditions that allow unfolding of the native RNA give large increases in the population of misfolded species. Our results suggest that DExD/H-box proteins act with sufficient breadth and efficiency to allow structured RNAs to populate a wider range of conformations than would be present at equilibrium. Thus, RNAs may face selective pressure to stabilize their active conformations relative to inactive ones to avoid significant redistribution by DExD/H-box proteins. Conversely, RNAs whose functions depend on forming multiple conformations may rely on DExD/H-box proteins to increase the populations of less stable conformations, thereby increasing their overall efficiencies.
All organisms encode a host of RNAs that must fold into functional structures and undergo extensive conformational transitions as they mediate essential cellular processes, including translation and, in eukaryotes, pre-mRNA processing and the maintenance of chromosome ends. Essentially all processes that are mediated by structured RNAs also require DExD/H-box proteins, which use cycles of ATP binding and hydrolysis to accelerate RNA conformational changes1,2. Although they are related to DNA helicases3, and some viral DExD/H-box proteins possess at least modest canonical RNA or DNA helicase activity4-6, many DExD/H-box proteins display very low activity in conventional helicase assays and are poorly processive, consistent with roles in disrupting local structural elements rather than unwinding long helices2,7.
There has been much progress using short model duplex RNAs and RNA-protein complexes to elucidate the basic capabilities of DExD/H-box proteins8-11. However, relatively little is known at the molecular level about how DExD/H-box proteins interact with structured RNAs to mediate conformational changes, and what determines which RNAs, and which of their conformations, are targeted for action. Whereas many DExD/H-box proteins are thought to function in the context of a defined RNA or RNA-protein complex, some proteins of the major subfamily, DEAD-box, function as general RNA chaperones by interacting less discriminately with structured RNAs to promote their folding12,13. This latter group includes the Neurospora crassa CYT-19 protein, which is required for efficient splicing of several mitochondrial group I introns and can facilitate folding of a diverse set of group I and group II introns in vitro or when expressed in Saccharomyces cerevisiae12-14.
These systems are valuable experimentally because the relatively simple group I RNAs are likely to provide insight into the mechanisms of DExD/H-box proteins in more complex systems. A particularly attractive candidate for detailed mechanistic studies of CYT-19 is the ribozyme derived from a group I intron of Tetrahymena thermophila, because its in vitro folding has been extensively characterized15-19. Further, because it folds preferentially to a long-lived misfolded conformation which then slowly re-folds to the native structure20-24, it is possible to generate populations of either predominantly native or predominantly misfolded ribozyme. Using a quantitative assay for ribozyme catalytic activity, we showed previously that CYT-19 interacts with the misfolded ribozyme, giving ATP-dependent re-folding to the catalytically active, native state25,26. Because the misfolded ribozyme is extensively structured, including all five long-range native tertiary contacts, and must unfold substantially to reach the native state27, CYT-19 apparently accelerates this folding reaction by promoting partial unfolding of the misfolded ribozyme.
Redistribution of native and misfolded ribozyme
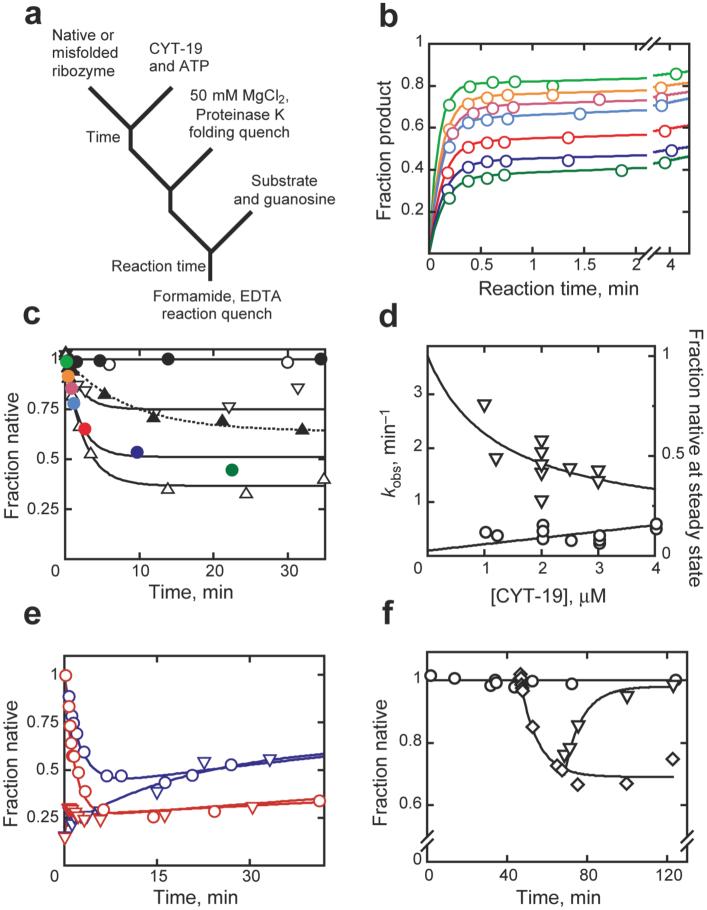
Here we probe the mechanism of CYT-19 action by exploring whether it recognizes structural features that are specific to the misfolded conformer or whether it can also mediate unfolding of the native ribozyme. We added CYT-19 and ATP to a preparation of ribozyme that was pre-folded to the native state (Fig. 1a). At various times thereafter, we inactivated CYT-19 by adding proteinase K and increasing Mg2+ concentration. We then determined the fraction of the ribozyme present in the native conformation, without interference from CYT-19 (Supplementary Fig. 1), by measuring the fraction of added oligonucleotide substrate (CCCUCUA5) that was rapidly cleaved by the ribozyme25. Our prior work showed that the substrate binds the native and misfolded species with similar rate constants but is only cleaved by the native ribozyme22, so that a burst of product is obtained, with the amplitude indicating the fraction of native ribozyme (Fig. 1b). This burst is followed by a slower phase of product accumulation, which reflects dissociation of the substrate from the misfolded ribozyme and subsequent binding and cleavage by the native ribozyme22,25.
