Abstract
Heritable silencing effects are gene suppression phenomena that can persist for generations after induction. In the majority of RNAi experiments conducted in Caenorhabditis elegans, the silencing response results in a hypomorphic phenotype where the effects recede after the F1 generation. F2 and subsequent generations revert to the original phenotype. Specific examples of transgenerational RNAi in which effects persist to the F2 generation and beyond have been described. In this study, we describe a systematic pedigree-based analysis of heritable silencing processes resulting from initiation of interference targeted at the C. elegans oocyte maturation factor oma-1. Heritable silencing of oma-1 is a dose-dependent process where the inheritance of the silencing factor is unequally distributed among the population. Heritability is not constant over generational time, with silenced populations appearing to undergo a bottleneck three to four generations following microinjection of RNA. Transmission of silencing through these generations can be through either maternal or paternal gamete lines and is surprisingly more effective through the male gametic line. Genetic linkage tests reveal that silencing in the early generations is transmitted independently of the original targeted locus, in a manner indicative of a diffusible epigenetic element.
RNA interference (RNAi) is a gene-specific silencing response of eukaryotic cells to double-stranded RNA (dsRNA) (Meister and Tuschl 2004). The dsRNA that triggers the RNAi response appears to act catalytically: a few molecules of dsRNA in most cases can elicit a response strong enough to mimic a genetic hypomorph mutation. The RNAi machinery makes use of trigger dsRNA through an intricate series of enzymatic steps. First, the trigger is “diced” to produce small dsRNA fragments. Following loading of the short dsRNAs into tight ribonucleoprotein assemblies called RNA-induced silencing complexes (RISC), one of the two strands from each RISC complex is cleaved and lost. The remaining trigger strand is used by the RISC complex in a search for complementary mRNA sequences in the cell, which are then destroyed by cleavage (“slicing”) (Bernstein et al. 2001). In some lower eukaryotes, the small RNAs derived from the original message are used to initiate the in vivo production of small RNA antisense to the targeted transcripts. This step leads to the amplification of the silencing response through RNA-directed RNA transcription.
The RNAi response in Caenorhabditis elegans is systemic and amplified. An injection of trigger in the gut or coelomic cavity can reach most tissues of the injected animal and its F1 progeny. Amplification of the RNAi signal appears to involve physical amplification of the initial silencing trigger population. This amplification is dependent on both the trigger and the presence of the target population. RNAi signal amplification is mediated by two RNA-dependent RNA polymerases (RdRP): RRF-1 for RNAi in the soma tissue (Sijen et al. 2001) and EGO-1 for silencing of targets in the germ tissue (Maine et al. 2005). The amplified RNAi response in C. elegans involves at least two structurally distinct populations of small guide RNAs. One population of guide RNAs appears to derive directly from cleavage of the exogenous (long) dsRNA. These “primary” siRNAs have structural features (including a 5′ phosphate) consistent with their proposed generation through cleavage of the long dsRNA innoculum by the dicer nuclease (Aoki et al. 2007; Pak and Fire 2007; Sijen et al. 2007). A second class of silencing-associated small RNAs in C. elegans are apparently produced by RdRP copying of mRNAs that have been targeted by RNAi (Aoki et al. 2007; Pak and Fire 2007; Sijen et al. 2007). These “secondary” effector RNAs have a 5′ triphosphate structure consistent with synthesis through de novo initiation by RNA-directed RNA polymerase activities.
In C. elegans, most RNAi effects persist in the injected parent and its F1 progeny. F1 animals receiving epigenetic signals and biochemical material from the injected parent can be considered in direct contact with the induction trigger. Once this contact is no longer present, the majority of RNAi effects are lost. Despite the general limitations of RNAi to one generation, the capacity for amplification of the RNAi signal might be expected to induce some type of heritable silencing effects in C. elegans. We initially consider three alternative mechanisms by which RNAi could initiate and maintain heritable silencing:
Heritable silencing could reflect a conservatively sustained population of primary siRNAs (derived from induction trigger) that is passed from generation to generation through the germline. This would support a model where heritable silencing would persist for as long as signals derived from the induction trigger were sustained.
