Abstract
Aneuploid cells are frequently observed in human tumors, suggesting that aneuploidy may play an important role in the development of cancer. In this review, I discuss the processes that may give rise to aneuploid cells in normal tissue and in tumors. Aneuploid cells may arise directly from diploid cells through errors in chromosome segregation, as a consequence of incorrect microtubule-kinetochore attachments, or through failure of the spindle checkpoint. A second route to formation of aneuploid cells is through a tetraploid intermediate, where division of tetraploid cells can yield very high rates of chromosome missegregation as a consequence of multipolar spindle formation. Diploid cells may become tetraploid through a variety of mechanisms, including endoreduplication, cell fusion, and cytokinesis failure. Although aneuploid cells may arise from either diploid or tetraploid cells, the fate of the resulting aneuploid cells may be distinct. It is therefore important to understand the different pathways that can give rise to aneuploid cells, and how the varied origins of these cells affect their subsequent ability to survive or proliferate.
Keywords: Aneuploidy, tetraploidy, chromosome nondisjunction, spindle checkpoint, cytokinesis
1. Introduction
The majority of cells in human tissues exist in a diploid state, with each cell possessing two homologous versions of each chromosome. Exceptions exist, however, as some normal tissues, including liver and heart, contain tetraploid genomes [1], whereas some normal cells in the central nervous system contain an aneuploid chromosome content [2], having gained or lost a few chromosomes. In contrast, cancer cells frequently contain nondiploid chromosome numbers (reviewed in [3]). In some cases, these aberrant chromosome contents are stable, suggesting the cell underwent a single cataclysmic event, but retains the cellular machinery to accurately duplicate and segregate its genome during subsequent divisions. In other aneuploid cancers, chromosome contents may change rapidly over rounds of cell division, reflecting ongoing defects in the ability of the cell to replicate or segregate its genome [4]. This form of genetic instability, in which the principle defect is a rapidly changing whole chromosome number, is referred to as “chromosome instability” or CIN, in contrast to other forms of genetic instability, such as microsatellite instability, which occur at the DNA sequence level. It is important to note that aneuploidy and CIN are distinct concepts. Aneuploidy is a state, which may or may not change as cells divide, whereas CIN implies a high rate of change in chromosome composition.
A variety of defects have been proposed to be responsible for CIN, including defects in the spindle checkpoint [5], defective chromosome cohesion [6], upregulation of cyclins [7], and erosion of telomeres [8], to name a few. However, it is becoming clear that the CIN phenotype might also be influenced by how cells respond to chromosome missegregation [9, 10]. If abnormal cells are eliminated efficiently, a cell population may be capable of maintaining a stable karyotype, even if cells missegregate chromosomes at a high frequency.
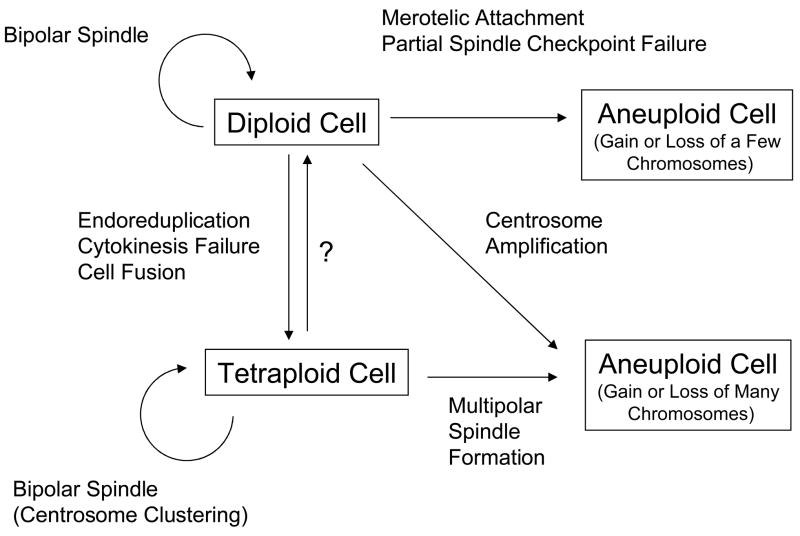
The potential relationships between diploid, aneuploid, and tetraploid cells are complex (Figure 1). In this review, I discuss some of the processes that give rise to aneuploid or tetraploid cells, and consider the potential relationships between these processes. Evidence suggests that the fate of aneuploid and tetraploid cells is likely to be distinct, and thus it is important to understand the different settings in which each of these cells is generated. Furthermore, different selective pressures may influence the survival or proliferation of aneuploid cells that arise through these different mechanisms. In the end, a fuller understanding of the causes and consequences of chromosome missegregation may enable us to detect malignant cells at an earlier stage, and may also provide new opportunities for therapeutic intervention.
Figure 1.
Summary of the potential relationships between diploid, tetraploid, and aneuploid cells, and some of the processes that may be involved. A process of conversion of tetraploid cells to diploid cells may exist but has not been well characterized (question mark).
2. Aneuploid Cells that Arise from Diploid Cells
There are several mechanisms that can give rise to aneuploid cells within a single round of cell division, including inappropriate attachment of chromosomes to the mitotic spindle, partial inactivation of spindle checkpoint proteins, and amplification of centrosomes. Here I discuss these mechanisms and the potential consequences of this form of missegregation for the development of cancer.
2. 1 Merotelic Attachment
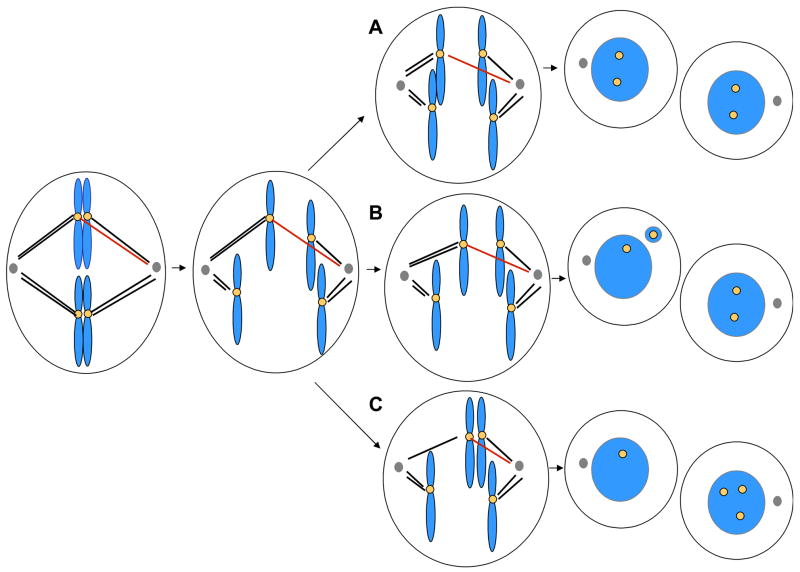
Merotelic kinetochore attachments have been proposed to represent a major source of aneuploidy in cultured mammalian cells (reviewed in [11]). Merotelic attachment is defined by the persistent attachment of microtubules from both spindle poles to a single kinetochore (Figure 2). Although a large fraction of merotelic attachments are resolved at the metaphase-anaphase transition [12], those that persist in anaphase may affect the fate of the segregating chromatid in one of three ways. Most often, the sister chromatid moves toward the appropriate pole, typically in the direction that contains more attached microtubules [13], without resulting in chromosome missegregation. Second, if the number of attached microtubules from both poles is nearly equivalent, the chromatid will not move in either direction at anaphase, producing a lagging chromatid. The chromatid may not be incorporated into either nucleus, yielding a micronucleus, in a process otherwise referred to as chromosome loss (Figure 2B). Less commonly, the lagging chromatid may be incorporated into the inappropriate daughter nucleus (Figure 2C), yielding a nondisjunction event.
Figure 2.