Figure 1.
Unfolding of native and misfolded Tetrahymena ribozyme. a, Reaction scheme. b, Substrate cleavage after incubation with 1 mM Mg2+, 2 μM CYT-19 and 2 mM ATP-Mg2+ for 0.25 (orange), 0.67 (red), 1 (cyan), 2.5 (magenta), 9.5 (blue), or 22 min (dark green). (Light green), no CYT-19. c, Native ribozyme unfolding (1 mM Mg2+). CYT-19 was 1 μM (∇), 2 μM (solid colored circles), or 3 μM without (σ) or with 2 mM ATP-Mg2+ (∆). Colored circles show burst amplitudes from corresponding curves (panel b). ○, no CYT-19; ●, 2 μM CYT-19, 2 mM ATP-Mg2+, 5 mM Mg2+. d, Rate constant (○) and steady-state value (∇) vs CYT-19 concentration. e, Approach to steady state from native (○) or misfolded (∇) ribozyme with 1.2 μM (blue) or 2 μM (red) CYT-19. f, Refolding to the native state (∇) after unfolding by CYT-19 (◇) and inactivation by proteinase K. ○, no CYT-19.
As expected, no net unfolding was observed under conditions shown previously to give complete accumulation of native ribozyme (5 mM Mg2+, Fig. 1c). However, under less stabilizing conditions (1 mM Mg2+), the fraction of native ribozyme decreased upon addition of high CYT-19 concentrations, indicating that CYT-19 can also unfold the native ribozyme (Fig. 1b-d; a subsequent slow increase reflects time-dependent inactivation of CYT-19 and inhibition by accumulating ADP, data not shown). The reaction was dependent on ATP, as its omission or replacement with ADP or AMP-PNP gave only low levels of residual activity (Fig. 1c & d and data not shown; higher concentrations of CYT-19, not shown, gave significant ATP-independent activity, presumably reflecting passive ‘strand capture’28). The ribozyme ultimately reached an apparent steady state between the native and alternative conformers, which was dependent on CYT-19 concentration (Fig. 1d). The observed rate constant for steady-state formation increased modestly with CYT-19 concentration, giving an efficiency of < 105 M-1 min-1. The same steady-state distribution was obtained whether starting from a population of predominantly native or misfolded ribozyme (Fig. 1e), indicating that the entire ribozyme population is subject to the action of CYT-19.
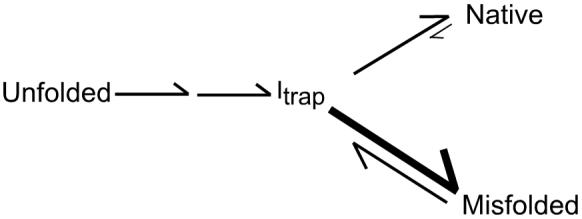
Additional insight into the action of CYT-19 on the native ribozyme and the nature of the steady-state redistribution came from the finding that the decrease in native ribozyme was accompanied by formation of the previously-characterized misfolded conformer. Upon inactivation of CYT-19 by proteinase K, the native ribozyme accumulated with the same rate constant as that for re-folding from the long-lived misfolded species and gave the same Mg2+ and urea concentration dependences (Fig. 1f and Supplementary Fig. 2), indicating that a large population of the misfolded conformer was either formed during the CYT-19 reaction or immediately upon its inactivation. These observations suggested a simple model in which CYT-19 partially unfolds both the native and misfolded ribozyme species, giving intermediates that subsequently fold along the same pathway that predominates in the absence of CYT-19. This pathway includes a late commitment point for folding from a trapped intermediate (Itrap) to the native and misfolded species, with preferential partitioning to the misfolded species18,22 (Scheme 1). This kinetic preference could allow the misfolded species to accumulate despite its lower stability than the native species.