Silencing may reflect a combined effect of “primary siRNAs” (RNAs derived directly from the induction trigger by Dicer cleavage) and “secondary siRNAs” (trigger RNAs resulting from target-dependent signal amplification through RdRP activity). In this model, heritable silencing might require only a catalytic contribution from the initial RNAi trigger. Long-term maintenance of such silencing would depend on the ability of secondary siRNA effectors to initiate new rounds of amplification.
Heritable silencing could conceivably reflect a molecular mechanism distinct from the initial RNA interference response. Among the potential alternative modes of action would be situations in which chromatin-targeted effects of ongoing RNA interference effects might selectively and stably shut off transcription of target genes.
Although RNAi persistence is generally limited to the injected animals and their progeny, longer-term heritability for silencing in C. elegans has been observed with several different target genes. Grishok et al. (2000) demonstrated inheritance of up to two generations after initiation of silencing. More recently, Grishok et al. (2005) and Vastenhouw et al. (2006) have proposed that long-term silencing induced by dsRNA exposure could reflect chromatin changes at the locus, preventing the transcription of the targeted gene. On the basis of its conservation in diverse eukaryotic species, amplified RNAi is clearly an important process for biological control. Thus it could be expected that several modes of inheritance might contribute to maintenance of the RNAi state.
In analyzing the potentially diverse contributions to heritable silencing, it is critical to consider the consequences of experimental selection as they affect the populations that are maintained. In particular, any protocol in which the silenced animals are dead or sterile (e.g., interfering with an essential gene) will produce a selective effect in which viability-selected populations are also selected for limited silencing efficacy. Conversely, with any protocol in which the nonsilenced animals are dead or sterile, one must consider that additional selection pressures inherent in the assay may substantially impact the results.
In this study, we make use of a pedigree-based assay for genetic suppression to examine the transmission character of heritable silencing initiated by a dsRNA trigger.
MATERIALS AND METHODS
Strains:
Strains were maintained at 16° on NGM agar plates seeded with Escherichia coli strain OP50 as described by Brenner (1974). We used the following mutant strains: LGIV—oma-1(zu405, zu405te33, zu405te36) (Detwiler et al. 2001; Lin 2003), dpy-20(e1282ts), and unc-24(e138) (Brenner 1974); LGV—him-5(e1467) (Hodgkin et al. 1979) and rde-1(ne300) (Tabara et al. 1999).
RNA synthesis:
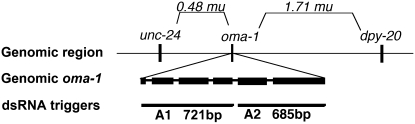
We used in vitro transcription to synthesize RNA corresponding to the oma-1 gene. Transcription was carried out on cloned genomic fragments of oma-1 DNA flanked with T3 and T7 promoters. Single-stranded RNA fragments were gel purified and annealed to render two long dsRNA fragments, a 721-nt (A1) fragment and a 685-nt (A2) fragment, spanning the majority of the coding region for oma-1 mRNA (see Figure 1).
Figure 1.—
Genomic region showing the oma-1 locus with linked morphological markers and the dsRNA triggers. We used two morphological markers, unc-24 and dpy-20, to follow the oma-1 locus during crosses. Locations of the triggers used for injections are shown. Long dsRNA triggers are shown aligned against the physical map of the oma-1 locus. Together, fragments A1 and A2 span the coding region of the oma-1 gene. A1 includes exons 1–4 and is 721 bases long. A2 includes exons 5 and 6 and is 685 bases long.
RNA delivery:
Of the several methods used to deliver dsRNA triggers to C. elegans (injection, feeding, soaking, and transgene-based expression), we chose injection because this method provided the greatest control over concentrations of dsRNA used for induction of RNAi. Feeding and soaking are frequently used in analysis of large numbers of samples, but generally lead to greater uncertainty of dose delivered and of the duration of delivery. Our starting concentration of dsRNA for RNAi induction was 50 ng/μl. Of the animals injected, we used for analysis only those animals where, by visual inspection, both gonad arms expanded under the pressure of dsRNA delivery. The delivered volume for these experiments is estimated (through optical means) to vary from 50 to 200 pl/gonad arm (Kimble and White 1981; Mello and Fire 1995).