Potential outcomes of merotelic attachment. Merotelic attachment occurs when microtubules from opposite spindle poles interact with the same kinetochore. In this figure, the red microtubules are merotelically attached. In anaphase, the merotelically attached chromatid may move to the proper pole (A), yielding two normal diploid cells. Alternatively, if the ratio of microtubules emanating from both poles is close to one, a lagging chromatid may be produced in anaphase, which may be incorporated into a micronucleus as the cell exits mitosis (B). The micronucleus may end up in either cell. Finally, it is possible that the lagging chromatid may in some cases be incorporated into the incorrect daughter nucleus, yielding a nondisjunction event (C). In the figure, the yellow circles indicate the staining pattern one would observe using centromeric FISH probes. Blue represents chromosomes in mitosis or chromatin in interphase cells. Microtubules are indicated as black or red lines; centrosomes are shown as gray circles.
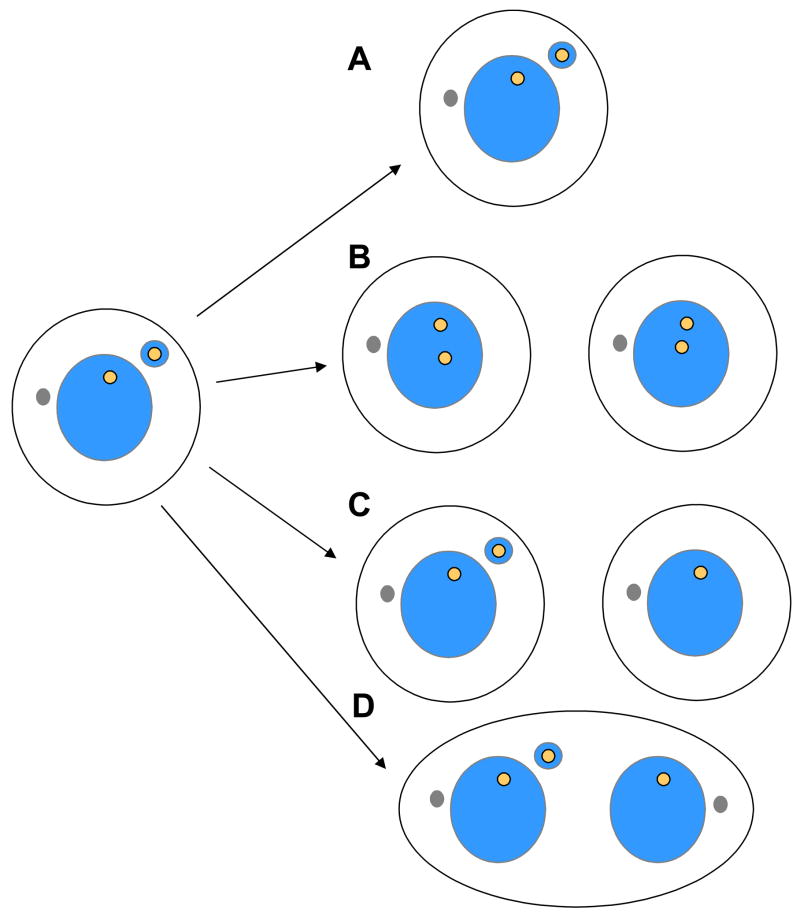
The potential fates of micronuclei that arise from merotelic kinetochore attachment have not been well characterized, but there are several possibilities (Figure 3). An important question is whether a chromatid located in a micronucleus will undergo appropriate DNA replication, and if so, whether it will condense normally and be segregated properly in the subsequent division. One study found that the X-chromosome in a lymphocyte micronucleus can undergo multiple rounds of replication without segregation [14]. Alternatively, the chromatid within the micronucleus may not replicate properly, perhaps due to impairment of the DNA replication machinery within the altered geometry of the micronucleus. Finally, it is posible that merotelic attachment might induce chromosome damage as a consequence of centromeric stretching, which has been observed in a subset of cases of merotelic attachment [13]. In either of these two situations, the cell containing a micronucleus may arrest in interphase secondary to activation of the DNA replication or DNA damage checkpoint. Alternatively, the cell may enter the next mitosis with an incompletely replicated micronucleus, which may subsequently induce chromosome bridging. In this model, micronucleus formation induced by merotelic attachment may set the stage for chromosome bridging to occur in the next mitosis, explaining why these events are frequently observed together within cell lines. Developing a better understanding of the fate of chromosomes in micronuclei, and of the cells that contain them, is an important area for future research.
Figure 3.
Potential fates of a cell containing a micronucleus. (A) A cell containing a micronucleus may fail to divide, perhaps as a consequence of activation of a DNA replication or DNA repair checkpoint pathway. It may also undergo apoptosis (not shown). (B) Alternatively, if the cell divides, the chromosome in the micronucleus may replicate normally. If it is also segregated properly in the next mitosis, two diploid cells may be produced. (C) The micronucleus may fail to be replicated and segregated. This may yield one daugter cell that retains the micronucleus, and one daughter cell that has lost the micronucleus. (D) The micronucleus may be replicated, but may not be segregated properly in the following mitosis, activating pathways that inhibit cytokinesis completion, producing a binucleated cell.
2.2 Partial Loss of Spindle Checkpoint Function
A second mechanism that can lead directly to loss or gain of a few chromosomes during mitosis is partial impairment of the spindle checkpoint. This checkpoint normally inhibits anaphase initiation until all chromosomes have achieved proper bipolar attachment on the metaphase plate (for review, see [15]). Complete loss of the spindle checkpoint is lethal [16–18], but mice lacking a single copy of checkpoint proteins such as Mad2, BubR1 or CENP-E are viable [18–21]. In all cases, reduction of expression of these genes leads to induction of aneuploidy, although the degree of aneuploidy, and also the prevalance of chromosome gains and losses, prove to be both gene- and dosage-specific. In most cases, the loss of spindle checkpoint proteins is associated with an increased rate of tumorigenesis, suggesting that aneuploidy may promote tumorigenesis. However, the relationship between aneuploidy and tumorigenesis may be more complex, as induction of aneuploidy can suppress tumorigenesis in certain circumstances [21].
2.2.1 Reduction of Mad2
Mad2 is a central component of the spindle checkpoint, and homozygous deletion of Mad2 in mice is lethal to the early embryo as a consequence widespread chromosome missegregation and induction of apoptosis [16]. The consequences of inactivation of a single copy of Mad2 were therefore evaluated [19]. Murine embryonic fibroblasts (MEFs) cultured from mice lacking a single copy of Mad2 showed premature sister-chromatid separation and a significant increase in the number of aneuploid cells [19]. However, rates of aneuploidy in circulating lymphocytes or other tissues were not evaluated in this study. Deletion of one copy of Mad2 from an otherwise genetically stable human colon cancer cell line was sufficient to functionally inactivate the spindle checkpoint and induce premature sister chromatid separation in the presence of nocodazole [19]. Furthermore, loss of a single copy of Mad2 was sufficient to produce an 80% increase in the frequency of aneuploid metaphases. Thus in both mouse cells and human cells, both copies of Mad2 are essential for ensuring accurate chromosome segregation and spindle checkpoint function.
Despite the elevated rates of chromosome missegregation observed in derived MEFs, mice lacking a single copy of Mad2 were found to develop normally [19]. However, these mice developed lung tumors at high rates after a long latency, but the incidence of other tumors, such as lymphomas was not increased [19]. This study was therefore the first to suggest that aneuploidy resulting from spindle checkpoint inactivation could increase the incidence of formation of certain types of tumors. However, if aneuploidy is increased in all tissues following loss of a single copy of Mad2, it is unclear why the tissue spectrum of induced tumors is so narrow. Perhaps elimination of a single copy of Mad2 increases the rate of aneuploidy in lung tissue more than others. Alternatively, lung tissue may have a distinct response to an increased rate of aneuploidy. The issues of gene-specific and tissue-specific effects have become recurring themes in similar studies of other checkpoint proteins, as described below.