Scheme 1.
Unfolding efficiency depends on RNA stability
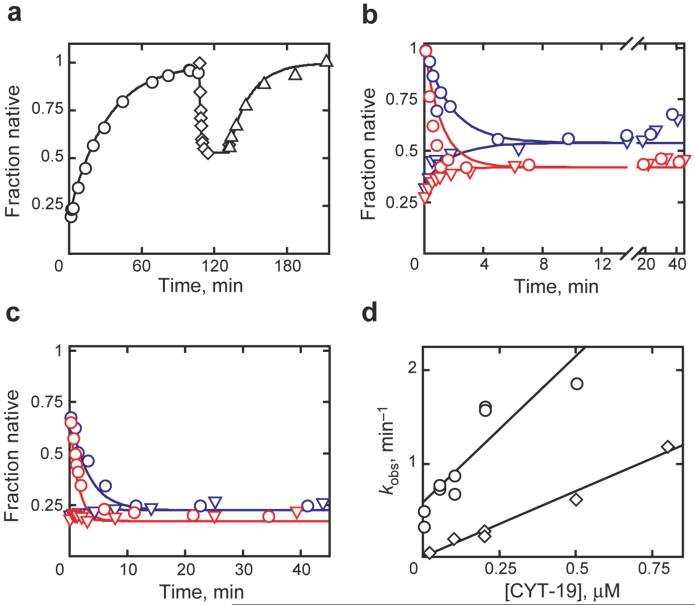
To explore this model further, we were interested in identifying features of the ribozyme that affect the efficiency of unfolding by CYT-19. Although CYT-19 is able to act upon the native ribozyme, the efficiency is estimated to be at least 50-fold lower than for the misfolded ribozyme (Fig. 1d and ref. 25). This difference raised the possibility that CYT-19 derives specificity from structural features of the misfolded species, but an alternative possibility was that the native ribozyme is unfolded less efficiently because it is more stable22,29. Therefore, we examined two ribozyme variants with decreased native stability (Fig. 2). First, we found that the native species of a ribozyme variant with a disrupted tertiary contact between the P4 helix and an A-rich internal loop in the peripheral helix P5a27,30 was unfolded efficiently by CYT-19 even at 5 mM Mg2+ (Fig. 3a and Supplementary Fig. 3), conditions that do not give detectable unfolding of the wild-type ribozyme. Again, the reaction was ATP-dependent (Supplementary Fig. 4) and, for a given CYT-19 concentration, the same steady state was reached when starting from populations of predominantly native or misfolded ribozyme (Fig. 3b).
Figure 2.
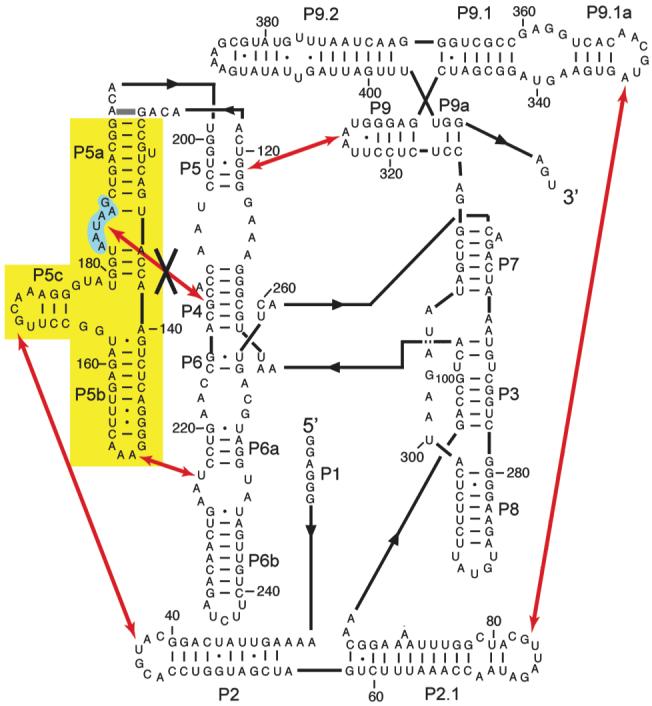
Secondary structure, long-range tertiary contacts, and mutations of the Tetrahymena ribozyme. The five long-range tertiary contacts are indicated with red arrows. In the P5abc deletion variant (EΔP5abc), the region shown in yellow is deleted and nucleotides 126 and 196 are directly connected (thick gray line above yellow region). In the P5a mutant, nucleotides 183-188 (shaded cyan) are each changed to uridine27. This mutation disrupts the tertiary contact indicated by the black ‘X’.
Figure 3.