Each of the injected animals, designated as I0 (injected generation 0), are the founders of a distinct pedigree of potentially affected animals, and each represents a different experiment, thereby allowing us to obtain data on variability between injections. The injections were conducted at room temperature (22°–23°) and animals immediately placed at the restrictive temperatures of 23° or 25°. To follow fertility in populations of animals, we picked animals and allowed them to self-fertilize on individual petri plates. We initially conducted experiments at 25°, the published restrictive temperature. Later we carried out a number of tests at 23°, a temperature more congenial to animal health than 25° (temperatures for each individual experiment are noted below).
Protocols for following silencing through multiple generations:
For any pedigree analysis with large potential populations, the experimenter must make key decisions at each generation in the choice of individuals to continue the study. Each C. elegans adult is capable of producing 300 self-progeny, resulting in a particular need for triage in following animals to characterize the population (consider that a single injected animal could have 90,000 grand progeny and 27,000,000 great-grand progeny). We describe the individual protocols used in this work as follows.
Highest silencing efficacy:
We initiated experiments with young adults of strain oma-1(zu405)dpy-20(e1282ts)IV;him-5(e1467)V kept at 16°. We injected these animals with dsRNA trigger A2 at a 50-ng/μl concentration, placed animals individually on plates, and incubated them at 23° for 3 days. All injected animals had viable progeny (indicating silencing of the oma-1 locus). We selected 14 F1's from each of three injected parents. We determined the frequency of animals with viable progeny and then selected 3 F1 families with the largest brood sizes to establish the subsequent populations. We repeated this process for the F2 and F3. For each experiment, the population size that we used to represent each generation was 42 animals. We use a pooled incidence calculation (Williams and Moffitt 2001, 2005) that accounts for weighting of individual plates of animals with different numbers of adults. This allows us to combine our results from plates initially having diverse numbers of animals. The pooled incidence operates on the number of animals per plate, the total number of animals with progeny (positive events), and the total number of plates (trials).
Intermediate silencing efficacy:
Pedigrees representing “intermediate silencing efficacy” populations at each generation had (1) at least one aunt with no viable progeny and (2) the selected individuals originated from medium broods (no less than 30 and no more than 80). We injected oma-1(zu405)dpy-20(e1282ts)IV;him-5(e1467)V with trigger A2 at a 50-ng/μl concentration. We placed injected animals individually on plates and incubated them at 23° for 3 days. We then scored the frequency at which each injected animal had viable progeny (our readout for silencing).
Bulk selection assays for oma-1 silencing:
We hypothesized that a population where individuals compete for resources (food and space) could, in a limited way, simulate the fitness expectations in the wild. This strategy would select for a subpopulations of animals most effective at long-term silencing. In this experiment, after injection, a group of viable animals were placed at the restrictive temperature for 24 days transferring every 3–4 days to fresh plates. We estimated that a population of viable and growing animals, under constant oma-1 silencing selection at 23° in an incubator, would reach the seventh generation by day 24. At this point, we used the protocol for “highest silencing efficacy selection” starting at what we estimate as generation F8. We picked animals individually (n = 100) and in groups of 10 animals/plate (n = 10). We found a transmission rate of 5.6% with a 95% confidence interval of 2.5–10.6%.
Tracking chromosomes exposed vs. not exposed to initial injection of dsRNA:
To track the origin of the oma-1 loci and the germ-cell type contributing the inherited silencing capacity, we used strains of C. elegans that, in addition to the oma-1(zu405) allele, carry a second morphological marker closely linked to oma-1 locus. The morphological mutation serves as a marker to distinguish the chromosomes inherited from the injected animals from those naive for the dsRNA injection. Three strains were used: unc-24(e138)oma-1(zu405)IV, oma-1(zu405)dpy-20(e1282ts), and oma-1(zu405)dpy-20(e1282ts)IV;him-5(e1467)V. The him-5(e1467) mutation increases the male incidence from the wild-type frequency of 1/500 to ∼1/6 (Hodgkin et al. 1979).