2.2.2 Reduction of BubR1
BubR1 is a protein kinase that plays an essential role in spindle checkpoint function. Like Mad2, homozygous deletion of BubR1 is lethal at early embryonic stages [17, 18], and thus the effects of partial loss of BubR1 have been evaluated. Mice with a single copy of BubR1 have been reported to have either no phenotype [18], or to have splenomegaly with increased circulating megakaryocytes [17] and an increased susceptibility to carcinogen-induced tumor formation [20]. In one study, MEFs isolated from the mice showed increased numbers of micronuclei but also a significant increase in the number of polyploid cells [17]. Further reduction in BubR1 levels, using a hypomorphic allele, resulted in a high incidence of aneuploidy in liver cells and derived MEFs, but did not yield an increase in the rate of spontaneous tumor formation [18]. Instead, these mice developed signs of premature aging [18, 22, 23], and MEFs derived from these mice showed profound premature cellular senescence [18]. BubR1 levels were found to decline in ovary, testis, and spleen as wild-type mice aged, suggesting that BubR1 may also be an important regulator of the aging process [24]. Thus despite the fact that both Mad2 and BubR1 are important components of the spindle checkpoint and are important for normal chromosome segregation, inactivation of these proteins can give rise to distinct phenotypes in the organism.
2.2.3 Reduction of Rae1 and Bub3
The protein complex Rae1-Nup98 has also been implicated in regulating anaphase onset, perhaps by inhibiting the degradation of securin by the Anaphase-Promoting Complex/Cyclosome (APC/C) [25]. Mice harboring a single copy of both Rae1 and Nup98 showed an increased rate of aneuploidy in splenocytes, and a premature chromatid separation phenotype [25]. Similarly, mice containing a single copy of Rae1 and the spindle checkpoint gene Bub3 [26] also showed a very high degree of chromosome missegregation. Like BubR1-deficient mice, these mice showed signs of early aging, but did not develop tumors [27].
The similarity in phenotypes between BubR1-deficient mice, and mice lacking Rae1 and Bub3, suggested that high rates of aneuploidy might be sufficient to trigger cell senescence and aging. However, rates of aging and aneuploidy did not correlate in a way that supported this hypothesis. Mice lacking a copy of Bub3 and Rae1 showed a high degree of aneuploidy in splenocytes (36%) at 5 months of age, yet showed no signs of premature aging [27]. In contrast, BubR1 hypomorphic mice showed a lower degree of aneuploidy in splenocytes (15%) at 5 months, yet exhibited signs of premature aging [18]. In support of these findings, MEFs from the Bub3/Rae1 mice showed a lower rate of senescence, despite being more aneuploid than MEFs derived from BubR1 hypomorphs. Therefore, it was concluded that aging is unlikely to be a direct consequence of aneuploidy itself, but may instead be a distinct consequence of spindle checkpoint inactivation. The trigger for senescence and premature aging in mice lacking BubR1 remains unclear, but appears to be p53-dependent [18].
2.2.3 Reduction of CENP-E
CENP-E is an essential kinesin-family motor protein that plays roles in chromosome-microtubule interactions [28–30], chromosome congression [31], and spindle checkpoint signaling [32, 33]. Cells lacking CENP-E exhibit defects in chromosome congression and spindle checkpoint function such that cells enter anaphase despite the presence of chromosomes remaining near the poles in 25% of cells [34]. Since CENP-E knockout mice are not viable [30], the effects of loss of CENP-E on the development of aneuploidy and tumor formation have been studied in heterozygous mice [21]. MEFs cultured from mice lacking a single CENP-E allele showed a 60% rate of aneuploidy by twelve days of culture, although the degree of aneuploidy of wild-type MEFs was surprisingly high (over 30%) [21]. Spectral karyotyping indicated that the aneuploid cells contained numerical but not structural chromosome changes, and showed no evidence of DNA damage. MEFs lacking a single copy of CENP-E grew at normal rates, but showed decreased activity in colony forming assays compared to wild-type cells.
Age-dependent accumulation of aneuploidy in lymphocytes was accelerated in CENP-E+/− animals [21]. More than 50% of lymphocytes from CENP-E+/− animals were aneuploid by 10 months of age, whereas lymphocytes from wild-type mice showed 25% aneuploid cells. In these experiments, chromosome losses predominated, with few chromosome gains observed, for reasons that are unclear. Spleen cells also showed an increase in the aneuploid fraction, again with chromosome losses predominating. Aneuploidy was also observed in cells in the colon at rates 4–6 fold higher in mice lacking a copy of CENP-E, compared to wild-type controls. By 19–21 months of age, 10% of CENP-E heterozygous mice contained a lymphoma in the spleen, whereas none were observed in wild-type mice. How loss of CENP-E promotes tumorigenesis is unclear, as the karyotype and the CENP-E status of the tumors was not reported [21]. However, the tumors may be polyploid as malignant cells contained nuclei that were 4- to 6-fold larger than normal lymphocytes [21]. CENP-E heterozygous mice also showed a three-fold increase in the rate formation of adenomas of the lung, but the karyotype and CENP-E status of the tumor cells was not reported, and thus it remains to be determined precisely how aneuploidy induced by loss of a single CENP-E allele might lead to tumor formation.
Surprisingly, the incidence of liver tumors was reduced in CENP-E heterozygotes compared to wild-type animals, although the difference was not statistically significant [21]. Furthermore, those liver tumors that did arise in CENP-E heterozygotes were significantly smaller than those in wild-type animals, suggesting that aneuploidy may slow the growth of these tumors. CENP-E heterozygotes also showed a trend towards resistance to carcinogen-induced lung tumors compared to wild-type mice. In CENP-E heterozygotes also lacking the tumor suppressor gene p19, tumors took significantly longer to form in CENP-E heterozygotes, suggesting that high rates of aneuploidy can slow the rate of formation of sarcomas and lymphomas that are prevalent in this tumor model. Together these findings indicate that a high rate of aneuploidy can, in certain contexts, antagonize the development or growth of tumors.
2.2.4 Reduction of Bub1
Bub1 is another spindle checkpoint protein that is essential for viability in mice, and thus the consequences of loss of Bub1 have been evaluated through analysis of hypomorphic alleles [10]. Splenocytes from five-month old mice showed increasing rates of aneuploidy, including chromosome gains and losses, from 1% aneuploid cells in wild-type mice, to 39% aneuploid cells in mice expressing the lowest levels of Bub1. A fraction of these cells also displayed a premature sister chromatid segregation (PMSCS) phenotype. A similar trend was observed with MEFs obtained from these mice, although PMSCS was not observed in this case.
Interestingly, cells that missegregated chromosomes died at a much higher rate in the presence of Bub1 than its absence. For example, in wild-type cells, 94% of cells that missegregated chromosomes showed cell death, whereas only 32% of such cells died if low amounts of Bub1 were present [10]. Micronuclei were also found to accumulate in the cells expressing low levels of Bub1, suggesting that Bub1 could be responsible for inducing a micronucleus-induced cell death pathway. Interestingly, cells with reduced levels of Bub1 showed no difference in sensitivity to DNA damaging agents or microtubule inhibitors [10], suggesting that the role of Bub1 could be quite specific.
Unlike the case for BubR1 mutant mice, there was no premature aging observed in Bub1 mutant mice [10]. However, mice expressing low levels of Bub1 were significantly more prone to spontaneous tumors than wild-type mice. Interestingly, mice expressing different levels of Bub1 developed different types of tumors: those expressing the lowest amount of Bub1 developed more sarcomas, lymphomas, and lung tumors, whereas mice expressing somewhat higher Bub1 levels developed sarcomas and liver tumors but not lymphomas or lung tumors [10]. Mice expressing even higher levels of Bub1 (Bub1 heterozygotes) showed a trend toward decreased tumor formation, particularly in liver and lung tissue. However, these mice were more susceptible to carcinogen-induced tumor formation. These data suggest that there are strong dose-dependent effects on both the tissue dependence and the prevalence of tumor formation. Whether these distinct patterns of tumor formation are a consequence of different levels of aneuploidy, or arise instead from the role that Bub1 may play in eliminating aneuploid cells, remains to be determined.