Unfolding of destabilized ribozyme variants. a, P5a variant with 0.8 μM CYT-19 (◇). Upon CYT-19 inactivation, native ribozyme accumulated (∆, 0.056 min-1) with the same rate constant within error as for refolding of the misfolded ribozyme (○, 0.032 min-1). b, Approach to steady state for native (○) or misfolded (∇) P5a variant with 0.5 μM (blue) or 1 μM (red) CYT-19. Curves depict kinetic simulations using experimentally-derived values (see Supplementary Fig. 6), not fits to the data. c, Approach to steady state for EΔP5abc ribozyme. Curves and symbols as in panel b except red (1.2 μM CYT-19). d, Unfolding of native (◇) and misfolded (○) EΔP5abc ribozyme. All reactions were 25 °C, pH 7.0, 5 mM Mg2+.
To test the model that CYT-19 produces intermediates that subsequently fold along the pathway that predominates in its absence, we compared the unfolding reactions to kinetic simulations (curves in Fig. 3b). The data were well-described using the experimental value for CYT-19-mediated unfolding of the native species and values for subsequent folding that were determined in the absence of CYT-19 (Supplementary Figs 5 & 6). Thus, CYT-19 apparently gives partial unfolding of the native and misfolded species and then allows the unfolded intermediates to fold again without significant interference in this latter process.
Next we examined a ribozyme variant with the entire P5abc peripheral element deleted (EΔP5abc, Fig. 2)31. This ribozyme populates the native and misfolded species nearly equally at equilibrium,29 leading to the prediction that the two species would be unfolded with comparable efficiency. CYT-19 readily acted on the native ribozyme to decrease the fraction native (1.4 × 106 M-1 min-1, Fig. 3c and d). However, when starting from a population of misfolded ribozyme, net re-folding was not observed; instead, a steady state was reached with the fraction of native ribozyme approximately equal to the small fraction that avoids the misfolded species and folds to the native state directly (Fig. 3c). This behavior supports the model because, if the native and misfolded species are rapidly unfolded with nearly equal efficiencies, even at modest CYT-19 concentrations the relative populations will be determined by the kinetic partitioning that occurs between them during folding. Nevertheless, the lack of native ribozyme accumulation prevented determination of the efficiency of CYT-19 for unfolding the misfolded ribozyme.
We circumvented this limitation by including a small excess of the group I intron-binding protein CYT-18 (ref. 32), which binds the native EΔP5abc ribozyme tightly33 but does not bind stably to the misfolded ribozyme under these conditions (A. Chadee and R.R., unpublished results), such that it traps any newly-formed native ribozyme. Net re-folding of misfolded EΔP5abc ribozyme to the native state was indeed observed. After correcting for the bias of the ribozyme to misfold again after CYT-19 action34, this measurement gave an efficiency for unfolding of the misfolded ribozyme of 3.1 × 106 M-1 min-1 (Fig. 3d). This value is ∼2-fold larger than that for unfolding of the native EΔP5abc ribozyme, similar to the equilibrium value of 1.4 (ref. 29). Thus, the relative stabilities of the native and misfolded species appear to play a central role in determining their efficiencies of unfolding by CYT-19, although we cannot exclude the possibility that the region of the ribozyme near P5abc is the preferred site of action by CYT-19 and it is the local stability of this region, rather than the global RNA stability, that governs CYT-19 efficiency. Again, kinetic simulation of the approaches to a steady-state mixture of native and misfolded ribozyme (Fig. 3c) gave good agreement with the data using the experimental values for CYT-19-mediated unfolding of the native and misfolded species and prior experimental values for folding of the EΔP5abc ribozyme in the absence of CYT-19 (ref. 34).
Model for kinetic redistribution by CYT-19
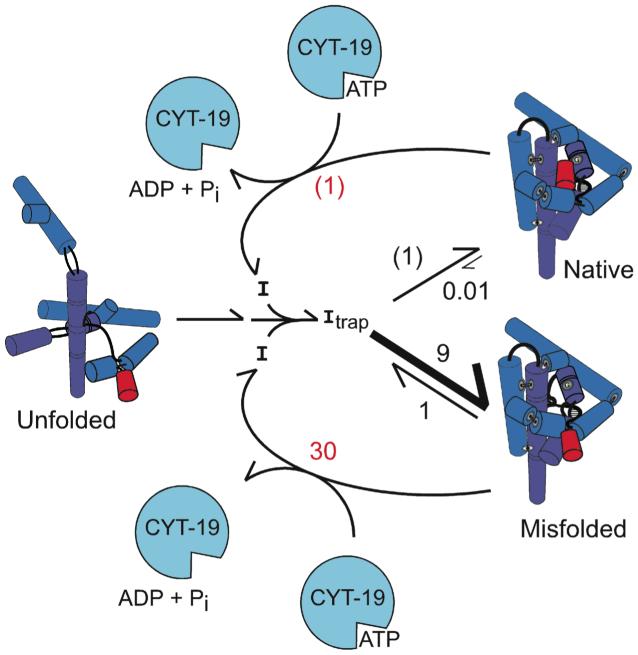
All of the results are consistent with the model shown in Fig. 4. CYT-19 unfolds both the native and misfolded species, with efficiencies that depend on their relative stabilities, and then allows folding to proceed along the same pathway that dominates in its absence22. This generates an ATP-dependent steady state in which the misfolded species, and presumably earlier folding intermediates, are populated to much larger extents than at equilibrium because their formation is favored kinetically during folding. It is intriguing that CYT-19 does not appear to affect subsequent folding of the ribozyme even though folding proceeds through intermediates that are, by definition, less stable than the native and misfolded species. Most likely, CYT-19 transiently unfolds these intermediates also, but the unfolding is not observed because the same intermediates are readily re-formed. Physical studies, probably at the single molecule level, will be necessary to explore the intermediates that result directly from CYT-19 action.