RESULTS
Assays for gene silencing using a conditional-lethal neomorphic mutation in the oma-1 gene:
We found the oocyte maturation oma-1 gene to be a suitable target for characterizing heritable silencing phenomena. The zu405 neomorphic allele of oma-1 is a semidominant conditional-lethal mutation. zu405 animals kept at 16° resemble wild type, while at temperatures >21°, the mutation renders all progeny of an adult animal inviable. The point mutation in zu405 results in an amino acid change (P240L) that eliminates a phosphorylation site required for the proper degradation of OMA-1 protein (Lin 2003). Failure to degrade OMA-1 results in embryonic lethality at 23° and above (Lin 2003). In the loss-of-function oma-1 background, animals have viable progeny, presumably by the function of oma-2, a second oocyte maturation gene (Detwiler et al. 2001).
To test the suitability of the oma-1 gene as the target for studying heritable silencing, we first evaluated the oma-1 gain-of-function and loss-of function phenotypes in the absence of dsRNA. To evaluate the penetrance of the gain-of-function allele zu405, we generated a large population of zu405 animals at 16° (the permissive temperature) and plated 100 animals individually and 200 animals in groups of 20 animals/plate. Both individually plated and grouped animals were then shifted to 23° and 25° (restrictive temperatures) at the L4 or young adult stage. We found that each animal or group of animals produced large numbers of eggs but that none of these eggs hatched.
As would be required for any suitable long-term assay for silencing, loss of oma-1 function is compatible with long-term propagation. Null alleles of oma-1 are fertile due to oma-2 function (Detwiler et al. 2001). To confirm the long-term viability in the absence of oma-1 activity, we have maintained oma-1 null mutant cultures through numerous passages under our standard growth conditions at 23° and have seen no evident deleterious effects on the populations (data not shown).
A potential source of false positives would be the genetic reversion of oma-1(zu405) to a loss-of-function mutant (which would be difficult to distinguish from robust heritable silencing). To address this concern, we carried out a continuous validation of the oma-1(zu405) strain grown at 16° by periodically shifting a population of animals to 23° and measuring the frequency with which animals could produce viable progeny. In 5 years of keeping the oma-1(zu405) stock (over 15 independent heritability experiments), we have seen no case of spontaneous reversion in this strain. These tests indicate that the oma-1(zu405) stock was genetically stable enough to proceed with long-term silencing assays.
oma-1(zu405) mutant rescue following dsRNA injection:
We found that the viability of oma-1(zu405) at the restrictive temperatures of 23° and 25° was dependent on the target specificity of dsRNA triggers and on the function of the RNAi pathway mediated by the rde-1 gene (see Table 1).
TABLE 1.
Suitability of oma-1(zu405) silencing assay in heritable silencing interference studies
| Test | dsRNA trigger | Recipient genotype | Animals tested | Biological effect |
|---|---|---|---|---|
| Effect of specific dsRNA | oma-1 (A1) | oma-1(zu405) | 5 | >50 progeny (5/5) |
| Effect of specific dsRNA | oma-1 (A2) | oma-1(zu405) | 5 | >50 progeny (5/5) |
| Effect of nonspecific dsRNA | gfp | oma-1(zu405) | 5 | Dead eggs only (5/5) |
| Effect of nonspecific dsRNA | unc-22 | oma-1(zu405) | 3 | Dead eggs only (3/3) |
| Effect of no dsRNA trigger | None | oma-1(zu405) | 3 | Dead eggs only (3/3) |
| Dependence on RNAi machinery | oma-1 (A1) | oma-1(zu405); rde-1(ne300) | 5 | Dead eggs only (5/5) |
We tested the response of animals carrying mutations in the oma-1 gene to determine the feasibility of the assay to study heritable silencing. Viable progeny at the restrictive temperature (23°) for animals oma-1(zu405) are the positive readout for silencing. Dead eggs only are negative readout for silencing.
Two triggers specific to the oma-1 transcript (A1 and A2 Figure 1) both rescue the embryos of injected parents. Triggers with specificity to gfp (n = 5) and unc-22 (n = 3) did not rescue the maternal-effect embryonic lethality of oma-1(zu405). To determine the dependence of the oma-1 silencing effect on the RNAi mechanism, we constructed the double-mutant strain oma-1(zu405)IV; rde-1(ne300)V, injected animals (n = 5) with trigger A2, and found no animals had viable progeny at 25°.