2.3 Centrosome Amplification
Another process that has the potential to directly yield aneuploid cells from diploid cells is the amplification of centrosomes. Although this process may be unlikely to occur in normal diploid cells, it may occur in tumor cells that overexpress regulators of centrosome duplication such as Aurora A [35–37], Polo kinases [38], cyclin-dependent kinases [39–41], and others [42–44]. Centrosome amplification during interphase may induce multipolar spindle formation in mitosis, leading to missegregation of multiple chromosomes. As mechanisms of centrosome amplification have been discussed in other reviews [45, 46], this topic will not be discussed further here.
2.4 Fates of Aneuploid Cells Arising from Diploid Cells
The studies described above indicate that aneuploid cells can accumulate at high frequencies in mice that are heterozygous in spindle checkpoint genes. However, the type of aneuploidy that predominates in each of the mutant backgrounds can be distinct. For example, aneuploid splenocytes and lymphocytes from CENP-E heterozygotes show almost exclusively chromosome loss [21], whereas aneuploid splenocytes from animals with reduced levels of Bub1, BubR1, or Rae1 and Bub3, show both chromosome loss and chromosome gain [18, 27]. These findings suggest that the latter group of proteins may help eliminate cells that have gained a chromosome, although Bub1 is the only protein that has been shown to affect the fate of cells with missegregation [10].
Although nontransformed mouse cells seem to be able to tolerate high degrees of aneuploidy, it is unclear whether the same is true of human cells. Induction of high rates of missegregation in a chromosomally stable human cancer line, using small molecules that increase the frequency of merotelic attachment, was not sufficient to generate cells with highly abnormal karyotypes [9]. Thus selective pressures against aneuploid cells may be stronger in human cells than in mouse cells.
Recent work in budding yeast indicates that chromosome gains may put cells at a selective disadvantage [47]. Haploid yeast that contain an extra copy of one of a number of different chromosomes show a series of common defects including delayed cell cycle progression, increased glucose uptake, and increased sensitivity to inhibitors of protein synthesis and folding. The effects of chromosome gains in a diploid background were more modest [47], and thus it is unclear whether a mammalian cell that has gained a chromosome through the processes described above would similarly be at a selective disadvantage compared to its diploid counterparts. Furthermore, the effect of loss of a specific chromosome in an otherwise diploid background was not addressed. Nevertheless, this work highlights the possibility that imbalanced protein expression may impose a fitness reduction on cells, perhaps explaining why high degrees of aneuploidy may suppress development or growth of tumors [21].
3. Sources of Tetraploid Cells
The data presented thus far support the notion that chromosome missegregation in diploid cells can lead directly to the formation of aneuploid cells. However, other data suggest that spontaneous chromosome missegregation may instead be associated with production of tetraploid cells, which may subsequently become aneuploid through further division. Because the behavior of tetraploid cells is likely to be distinct from diploid or near-diploid aneuploid cells, it is important to understand the potential relationships between these processes. Tetraploid cells may arise from diploid cells through a number of different mechanisms, including cell fusion, endoreduplication, and cytokinesis failure (reviewed in [1],[3]). Here I focus on the relationship between chromosome missegregation and cytokinesis failure, providing some potential mechanistic explanations for links between these processes.
3.1 A Correlation Between Chromosome Nondisjunction and Cytokinesis Failure
A correlation between chromosome nondisjunction and cytokinesis failure emerged from the comparison of nondisjunction rates in mitotic cells and spontaneously-arising binucleated cells [48]. In telomerase-immortalized keratinocytes, we observed that the rate of chromosome nondisjunction was 0.05–0.1% per chromosome per division in the entire mitotic population, but was 80–160 fold higher in spontaneously-arising binucleated cells. Similar results were observed in HeLa cells and immortalized prostate epithelial cells. Live cell imaging experiments indicated that the majority of binucleated cells arose through a process of cytokinesis failure, at very late stages of division, near the time of abscission. Elevated rates of chromosome missegregation in these cells were directly confirmed by combining live-cell imaging followed by FISH.
A possible explanation for these findings was the presence of chromatin in the cleavage furrow, which might impair cytokinesis completion, perhaps by strengthening the cytoplasmic bridge, as first proposed by Mullins and Biselle [49]. However, clear evidence of chromatin bridging was observed in less than 20% of cells that failed cytokinesis [48]. Although very thin chromatin bridges might not have been detected in these experiments, these findings suggested that mechanical strengthening of the cleavage furrow by chromatin was unlikely to explain cytokinesis failure in most cases. Lagging chromosomes were also present in 30% of cells that became binucleated [48]. However, an earlier study indicated that chromosome lagging that occurred as a consequence of merotelic attachment was associated with delay in the late stages of cytokinesis but not with cytokinesis failure [50]. We observed that the nondisjunction frequency, rather than the frequency of bridging and lagging chromosomes, was more closely correlated with the rate of cytokinesis failure, in both HeLa cells and immortalized diploid keratinocytes [48]. Therefore, we proposed that there may be important mechanistic connections between the processes that induce chromosome nondisjunction and the pathways that regulate cytokinesis.
Genetic approaches discussed in the first section of this review indicate that partial spindle checkpoint inactivation leads to the direct formation of aneuploid cells rather than tetraploid cells [51]. Thus it is clear that nondisjunction is not obligatorily coupled to cytokinesis failure in all circumstances. It is therefore unlikely that cells have mechanisms to directly “sense” the presence of missegregated chromosomes, and induce cleavage failure as a result. Furthermore, these findings suggest that the spontaneous nondisjunction events that we observed in human cell lines are unlikely to be a consequence of spindle checkpoint failure. Instead, spontaneous nondisjunction is likely to be caused by distinct mechanisms that are associated with cytokinesis failure rather than with cytokinesis completion. Here I describe some mechanisms that could explain the correlation between nondisjunction and cytokinesis failure.
3.2 The “NoCut” Pathway
A fraction of cells that undergo chromosome nondisjunction and cytokinesis failure in human cell lines show the presence of a bridging chromosome that spans the cleavage furrow, or lagging chromosomes that remain very close to the cleavage furrow [48]. What are the potential mechanisms by which bridging chromosomes could induce cytokinesis failure? A study in budding yeast identified a pathway, referred to as the “NoCut” pathway, that delays the final stage of cytokinesis, termed abscission, in cells that have mitotic spindle defects [52]. Absence of this pathway led to formation of DNA double strand breaks in a manner that depended on function of the cytokinesis machinery. This finding supports the idea that the cytokinesis machinery is indeed capable of severing DNA that may be present in the furrow. The “NoCut” pathway depends on the Aurora kinase family member Ipl1, and also on two proteins called Boi1 and Boi2. However, it remains unclear how Ipl1 becomes activated by chromatin or midzone defects, how Ipl1 regulates Boi1/2, or how these proteins might block cytokinesis completion.
Does a similar pathway regulate cytokinesis completion in other organisms? In fission yeast, a large number of “cut” mutants fail in chromosome segregation yet complete cytokinesis [53]. Fission yeast may therefore not have a “NoCut” pathway, or these cells may eventually adapt to inhibitory signals, completing cytokinesis despite activation of a similar pathway. In either case, the results suggest that in both budding and fission yeast that the cytokinesis machinery is capable of completing cytokinesis despite the presence of DNA, indicating that chromatin itself is not likely to mechanically inhibit cytokinesis completion.
In vertebrate cells, it remains unclear whether an Aurora-regulated pathway might operate in the same manner. Unlike budding or fission yeast, where Aurora kinase orthologs are dispensable for cytokinesis, Aurora B is essential for cytokinesis in metazoans (reviewed in [54]). Aurora B plays important roles in both early and late stages of cytokinesis, by phosphorylating a wide variety of proteins essential for different steps of the process. Application of an Aurora B inhibitor even at very late stages of cytokinesis will induce furrow regression [55], suggesting that Aurora B is positively required for abscission in mammalian cells. Alternatively, it has been suggested that Aurora A could negatively regulate cytokinesis completion in mammalian cells in a manner similar to that observed in budding yeast [52]. Overexpression of Aurora A has been shown to inhibit cytokinesis, although this effect surprisingly does not seem to require Aurora A kinase activity [37]. Thus while it remains possible that chromatin in the cleavage furrow could inhibit abscission through a “NoCut”-like pathway in mammalian cells, the molecular details are likely to be different from those in budding yeast.