Figure 4.
Model for chaperone activity. CYT-19 generates ribozyme intermediates (I) and then allows them to fold again, thus increasing the population of less stable intermediates that are kinetically favored. Values are for the P5a variant, normalized by native ribozyme unfolding (red; (1) indicates 5 × 105 M-1 min-1, Supplementary Fig. 3), or ‘direct’ native state formation (black; (1) indicates 0.4 min-1, Supplementary Fig. 5). The wild-type ribozyme behaves similarly at low Mg2+ concentration, but at higher Mg2+ concentration the native species is sufficiently stable that CYT-19 unfolds it poorly and therefore can accelerate its formation from the kinetically-trapped misfolded species.
It was proposed prior to the discovery of RNA chaperones that proteins might be required to accelerate formation of the most stable conformations of RNA by facilitating transitions that require unfolding35,36, and these general ideas were made more specific a decade later in a model in which CYT-19 actively unfolds misfolded group I RNAs, allowing them multiple chances to fold properly37. Whereas it was assumed for simplicity that the native species would be impervious to further chaperone action and would therefore accumulate, here we find that CYT-19 can have the opposite effect, decreasing the fraction of native ribozyme even though it is the most stable species. The mechanism of CYT-19 action is presumably the same as proposed earlier, but because it acts on the native as well as misfolded species it changes the distribution from equilibrium to kinetic control, allowing increased population of intermediates that can form rapidly, even if they are less stable.
Implications for structured RNAs
Our results suggest that, in vivo, DExD/H-box proteins can act with sufficient breadth and efficiency to allow structured RNAs to populate a wider range of conformations than would be present at equilibrium. This redistribution of intermediates is analogous to an effect demonstrated for DEAD-box proteins using model RNA duplexes of varying stability11, and it is reasonable to imagine that depletion of the native state, rather than accumulation, is a general issue for RNA38. Indeed, co-expression of an unrelated RNA chaperone protein was shown to decrease in vivo self-splicing of several mutant group I RNAs with reduced thermostability by unfolding their native structures39. The underlying reason for this behavior is probably that RNA chaperones are unable to distinguish unambiguously between native and misfolded RNAs. Although an analogous challenge exists for protein chaperones, they can achieve a strong bias for interacting productively with misfolded species by recognizing exposed hydrophobic surfaces (ref. 40 and references therein). In contrast, native and misfolded RNAs can be highly similar in global structure and perhaps identical on their surfaces27.
Thus, RNAs that are required to populate one native structure may face selective pressure to minimize the extent to which their native structures are disrupted by DExD/H-box chaperones. Presumably, one important strategy is to ‘hyper-stabilize’ the native structures relative to alternative structures, beyond the level necessary simply to ensure their accumulation at equilibrium. Such an effect may contribute to the observation that the native state of the wild-type Tetrahymena ribozyme is ∼6 kcal/mol more stable than the misfolded conformation under standard in vitro conditions29, as this large energy gap results in the native species being unfolded by CYT-19 so infrequently compared to the misfolded species that the kinetic preference for misfolding is overcome, and CYT-19 action gives accumulation of the native state25. For many RNAs, proteins that associate to form functional complexes also presumably contribute to this energy gap.
There may be additional strategies available to structured RNAs to minimize action of DExD/H-box proteins on their native states. CYT-19 is strongly biased to unwind a duplex when it is unable to form tertiary contacts with the body of the Tetrahymena ribozyme25, suggesting that tight packing of a native structured RNA would direct CYT-19 to interact more efficiently with extended or loosely-packed misfolded structures. The quality control protein Ro selectively binds single-stranded ends of structured RNAs41, so native RNAs can presumably evade its action by protecting their ends. A further strategy, available for certain RNAs, is for the native structure to rapidly undergo an irreversible process, such as self-splicing, allowing mass action to drive the equilibrium toward the native form even if it is efficiently unfolded by chaperones.