To determine if the oma-1 selection was required for silencing, we placed injected animals at the permissive temperature of 16° for 7 days. We then selected animals from the three largest F1 broods to determine the viability of F2 animals. We found that 100% F1 animals (n = 42) had viable progeny at 23°, indicating that the selection of oma-1 in the F1 generation was not required for the silencing effect to persist in the F2 generation. Selecting for brood size as a criterion was sufficient to select for F2 viability.
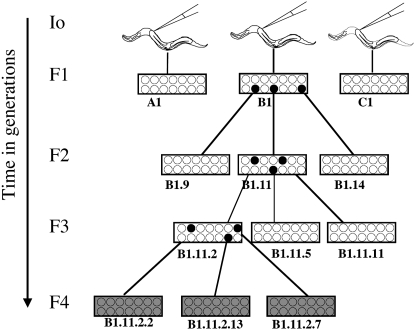
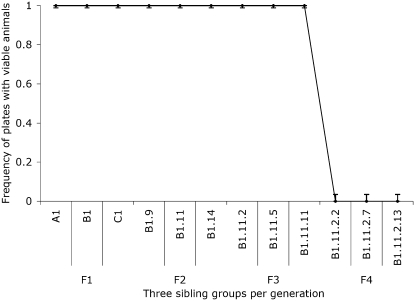
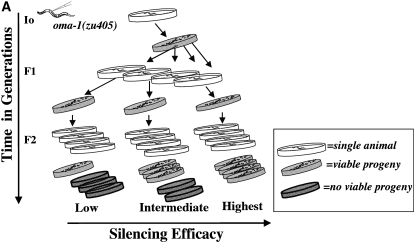
A bottleneck in transmission of silencing character:
We saw a reproducible and distinctive pattern of rescue following oma-1 dsRNA injection and “highest silencing efficacy” pedigree selection (see materials and methods). In generations F1, F2, and F3, all selected animals had viable progeny (42/42 in each generation for each experiment), while a comparable set of selected animals in the F4 had zero viable progeny (see Figures 2 and 3). To experimentally investigate the striking drop in F4 frequency with greater precision, we needed to collect worms in greater numbers to detect lower frequencies of transmission. From the analyzed F3 sibling-group plates we opted to transfer a large group of F4 animals by chunking a centimeter square of an F3 plate onto a plate with fresh OP50 bacteria (F3 plates had been starved by this point, each plate containing hundreds or thousands of animals in the agar). A small chunk of agar from a starved plate was used to transfer large numbers of animals (from plates on which the F3 animals had been picked 6 days earlier). We estimated that larvae on these plates were at generation F4 or F5. Picking individual animals (200 single-animal plates) yielded 5 with viable progeny. In plates where we grouped animals 10 to a plate, we found 1 of 20 plates had viable progeny. We found that the late generation populations had broods of <10 animals while the early generations had average brood sizes of 90–110. In addition to smaller broods, we found that late-generation surviving worms had an unhealthy appearance (sluggish and morphologically abnormal).
Figure 2.—
Pedigree selection scheme for determining of persistence of heritable silencing. Young adult hermaphrodites of the strain oma-1(zu405), dpy-20(e1282ts)IV; him-5(e1467)V were injected with dsRNA trigger and allowed to recover at room temperature. Individual I0 animals were plated onto petri dishes containing fresh lawns of OP-50 bacteria and grown at 23° for 3 days, when they were scored for viable progeny. Criteria for selecting animals to pedigree were (1) groups where all observed siblings had viable progeny and (2) from the sibling group, the individuals with largest brood sizes. Both these criteria were our indicators of a strong silencing response. We designated I0's (labeled “A1,” “B1,” and “C1”) and individually plated 14 F1 animals from each. Three days later, we scored F1 animals for viable progeny. We chose B1 as the best sibling group and picked plates B1.9, B1.11, and B1.14 as our source for L4 larvae (14 F2 animals each). Three days later we scored F2 for viable progeny. We chose B1.11 as the best sibling group, and plates B1.11.2, B1.11.5, and B1.11.11 as our source for L4 F3 animals each. For F4's, we chose F3 sibling group B1.11.2 and picked 14 animals from plates B1.11.2. 2, B1.11.2.7, and B1.11.2.13. We repeated this pedigree selection protocol using each of the two long dsRNA triggers, A1 and A2 (see Figure 1). For each generation and both triggers we scored 42 animals. We depict viability of progeny in sibling groups with a white background and inviability with a gray background. Solid circles represent plates selected at each generation to further pedigrees.