3.3 Inactivation of a Component that is Required for Both Chromosome Segregation and Cytokinesis
Studies of the mechanisms of cytokinesis have revealed that many of the components involved in cytokinesis are also required for accurate chromosome segregation [54]. Many of these proteins localize to chromosomes, kinetochores, centrosomes, or the mitotic spindle during mitosis, and then relocalize to the cleavage furrow or midbody during cytokinesis. Examples include Polo kinase (Plk1) and the chromosome passenger complex, which consists of Aurora B, INCENP, survivin, and borealin (reviewed in [56]). Therefore, interference with the expression or regulation of these components may lead to both chromosome missegregation and failure of cytokinesis.
Curiously absent from the collection of mitotic regulators that are required for both accurate chromosome segregation and cytokinesis are the core spindle checkpoint components such as the Mad and Bub proteins. The lack of a requirement for these proteins in cytokinesis is supported by the finding that their genetic inactivation in mice is not associated with production of tetraploid cells, as discussed earlier. Although these results could be interpreted to mean that the spindle checkpoint plays no role in the regulation of cytokinesis, some data suggest instead that spindle checkpoint proteins could negatively regulate cytokinesis, as discussed below.
3.3.1 DNA Damage and Cytokinesis Failure
The activation of DNA damage pathways may provide one possible explanation for the link between chromosome missegregation and cytokinesis failure. In this model, activation of DNA damage pathways would coordinately inhibit components required for both chromosome segregation and cytokinesis completion. Such a pathway may act to prevent segregation of sister chromatids that contain regions of damaged or incompletely replicated chromatin, and simultaneously inhibit cytokinesis to prevent damage to the missegregated chromosome.
Several lines of evidence suggest that cytokinesis could be regulated in response to DNA damage. For example, some proteins involved in DNA repair, such as BRCA2, are also required for cytokinesis. BRCA2 is required for recombination-based repair of DNA double-strand breaks [57]. However, BRCA2-deficient cells also show centrosome amplification that may be a consequence of defective cytokinesis [58]. BRCA2 localizes to the midbody, a structure required for completion of cytokinesis, and inactivation of BRCA2 in murine embryonic fibroblasts and HeLa cells interferes with cytokinesis [59]. From a mechanistic perspective, it remains unclear how BRCA2 might promote normal cytokinesis, or how its cytokinesis function might be regulated in response to DNA damage. However, proteins that interact with BRCA2, such as BCCIP, have recently been shown to be required for efficient cytokinesis as well [60], supporting a functional role for BRCA2 in cytokinesis.
Other proteins involved in the DNA damage responses also bind to proteins required for cytokinesis. For example, the DNA damage checkpoint kinase Rad53 has been shown to associate with septins, proteins that are important for cytokinesis in budding yeast [61]. In mammalian cells, Ku70, a DNA-binding protein required for DNA damage repair, forms a complex with ARF6, a small GTPase that regulates vesicle trafficking events necessary for completion of cytokinesis [62].
Transcriptional controls may provide another mechanism for inhibiting cytokinesis in response to DNA damage. The expression of several cytokinesis proteins, including Plk1, ECT2, anillin, and survivin, is repressed when DNA is damaged, in a manner that depends on an intact Rb pathway [63]. Other studies suggest that expression of cytokinesis proteins may be inhibited by activation of the p53 pathway [64]. For example, ECT2 expression is repressed by p53 via protein methyltransferases, suggesting that cytokinesis could be more likely to fail under conditions of p53 activation [65].
Post-translational modifications may also regulate cytokinesis in response to DNA damage. For example, Aurora B becomes highly poly-ADP-ribosylated when DNA is damaged, a modification that inhibits its kinase activity [66]. Because Aurora B activity is essential for chromosome segregation and cytokinesis, induction of DNA damage could lead to errors in chromosome segregation and failure of cytokinesis. Interestingly, if Aurora B activity is downregulated in cells that contain DNA damage, this would also provide an explanation for a relationship between lagging chromosomes in anaphase and delayed completion of cytokinesis. Because Aurora B is required for disassembly of merotelic attachments [67, 68], cells with DNA damage and reduced Aurora B activity may be less able to disassemble merotelic attachments, increasing the number of lagging chromosomes. Inactivation of Aurora B would also increase the likelihood of cytokinesis failure, given the requirement for Aurora B kinase in cytokinesis completion.
3.3.2 Delayed DNA Replication and Cytokinesis Failure
An interesting connection between delayed DNA replication, chromosome missegregation, and cytokinesis failure has emerged from the work of Thayer and colleagues. Using a microcell-based chromosome delivery technique, it was found that a single chromosome from a human rhabdomyosarcoma could induce chromosome instability when introduced into genetically stable mouse myoblasts [69]. This chromosome contained a translocation, isochromosome 3q, that occurs frequently in a hematological disorder called persistent polyclonal B-cell lymphocytosis [70]. This disorder is characterized by elevated levels of circulating B cells [71], many of which are binucleated, suggesting that presence of this chromosome could be associated with cytokinesis failure. Characterization of mouse cells carrying human isochromosome 3q revealed that this chromosome showed delayed condensation during mitosis [72]. Interestingly, the undercondensed chromosome could only be detected in metaphase spreads of cells that were not treated with colcemid [72], perhaps because prolonged mitotic arrest in colcemid permitted the chromosome to condense. The chromosome exhibiting the delay in mitotic condensation (DMC) phenotype lacked histone H3 phosphorylation in mitosis, whereas condensed chromosomes in the same cell showed no defect in H3 phosphorylation [72]. These findings suggested that the delay in chromosome condensation did not reflect a general inhibition of Aurora B-mediated histone phosphorylation, but was instead a chromosome-specific phenomenon. The DMC phenotype was not restricted to isochromosome 3q, as chromosomes from other human tumors, containing distinct translocations, showed a similar DMC phenotype when introduced into mouse cells. Furthermore, five of seven human cancer cell lines, and five of thirteen primary tumor samples, contained chromosomes that exhibited a DMC phenotype, suggesting that the phenomenon may be widespread in tumor cells [72].
When timing of DNA replication was examined in these cells, it was found that chromosomes exhibiting DMC also showed delayed replication timing (DRT), whereas other chromosomes in the cell showed normal replication timing [72]. Both initiation and completion of DNA replication were delayed, by 2–3 hours, with DMC chromosomes continuing to replicate DNA into mitosis [73]. Delayed replication occurred across the entire chromosome, suggesting that the presence of the translocation somehow globally disrupts timing of replication across the chromosome, but in a chromosome-autonomous fashion. In support of the idea that the chromosomal translocation itself delays replication, it was found that exposing cell lines, primary blood lymphocytes, or mice to ionizing radiation (IR) resulted in the generation of chromosomes with DRT/DMC in as many as 25% of surviving cells [74]. Although DRT/DMC occurred frequently, on approximately 5% of chromosomes with interchromosomal translocations, it was not detected on the majority of chromosomes with translocations, or on non-rearranged chromosomes, suggesting that DRT/DMC occurs only in the context of specific chromosomal exchanges [74].
To better understand this phenomenon, Thayer and colleagues generated cell lines containing specific translocations using the Cre/loxP system, and found that 10% of the translocations generated cells with a DRT/DMC phenotype [75]. Their observations suggest that the replication timing of certain chromosome translocations is regulated in cis by a mechanism that results in delayed replication along the entire length of the chromosome. How translocations lead to delayed replication timing remains unclear. One possibility is that translocation leads to loss of a genetic element that is required for timely replication of the chromosome. Alternatively, specific chromosomal exchanges may generate dominant interfering elements that act in cis to delay normal chromosome replication timing by some unknown mechanism [75].