Last, the ability of DExD/H-box proteins to increase the relative populations of less stable RNA structures is also likely to present opportunities for a broad range of RNAs whose functions require formation of multiple structures. A striking example is the spliceosome, which relies on several DExD/H-box proteins to facilitate conformational changes during its reaction cycle42,43. Some of these transitions are likely to be thermodynamically unfavorable, as exemplified by the required separation of the extensively-base-paired U4 and U6 snRNAs, and the action of DExD/H-box proteins may be necessary to prevent the more stable complex from dominating the steady-state population. MicroRNAs, which regulate gene expression by forming base-paired complexes with mRNA targets, may also use the action of chaperones to increase the sampling of alternative complexes, allowing a broader spectrum of physiological targets than would be expected from the relative stabilities of the target complexes44. This action of DExD/H-box proteins may also assist in the evolution of structured RNAs by allowing them to sample alternative structures, some of which could fortuitously possess beneficial activities45 and would then be subject to further selection for stability and activity.
Methods Summary
Materials
Ribozymes were transcribed in vitro and purified using a Qiagen RNeasy column as described25. Oligonucleotide substrate (CCCUCUA5, Dharmacon Research, Lafayette, CO) was 5′ end-labeled with [γ-32P]ATP by using T4 polynucleotide kinase and purified by nondenaturing polyacrylamide gel electrophoresis46. CYT-19 was expressed and purified essentially as described26.
Activity assay to monitor CYT-19-mediated unfolding
The fraction of native ribozyme was determined by monitoring substrate cleavage activity (Fig. 1a)25. For reactions monitoring unfolding of the native ribozyme, the ribozyme was pre-folded to the native state in the presence of 10 mM Mg2+ (50 °C, 30 min). For reactions monitoring unfolding of misfolded ribozyme and subsequent re-folding to the native state, the ribozyme was incubated briefly with Mg2+ (5-10 min at 25 °C) to trap the misfolded species22. For all reactions, unfolding was initiated by addition of CYT-19 in the presence of 2 mM ATP-Mg2+. At various times thereafter, aliquots were quenched for further CYT-19 action by adding 1 mg/ml Proteinase K and 50 mM MgCl2 (ref. 25). To allow subsequent determination of the native fraction by activity, this quench solution included 500 μM guanosine, and for reactions of the EΔP5abc ribozyme it included 250 nM P5abc RNA (ref. 47). After incubating for an additional 5-30 min, enough time for partially folded intermediates to complete folding, predominantly to the misfolded state, but not enough time for significant re-folding of misfolded ribozyme to the native state22,27, the fraction of native ribozyme was determined by adding trace 32P-labeled substrate and measuring the fraction cleaved rapidly in a burst (Fig. 1b)22,27. To give the fraction of native ribozyme, burst amplitudes were divided by 0.80-0.85 to correct for a small fraction of inactive and presumably damaged ribozyme (15-20%, ref. 29).
Error estimates
Each experiment was performed at least twice. Variability between duplicates was typically 30-50% (Fig. 1d, 3d, and Supplementary Fig. 3). In some cases, differences of up to 3-fold were observed between duplicate experiments performed with different preparations of CYT-19.
Supplementary Material
Acknowledgments
We thank Pilar Tijerina and Jacob Grohman for purification of CYT-19, Robert Coon for purification of CYT-18, Dan Herschlag and Alan Lambowitz for discussions and comments on the manuscript, and Ken Johnson for sharing an unpublished version of the simulation program Kinetic Explorer. This work was funded by grants from the Welch Foundation and the National Institutes of Health (to R.R.).
Appendix
Methods
Purification of CYT-19
CYT-19 was purified as described26 or with minor modifications. In the modified protocol, CYT-19 was expressed from Escherichia coli strain BL21 as a fusion with the maltose-binding protein (MBP). Induction was performed at 20 °C for 16 h in the presence of 1 mg/ml isopropyl-β-D-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation, resuspended in 20 mM Tris-HCl pH 7.5, 500 mM KCl, 1 mM EDTA, and 2 mM DTT, and lysed by incubating for 20 min on ice in the presence of 1 mg/ml lysozyme followed by three sonication bursts of 15 s each (power setting six with a double-stepped microtip; Branson Sonifier S-450-A; VWR Scientific). The lysate was cleared by centrifugation (20,000g for 30 min at 4 °C), polyethyleneimine (PEI) was added slowly to 0.4%, and precipitated material was removed by centrifugation (18,500g for 15 min at 4 °C). The supernatant was loaded onto a 5 ml amylose column (high-flow resin, New England Biolabs) and washed with 3 column volumes of buffer containing 20 mM Tris-HCl pH 7.5, 500 mM KCl, 1 mM EDTA, and 2 mM DTT. The column was then washed with 10 column volumes of the same buffer containing 1.5 M KCl and re-washed with 10 column volumes of the same buffer containing 500 mM KCl. The fusion protein was eluted with the same buffer containing 10 mM maltose. Peak fractions were pooled (5 ml total) and incubated overnight at 4 °C in the presence of 40 μg/ml tobacco etch virus (TEV) protease to cleave the MBP tag. To remove the cleaved tag, the preparation was loaded on a 2 ml ceramic hydroxyapatite column (CHT, Bio-Rad Laboratories, Hercules, CA). This step differed from the earlier protocol26, which used a gravity-flow hydroxyapatite column followed by a second application to the amylose column. The CHT column was washed with 40 column volumes of 20 mM potassium phosphate pH 7.3, 200 mM NaCl, and CYT-19 was eluted with 500 mM potassium phosphate pH 7.3. Peak fractions were pooled (3 ml), dialyzed overnight at 4 °C against 50 volumes of storage buffer (20 mM Tris-HCl pH 8.5, 500 mM KCl, 1 mM EDTA, 0.2 mM DTT, and 50% (v/v) glycerol), and stored at -80 °C.