Figure 3.—
Multigenerational assays for oma-1(zu405) silencing. Following the protocol described in Figure 2 leads to robust silencing in the F1, F2, and F3 generations followed by a severe drop to zero silencing in the F4 generation. At each generation, we plotted three subpopulations of 14 animals each. All animals analyzed had viable progeny (100% observed in F1, F2, and F3), and all F4 animals had no viable progeny. The error bars represent one standard deviation for each sibling group.
Although initially observed with dsRNA trigger A1, the F4 bottleneck pattern was not limited to this trigger. Figure 3 shows comparable observations with nonoverlapping trigger dsRNA trigger A2. As with the A1 trigger, in experiments with A2 trigger, all selected F1, F2, and F3 animals have viable progeny, while zero of the 42 examined F4 descendants had viable progeny.
Dilution of induction trigger affects the persistence of heritable silencing:
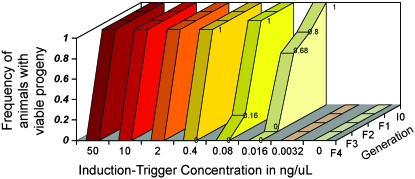
We tested the induction trigger in a fivefold dilution series and examined the effects on heritable silencing. Final concentrations of 50, 10, 2, 0.40, 0.08, 0.016, 0.0032, and 0 ng/μl were used. Figure 4 shows three patterns of response:
At higher concentrations (50, 10, 2, and 0.4 ng/μl) we observed a robust RNAi response (100% of animals tested silenced the oma-1 target to the F3 generation), followed by abrupt loss of silencing in the F4 generation (100% of animals tested were not silenced).
At concentrations of 0.08 and 0.016 ng/μl, we observed silencing, albeit with reduced efficacy: for 0.08 ng/μl, silencing dropped to 16% at F3 and 0% at F4; and for 0.016 ng/μl, silencing dropped to 80% at F1, 68% at F2, and 0% at F3.
The most extreme dilution (0.0032 ng/μl) and the negative control (0 ng/μl) showed no silencing response.
Figure 4.—
Dependence of heritable silencing on injected trigger concentration. We tested a fivefold serial dilution of the dsRNA induction trigger using the same pedigree selection scheme described in Figure 2. At each concentration, we picked three injected animals to select 14 F1 animals, placed them on individual plates, and incubated for 3 days at 23°. We used the same criteria to select the descendant populations. We found animals injected with concentrations of 50 to 0.4 ng/μl had an equivalent silencing response: all animals scored in generations F1, F2, and F3 had viable progeny, while all animals of the F4 generation had no viable progeny. Silencing efficacy of concentrations of 0.08 and 0.016 ng/μl were less effective at both the overall silencing frequency (<100%) and in the persistence of the silencing response. Animals injected with 0.0032 ng/μl showed no silencing response.
Silencing capacity is preferentially packaged into early born progeny:
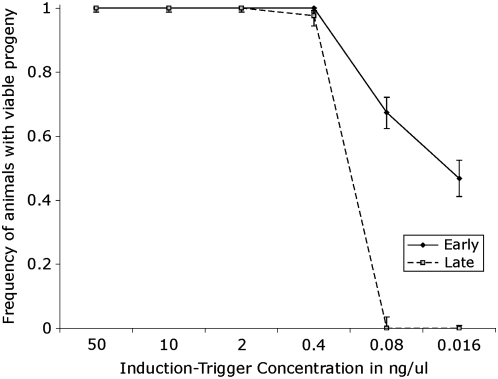
We measured oma-1 rescue capacity as a function of the birth order on the basis of the assumption that, under a limited amount of trigger, progeny might receive unequal quantities of injected trigger and this could result in a decreased in silencing efficacy. The birth order assay distinguished early born (born within 24 hr of injection) and late born (born in the second 24 hr after injection) F1 progeny. We collected F1's from three injected animals at each concentration that we tested. I0 animals injected with high trigger concentrations (50–0.4 ng/μl) had many F1 progeny (∼50 progeny each 24 hr period). We selected 14 F1 animals from both the early and late-born populations. At these concentrations, we saw no difference in silencing efficacy between late and early progeny. At trigger concentrations of 0.08 and 0.016 ng/μl, we picked all available F1 animals from both early and late-born populations. We found that at concentrations of 0.08 ng/μl and 0.016 ng/μl, the early born F1 animals had a significantly greater silencing efficacy than their later born siblings (see Figure 5).