Several lines of evidence suggest that cells containing chromosomes with DRT/DMC are prone to cytokinesis failure. Introduction of isochromosome 3q into mouse myoblasts induces polyploidy and centrosome amplification that is consistent with cytokinesis failure [69]. Whereas parental C2C12 cells contained one or two centrosomes in 98% of cells, hybrid cells that contain a DRT/DMC chromosome contained >2 centrosomes in 62% of interphase cells [73]. Consistent with this finding, multipolar spindles were present in 10% of cells containing the DRT/DMC chromosome, but not in the parental cells [73]. In the analysis of mice treated with ionizing radiation, cells containing chromosome translocations associated with the DRT/DMC phenotype were typically tetraploid [74], suggesting that these cells were prone to cytokinesis failure.
Precisely how the DRT/DMC phenotype may be related to cytokinesis failure is not yet clear; however, components of the replication checkpoint may be involved. During interphase, chromosomes with a DRT/DMC phenotype stain positively for phosphorylated Chk1, an indicator of ATR-dependent replication checkpoint activation on these chromosomes [73]. During mitosis, recruitment of Aurora B to DRT/DMC chromosomes is delayed, although INCENP recruitment is not [73]. The attachment of DRT/DMC chromosomes to the metaphase plate is also delayed. Despite the delayed recruitment of Aurora B to these chromosomes, Mad2 appears to be recruited efficiently to kinetochores, suggesting that they can generate a spindle checkpoint signal [73]. It is possible that delayed recruitment of Aurora B to DRT/DMC chromosomes could lead to the formation of lagging chromosomes during mitosis, and may also impact the efficiency of cytokinesis completion as described earlier.
Another possibility is that there may be direct mechanistic connections between the DNA replication machinery and cytokinesis, such that the presence of late DNA replication directly interferes with cytokinesis completion. In vertebrate cells, Orc6, a component of the Origin Recognition Complex, localizes to kinetochores in early mitosis and to the cleavage furrow and midbody during cytokinesis [76]. Elimination of Orc6 induces mutlipolar spindles and formation of multinucleated cells in both human cells [76] and Drosophila [77], suggesting this function is conserved. In Drosophila, Orc6 interacts with a septin protein that may be important for cytokinesis. Domains of Orc6 required for DNA replication and cytokinesis appear separable, suggesting that Orc6 has evolved a domain that participates specifically in cytokinesis [77].
3.4 Telomere shortening and cytokinesis failure
Telomere shortening or dysfunction is a well-established cause of genomic instability [78], and may be associated with cytokinesis failure. Telomere erosion may lead to the fusion of chromosome ends, producing dicentric chromosomes. These chromosomes may yield bridging chromosomes during anaphase [8], which may inhibit cytokinesis completion by activating a “NoCut”-like pathway. Alternatively, these chromosomes may break during anaphase [8], leading to activation of the DNA damage pathway that may interfere with cytokinesis completion through some of the mechanisms described earlier. The potential consequences of telomere loss on cellular senescence and genetic instability have been well-covered in other reviews [78–80] and therefore will not be discussed further here.
3.5 Prolonged Spindle Checkpoint Activation and Cytokinesis Failure
Whereas partial inactivation of the spindle checkpoint may give rise to near-diploid aneuploid cells, as described earlier, most tumor cell lines have an intact spindle checkpoint, including those that exhibit chromosome instability [9, 81]. In fact, some tumors may overexpress spindle checkpoint proteins in a manner that interferes with chromosome segregation and cytokinesis, explaining the correlation between nondisjunction and cytokinesis failure. For example, expression of the spindle checkpoint protein Mad2 is regulated by E2F family proteins, which are hyperactivated in cancer cells as a consequence of inactivation of the Rb pathway [82]. Cells with inactivated Rb contain elevated Mad2, which has been associated with chromosome missegregation and cytokinesis failure [82]. Furthermore, overexpression of Mad2 is sufficient to induce chromosome missegregation but yields a large fraction of tetraploid cells [83]. In mouse models, overexpression of Mad2 is sufficient to induce a wide spectrum of tumors in a highly penetrant fashion [83].
Why is Mad2 overexpression associated with chromosome missegregation and cytokinesis failure? Elevated Mad2 levels may perturb the normal timing of mitotic proteolysis that is essential for proper anaphase and cytokinesis. By inhibiting the APC/C, elevated Mad2 levels may delay the degradation of multiple APC/C substrates, potentially interfering with activation of separase, which is essential for efficient separation of sister chromatids [84, 85]. Furthermore, failure to fully inactivate mitotic cyclin dependent kinases (CDKs) may interfere with cytokinesis initiation or completion, as CDK activity negatively regulates cytokinesis at several steps [86]. Interestingly, mitotic exit in the face of a persistently activated spindle checkpoint has been shown to perturb the degradation of substrates of APC/C-CDH1 as well, including proteins such as TPX2 [87]. Thus failure to properly inactivate the checkpoint could lead to a spectrum of changes that together perturb both chromosome segregation and cytokinesis. A second explanation for the effects of sustained checkpoint activation is that DNA may become damaged during prolonged mitotic arrest [88]. This may lead to the activation of the DNA damage response that may then induce chromosome missegregation and inhibit cytokinesis through some of the mechanisms described earlier.
4. Aneuploid Cells Arising from Tetraploid Cells
Tetraploid cells may undergo one of several distinct fates, which may depend on how they arose in the first place (Figure 4). Cells may undergo apoptosis or cell cycle arrest, or they may continue to divide. Cells that divide may do so with a bipolar spindle, generating equivalent daughter cells, or they may divide with a multipolar spindle, in which case highly aneuploid daughter cells may result.
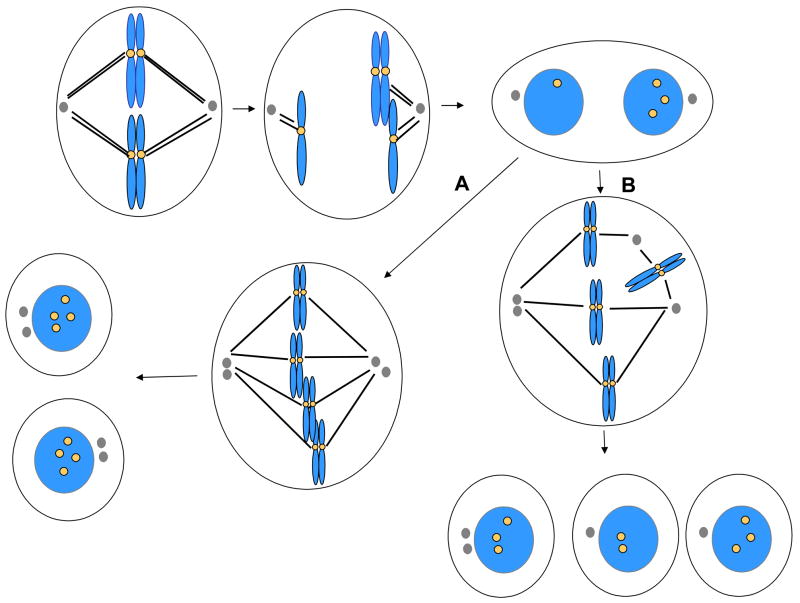
Figure 4.
Nondisjunction and cytokinesis failure, and fates of binucleated cells. Certain forms of chromosome nondisjunction are associated with cytokinesis failure, yielding a binucleated cell. These cells may arrest, or they may divide with a bipolar spindle, yielding two tetraploid, mononuclear cells with equivalent genomes (A). This process requires that cells be capable of clustering centrosomes as indicated. If centrosomes do not cluster (B), then these cells may divide with a multipolar spindle, yielding several aneuploid cells. In some cases only one or two daughter cells may be produced due to cytokinesis failure in cells containing multipolar spindles.