Activity assay to monitor CYT-19-mediated unfolding
The ribozyme was first incubated to give the desired starting conformation. Wild-type and variant ribozymes (2 μM) were incubated in the presence of 10 mM Mg2+ to give predominantly the native conformation (50 °C for 30 min)48 or predominantly the misfolded conformation (25 °C, 10 min, which gives ∼90% misfolded ribozyme and 10% native ribozyme22). For each reaction, the starting fraction of native ribozyme was determined by adding trace 32-P-labeled substrate to an aliquot and processing it as described below. These reactions confirmed that the incubations gave populations of predominantly native or misfolded ribozyme as expected. Incubation of the EΔP5abc ribozyme at 50 °C gave only ∼60% native ribozyme, with the remainder misfolded, also as expected from previous work29.
To initiate unfolding, CYT-19 was added at 25 °C. For control reactions in the absence of CYT-19, an equivalent volume of the storage buffer for CYT-19 was added so that all reactions were performed under the same solution conditions. Final concentrations were 200 nM ribozyme, 50 mM Na-MOPS, pH 7.0, 5 mM Mg2+, 2 mM Mg2+-ATP (all Mg2+ added as MgOAc). From addition of CYT-19 or equivalent buffer, all reactions also contained 2 mM Tris-HCl pH 8.5, 50 mM KCl, 0.1 mM EDTA, 0.02 mM DTT, and 5% (v/v) glycerol.
At various times, aliquots were ‘quenched’ by diluting 3-fold into a solution containing high MgCl2 concentration (50 mM final concentration), which slows re-folding of the misfolded ribozyme to immeasurable levels22 and blocks CYT-19 activity (see Supplementary Fig. 1). This solution also included 50 mM Na-MOPS, pH 7.0, 500 μM guanosine, and for reactions of the EΔP5abc ribozyme, it included 250 nM P5abc RNA.
After a further incubation of 5-30 min at 25 °C, the fraction of native ribozyme was determined by adding a trace amount of 32P-labeled substrate (CCCUCUA5). Because the substrate binds both the misfolded and native species of the ribozyme but is cleaved only by the native ribozyme, a burst of product formation is observed with the amplitude providing a measure of the fraction of native ribozyme. For each ribozyme variant, in initial experiments aliquots of this cleavage reaction were quenched at various times thereafter by adding two volumes of a solution containing 70% formamide and 100 mM EDTA, thus giving a complete reaction time course. For both the P5a variant and the EΔP5abc ribozyme upon addition of P5abc34, cleavage under these conditions was rapid (>10 min-1), similar to the wild-type ribozyme, and thus the amplitude of the burst was obtained in subsequent experiments by performing a single reaction time of 1 min (ref. 25). This time is sufficient for complete cleavage by the native ribozyme but not for significant dissociation of substrate from the misfolded ribozyme22. For reactions of the EΔP5abc ribozyme in complex with CYT-18 protein, cleavage was somewhat slower (∼1-2 min-1, data not shown), and therefore a reaction time point of 2 min was used. For all reactions, quenched time points were processed by separating labeled substrate and the shorter oligonucleotide product by 20% denaturing polyacrylamide gel electrophoresis (PAGE), and quantitating the reaction progress using a phosphorimager.
Measurement of folding parameters for P5a mutant in the absence of CYT-19
For use in simulations of CYT-19-mediated unfolding and subsequent re-folding of the P5a mutant, the initial folding parameters were measured using ribozyme activity as described previously20. Tertiary folding (660 nM ribozyme) was initiated by adding Mg2+ in the presence of 750 μM guanosine and a small excess of 32P-labeled substrate (1 μM). At various times, reactions were quenched by adding two volumes of a solution containing 70% formamide and 100 mM EDTA, and labeled substrate and product were separated by 20% denaturing PAGE and analyzed as above. The progress of the cleavage reaction reflects the progress of folding, as control experiments (not shown) indicated that substrate cleavage by this ribozyme variant is at least 10 min-1 with Mg2+ concentrations ≥ 10 mM, substantially faster than folding under the same conditions. Thus, folding is rate-limiting for cleavage, so the progress of cleavage gives the rate constant for folding and the fraction of the ribozyme that folds directly to the native state, avoiding misfolding (see Scheme 1).
References From Methods
- 48.Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
References
- 1.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 2.Jankowsky E, Fairman ME. RNA helicases - one fold for many functions. Curr. Opin. Struct. Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 4.Shuman S. Vaccinia virus RNA helicase: an essential enzyme related to the DE-H family of RNA-dependent NTPases. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10935–10939. doi: 10.1073/pnas.89.22.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 6.Pang PS, Jankowsky E, Planet PJ, Pyle AM. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linder P. Dead-box proteins: a family affair-active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers GW, Jr., Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 9.Fairman ME, et al. Protein displacement by DExH/D RNA helicases without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 10.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat. Struct. Mol. Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 11.Yang Q, Fairman ME, Jankowsky E. DEAD-box-protein-assisted RNA structure conversion towards and against thermodynamic equilibrium values. J. Mol. Biol. 2007;368:1087–1100. doi: 10.1016/j.jmb.2007.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 13.Huang HR, et al. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl. Acad. Sci. U.S.A. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 16.Zarrinkar PP, Williamson JR. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 17.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 18.Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 19.Russell R, Millett IS, Doniach S, Herschlag D. Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nat. Struct. Biol. 2000;7:367–370. doi: 10.1038/75132. [DOI] [PubMed] [Google Scholar]
- 20.Russell R, Herschlag D. New pathways in folding of the Tetrahymena group I RNA enzyme. J. Mol. Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- 21.Pan J, Deras ML, Woodson SA. Fast folding of a ribozyme by stabilizing core interactions: Evidence for multiple folding pathways in RNA. J. Mol. Biol. 2000;296:133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 22.Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: Commitment to form the native conformation is late in the folding pathway. J. Mol. Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 23.Treiber DK, Williamson JR. Concerted kinetic folding of a multidomain ribozyme with a disrupted loop-receptor interaction. J. Mol. Biol. 2001;305:11–21. doi: 10.1006/jmbi.2000.4253. [DOI] [PubMed] [Google Scholar]
- 24.Russell R, et al. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tijerina P, Bhaskaran H, Russell R. Non-specific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grohman JK, et al. Probing the mechanisms of DEAD-box proteins as general RNA chaperones: The C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry. 2007;46:3013–3022. doi: 10.1021/bi0619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell R, et al. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J. Mol. Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 28.Del Campo M, et al. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol. Cell. 2007 doi: 10.1016/j.molcel.2007.07.028. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson TH, Tijerina P, Chadee AB, Herschlag D, Russell R. Structural specificity conferred by a group I RNA peripheral element. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10176–10181. doi: 10.1073/pnas.0501498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battle DJ, Doudna JA. Specificity of RNA-RNA helix recognition. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11676–11681. doi: 10.1073/pnas.182221799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joyce GF, van der Horst G, Inoue T. Catalytic activity is retained in the Tetrahymena group I intron despite removal of the large extension of element P5. Nucleic Acids Res. 1989;17:7879–7889. doi: 10.1093/nar/17.19.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambowitz AM, Caprara MG, Zimmerly S, Perlman PS. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor; New York: 1999. pp. 451–485. [Google Scholar]
- 33.Mohr G, Caprara MG, Guo Q, Lambowitz AM. A tyrosyl-tRNA synthetase can function similarly to an RNA structure in the Tetrahymena ribozyme. Nature. 1994;370:147–150. doi: 10.1038/370147a0. [DOI] [PubMed] [Google Scholar]
- 34.Russell R, Tijerina P, Chadee AB, Bhaskaran H. Deletion of the P5abc peripheral element accelerates early and late folding steps of the Tetrahymena group I ribozyme. Biochemistry. 2007;46:4951–4961. doi: 10.1021/bi0620149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karpel RL, Miller NS, Fresco JR. Mechanistic studies of ribonucleic acid renaturation by a helix-destabilizing protein. Biochemistry. 1982;21:2102–2108. doi: 10.1021/bi00538a019. [DOI] [PubMed] [Google Scholar]
- 36.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 37.Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 38.Mahen EM, Harger JW, Calderon EM, Fedor MJ. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Mol. Cell. 2005;19:27–37. doi: 10.1016/j.molcel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 39.Grossberger R, et al. Influence of RNA structural stability on the RNA chaperone activity of the Escherichia coli protein StpA. Nucleic Acids Res. 2005;33:2280–2289. doi: 10.1093/nar/gki515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Z, Rye HS. GroEL-mediated protein folding: making the impossible, possible. Crit. Rev. Biochem. Mol. Biol. 2006;41:211–239. doi: 10.1080/10409230600760382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–539. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 43.Schwer B. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 2001;8:113–116. doi: 10.1038/84091. [DOI] [PubMed] [Google Scholar]
- 44.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38:S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 45.Schultes EA, Bartel DP. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science. 2000;289:448–452. doi: 10.1126/science.289.5478.448. [DOI] [PubMed] [Google Scholar]
- 46.Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 47.van der Horst G, Christian A, Inoue T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc. Natl. Acad. Sci. U.S.A. 1991;88:184–188. doi: 10.1073/pnas.88.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.