Long-term time-lapse imaging has been used to characterize the fate of binucleated cells that arise through spontaneous cytokinesis failure [48]. HeLa cells that fail cytokinesis divide with high frequency, although a small fraction undergo apoptosis. The vast majority of the cells divide with a multipolar spindle, producing progeny that are often inviable [48]. In contrast, the majority of diploid immortalized keratinocytes that fail cytokinesis arrest in interphase as binucleated cells. Of the limited number that go on to divide, most divide with a bipolar spindle yielding two mononuclear tetraploid daughter cells. However, some cells divide with multipolar spindles, producing aneuploid progeny. The distinct behaviors of binucleated cells in these two cell lines may be related to the status of the p53 pathway, which is inactivated in HeLa cells. Other factors that may limit the proliferation of polyploid cells have been reviewed recently [89]. Tetraploid cells appear to have a greater degree of genetic instability than diploid cells, which may explain why tetraploid cells are capable of inducing tumor formation whereas matched diploid cells are not [90]. Again, the p53-pathway may be important in limiting proliferation of tetraploid cells, as tetraploid mouse mammary epithelial cells can only be propagated in culture if the p53 gene is deleted [90].
5. Conclusions
As outlined in this review, aneuploid cells can be produced through a variety of mechanisms. Merotelic attachment of microtubules to kinetochores, which is not sensed by the spindle checkpoint, can directly yield aneuploid cells from diploid cells. Although the role of merotely in generating aneuploid cells in vivo has not yet been evaluated, merotelic attachment is a major source of aneuploid cells in culture [91]. Mutations that partially inactivate the spindle checkpoint represent another potential source of aneuploidy in mice and humans [92], but it is not yet clear whether spindle checkpoint failure represents a physiological source of aneuploidy in wild-type cells in vivo. Most cell lines exhibiting chromosome instability appear to have an intact spindle checkpoint [9, 81], and may in fact exhibit high rates of missegregation as a consequence of defects in components required for chromosome cohesion [6]. In this context, an intact spindle checkpoint may be essential to suppress an otherwise catastrophic rate of chromosome missegregation that would result from the combined effects of checkpoint inactivation and cohesion defects.
Although defects in the mitotic machinery can directly yield aneuploid cells, it is becoming clear that perturbation of earlier steps in the cell cycle may also be capable of yielding aneuploid cells. There are new molecular connections emerging between the pathways that control DNA replication and repair with proteins that regulate chromosome segregation and cytokinesis. Defects in chromosome replication, or induction of DNA damage, may directly hinder both chromosome segregation and cytokinesis completion. Thus in many cases chromosome missegregation may not directly yield aneuploid cells, but may instead be associated with tetraploid cell formation as a consequence of coordinate inhibition of chromosome segregation and cytokinesis. As the resulting tetraploid cells are likely to have a distinct physiology from near-diploid aneuploid cells, it will be important to understand the pathways that regulate the proliferation and survival of tetraploid cells to develop a truly comprehensive picture of the consequences of chromosome missegregation.
Acknowledgments
Work in the laboratory is supported by the NIH (GM 66492), the Susan G. Komen Foundation, and the Dana-Farber Harvard Cancer Center SPORE in Breast Cancer (CA089393). I thank the anonymous reviewers for helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 2.Rehen SK, McConnell MJ, Kaushal D, Kingsbury MA, Yang AH, Chun J. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc Natl Acad Sci U S A. 2001;98:13361–13366. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 5.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 6.Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C, Hieter P. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 8.Stewenius Y, Gorunova L, Jonson T, Larsson N, Hoglund M, Mandahl N, Mertens F, Mitelman F, Gisselsson D. Structural and numerical chromosome changes in colon cancer develop through telomere-mediated anaphase bridges, not through mitotic multipolarity. Proceedings of the National Academy of Sciences. 2005;102:5541–5546. doi: 10.1073/pnas.0408454102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimini D, Degrassi F. Aneuploidy: a matter of bad connections. Trends Cell Biol. 2005;15:442–451. doi: 10.1016/j.tcb.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Salmon ED, Cimini D, Cameron LA, DeLuca JG. Merotelic kinetochores in mammalian tissue cells. Philos Trans R Soc Lond B Biol Sci. 2005;360:553–568. doi: 10.1098/rstb.2004.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 14.Leach NT, Jackson-Cook C. Micronuclei with multiple copies of the X chromosome: do chromosomes replicate in micronuclei? Mutat Res. 2004;554:89–94. doi: 10.1016/j.mrfmmm.2004.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 16.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Liu T, Fang Y, Xie S, Huang X, Mahmood R, Ramaswamy G, Sakamoto KM, Darzynkiewicz Z, Xu M, Dai W. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–1285. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- 18.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 19.Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 20.Dai W, Wang Q, Liu T, Swamy M, Fang Y, Xie S, Mahmood R, Yang YM, Xu M, Rao CV. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- 21.Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto T, Baker DJ, d'Uscio LV, Mozammel G, Katusic ZS, van Deursen JM. Aging-associated vascular phenotype in mutant mice with low levels of BubR1. Stroke. 2007;38:1050–1056. doi: 10.1161/01.STR.0000257967.86132.01. [DOI] [PubMed] [Google Scholar]
- 23.Hartman TK, Wengenack TM, Poduslo JF, van Deursen JM. Mutant mice with small amounts of BubR1 display accelerated age-related gliosis. Neurobiol Aging. 2007;28:921–927. doi: 10.1016/j.neurobiolaging.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Baker DJ, Chen J, van Deursen JM. The mitotic checkpoint in cancer and aging: what have mice taught us? Curr Opin Cell Biol. 2005;17:583–589. doi: 10.1016/j.ceb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Jeganathan KB, Malureanu L, van Deursen JM. The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature. 2005;438:1036–1039. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- 26.Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, van Deursen JM. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172:529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 1991;10:1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McEwen BF, Chan GK, Zubrowski B, Savoian MS, Sauer MT, Yen TJ. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell. 2001;12:2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell. 2002;3:351–365. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- 31.Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao Y, Abrieu A, Cleveland DW. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 2003;114:87–98. doi: 10.1016/s0092-8674(03)00475-6. [DOI] [PubMed] [Google Scholar]
- 33.Mao Y, Desai A, Cleveland DW. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J Cell Biol. 2005;170:873–880. doi: 10.1083/jcb.200505040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weaver BA, Bonday ZQ, Putkey FR, Kops GJ, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim SK, Gopalan G. Antizyme1 mediates AURKAIP1-dependent degradation of Aurora-A. Oncogene. 2007;26:6593–6603. doi: 10.1038/sj.onc.1210482. [DOI] [PubMed] [Google Scholar]
- 36.Lim SK, Gopalan G. Aurora-A kinase interacting protein 1 (AURKAIP1) promotes Aurora-A degradation through an alternative ubiquitin-independent pathway. Biochem J. 2007;403:119–127. doi: 10.1042/BJ20061272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO Journal. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 39.Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- 41.Duensing A, Ghanem L, Steinman RA, Liu Y, Duensing S. p21(Waf1/Cip1) deficiency stimulates centriole overduplication. Cell Cycle. 2006;5:2899–2902. doi: 10.4161/cc.5.24.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangold U, Hayakawa H, Coughlin M, Munger K, Zetter BR. Antizyme, a mediator of ubiquitin-independent proteasomal degradation and its inhibitor localize to centrosomes and modulate centriole amplification. Oncogene. 2008;27:604–613. doi: 10.1038/sj.onc.1210685. [DOI] [PubMed] [Google Scholar]
- 43.Hergovich A, Lamla S, Nigg EA, Hemmings BA. Centrosome-associated NDR kinase regulates centrosome duplication. Mol Cell. 2007;25:625–634. doi: 10.1016/j.molcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 44.Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doxsey S. Duplicating dangerously: linking centrosome duplication and aneuploidy. Mol Cell. 2002;10:439–440. doi: 10.1016/s1097-2765(02)00654-8. [DOI] [PubMed] [Google Scholar]
- 46.Srsen V, Merdes A. The centrosome and cell proliferation. Cell Div. 2006;1:26. doi: 10.1186/1747-1028-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torres EM2, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 48.Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- 49.Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cimini D, Fioravanti D, Salmon ED, Degrassi F. Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J Cell Sci. 2002;115:507–515. doi: 10.1242/jcs.115.3.507. [DOI] [PubMed] [Google Scholar]
- 51.Weaver BA, Silk AD, Cleveland DW. Cell biology: nondisjunction, aneuploidy and tetraploidy. Nature. 2006;442:E9–10. doi: 10.1038/nature05139. discussion E10. [DOI] [PubMed] [Google Scholar]
- 52.Norden C, Mendoza M, Dobbelaere J, Kotwaliwale CV, Biggins S, Barral Y. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 2006;125:85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 53.Hirano T, Funahashi SI, Uemura T, Yanagida M. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 1986;5:2973–2979. doi: 10.1002/j.1460-2075.1986.tb04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: From parts list to mechanisms. Annual Review of Biochemistry. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 55.Guse A, Mishima M, Glotzer M. Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr Biol. 2005;15:778–786. doi: 10.1016/j.cub.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 56.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nature Reviews Molecular Cell Biology. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 57.Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 58.Tutt A, Gabriel A, Bertwistle D, Connor F, Paterson H, Peacock J, Ross G, Ashworth A. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9:1107–1110. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 59.Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–879. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- 60.Meng X, Fan J, Shen Z. Roles of BCCIP in chromosome stability and cytokinesis. Oncogene. 2007;26:6253–6260. doi: 10.1038/sj.onc.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smolka MB, Chen SH, Maddox PS, Enserink JM, Albuquerque CP, Wei XX, Desai A, Kolodner RD, Zhou H. An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth. J Cell Biol. 2006;175:743–753. doi: 10.1083/jcb.200605081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schweitzer JK, D'Souza-Schorey C. A requirement for ARF6 during the completion of cytokinesis. Exp Cell Res. 2005;311:74–83. doi: 10.1016/j.yexcr.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 63.Jackson MW, Agarwal MK, Yang J, Bruss P, Uchiumi T, Agarwal ML, Stark GR, Taylor WR. p130/p107/p105Rb-dependent transcriptional repression during DNA-damage-induced cell-cycle exit at G2. J Cell Sci. 2005;118:1821–1832. doi: 10.1242/jcs.02307. [DOI] [PubMed] [Google Scholar]
- 64.Date DA, Jacob CJ, Bekier ME, Stiff AC, Jackson MW, Taylor WR. Borealin is repressed in response to p53/Rb signaling. Cell Biology International. 2007;31:1470–1481. doi: 10.1016/j.cellbi.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scoumanne A, Chen X. The epithelial cell transforming sequence 2, a guanine nucleotide exchange factor for Rho GTPases, is repressed by p53 via protein methyltransferases and is required for G1-S transition. Cancer Res. 2006;66:6271–6279. doi: 10.1158/0008-5472.CAN-06-0121. [DOI] [PubMed] [Google Scholar]
- 66.Monaco L, Kolthur-Seetharam U, Loury R, Murcia JM-d, de Murcia G, Sassone-Corsi P. Inhibition of Aurora-B kinase activity by poly(ADP-ribosyl)ation in response to DNA damage. Proceedings of the National Academy of Sciences. 2005;102:14244–14248. doi: 10.1073/pnas.0506252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 68.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 69.Smith L, Liu SJ, Goodrich L, Jacobson D, Degnin C, Bentley N, Carr A, Flaggs G, Keegan K, Hoekstra M, Thayer MJ. Duplication of ATR inhibits MyoD, induces aneuploidy and eliminates radiation-induced G1 arrest. Nat Genet. 1998;19:39–46. doi: 10.1038/ng0598-39. [DOI] [PubMed] [Google Scholar]
- 70.Mossafa H, Tapia S, Flandrin G, Troussard X. Chromosomal instability and ATR amplification gene in patients with persistent and polyclonal B-cell lymphocytosis (PPBL) Leuk Lymphoma. 2004;45:1401–1406. doi: 10.1080/10428194042000191738. [DOI] [PubMed] [Google Scholar]
- 71.Gordon DS, Jones BM, Browning SW, Spira TJ, Lawrence DN. Persistent polyclonal lymphocytosis of B lymphocytes. N Engl J Med. 1982;307:232–236. doi: 10.1056/NEJM198207223070407. [DOI] [PubMed] [Google Scholar]
- 72.Smith L, Plug A, Thayer M. Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc Natl Acad Sci USA. 2001;98:13300–13305. doi: 10.1073/pnas.241355098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang BH, Smith L, Huang J, Thayer M. Chromosomes with delayed replication timing lead to checkpoint activation, delayed recruitment of Aurora B and chromosome instability. Oncogene. 2007;26:1852–1861. doi: 10.1038/sj.onc.1209995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Breger KS, Smith L, Turker MS, Thayer MJ. Ionizing Radiation Induces Frequent Translocations with Delayed Replication and Condensation. Cancer Res. 2004;64:8231–8238. doi: 10.1158/0008-5472.CAN-04-0879. [DOI] [PubMed] [Google Scholar]
- 75.Breger KS, Smith L, Thayer MJ. Engineering translocations with delayed replication: evidence for cis control of chromosome replication timing. Hum Mol Genet. 2005;14:2813–2827. doi: 10.1093/hmg/ddi314. [DOI] [PubMed] [Google Scholar]
- 76.Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- 77.Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci USA. 2003;100:9150–9155. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stindl R. Defining the steps that lead to cancer: Replicative telomere erosion, aneuploidy and an epigenetic maturation arrest of tissue stem cells. Med Hypotheses. 2008 doi: 10.1016/j.mehy.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 79.Cheung AL, Deng W. Telomere dysfunction, genome instability and cancer. Front Biosci. 2008;13:2075–2090. doi: 10.2741/2825. [DOI] [PubMed] [Google Scholar]
- 80.Bailey SM, Murnane JP. Telomeres, chromosome instability and cancer. Nucleic Acids Res. 2006;34:2408–2417. doi: 10.1093/nar/gkl303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tighe A, Johnson VL, Albertella M, Taylor SS. Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep. 2001;2:609–614. doi: 10.1093/embo-reports/kve127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, Lowe SW, Cordon-Cardo C. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 83.Sotillo R, Hernando E, Di?az-Rodri?guez E, Teruya-Feldstein J, Cordo?n-Cardo C, Lowe SW, Benezra R. Mad2 Overexpression Promotes Aneuploidy and Tumorigenesis in Mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chestukhin A, Pfeffer C, Milligan S, DeCaprio JA, Pellman D. Processing, localization, and requirement of human separase for normal anaphase progression. Proc Natl Acad Sci USA. 2003;100:4574–4579. doi: 10.1073/pnas.0730733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shepard JL, Amatruda JF, Finkelstein D, Ziai J, Finley KR, Stern HM, Chiang K, Hersey C, Barut B, Freeman JL, Lee C, Glickman JN, Kutok JL, Aster JC, Zon LI. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes Dev. 2007;21:55–59. doi: 10.1101/gad.1470407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolf F, Sigl R, Geley S. '... The end of the beginning': Cdk1 thresholds and exit from mitosis. Cell Cycle. 2007;6:1408–1411. [PubMed] [Google Scholar]
- 87.Brito DA, Rieder CL. Mitotic Checkpoint Slippage in Humans Occurs via Cyclin B Destruction in the Presence of an Active Checkpoint. Current Biology. 2006;16:1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dalton WB, Nandan MO, Moore RT, Yang VW. Human Cancer Cells Commonly Acquire DNA Damage during Mitotic Arrest. Cancer Res. 2007;67:11487–11492. doi: 10.1158/0008-5472.CAN-07-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ganem NJ, Pellman D. Limiting the proliferation of polyploid cells. Cell. 2007;131:437–440. doi: 10.1016/j.cell.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 90.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 91.Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jacquemont S, Boceno M, Rival JM, Mechinaud F, David A. High risk of malignancy in mosaic variegated aneuploidy syndrome. Am J Med Genet. 2002;109:17–21. doi: 10.1002/ajmg.10281. [DOI] [PubMed] [Google Scholar]