Figure 5.—
Comparison of silencing efficiency between early and late-born progeny. We injected animals and selected F1 progeny by birth order and determined their silencing capacity. We segregated animals from the same brood as (1) early born animals (born the first 24 hr after the injection) and (2) late-born animals (born the second 24 hr after injection). We found injected concentrations of 50, 10, 2, and 0.4 ng/μl show no significant difference in silencing between early and late-born siblings. In contrast, at concentrations of 0.08 and 0.016 ng/μl, there is a significant difference between the early born animals (solid line) and late-born animals (dashed line). Early born progeny of injection concentrations of 0.08 and 0.016 ng/μl had silencing frequencies of 67.4 and 46.7%, respectively, while the late-born progeny for both concentrations has a silencing frequency of 0%. Bars represent 1 SD.
Testing for association between silencing, gamete type, and the oma-1 locus:
Silencing could conceivably be inherited and maintained at any given stage as a diffuse signal separable from the chromosome (e.g., a diffusible small RNA) or as a stably chromosome-associated feature (e.g., with linkage to the original oma-1 locus). To explore these hypotheses, we followed the transmission of the exposed locus in genetic crosses in which the oma-1 locus was linked to a genetic marker. If the silencing signal were linked to the original locus, we would expect that only the grand-progeny (F3) inheriting both chromosomal loci from the originally exposed grandparent would be silenced. In contrast, a diffusible or transferable signal might be inherited independently from the original oma-1(zu405) locus.
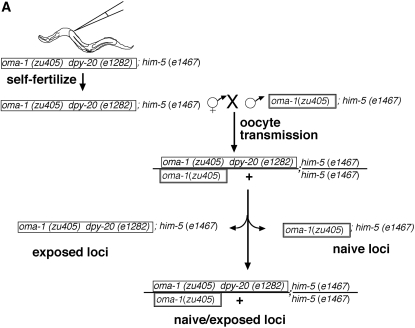
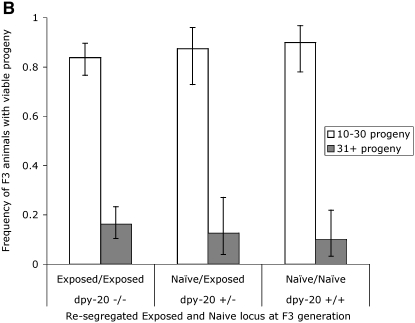
We used a recessive morphological marker to mark the origin of the chromosome and distinguish between exposed and nonexposed loci. The morphological markers that we used are shown in Figure 1. Marker unc-24 is 0.48 and dpy-20 is 1.71 MU from the oma-1 locus. We injected trigger A2 into oma-1(zu405)dpy-20(e1282ts)IV; him-5(e1467)V (see Figure 6A). We picked groups of 5 F1 hermaphrodites at the L4 larvae stage and mated them with 5 nonexposed oma-1(zu405)IV;him-5(e1467)V males. F1 hermaphrodites are dumpy and are visibly distinct from the non-dumpy oma-1(zu405)IV;him-5(e1467)V males. Three days later, we screened plates for F2 heterozygous, non-dumpy progeny (the cross-progeny). We plated individually the non-dumpy heterozygotes and incubated for 3 days. We picked 59 dumpy and 44 non-dumpy F3 animals to individual plates and we recorded (1) the F3 (parent) phenotype, (2) the F3 brood size (to infer silencing efficacy), and (3) the phenotype of the progeny, to infer the F3 parent genotype. Dumpy animals are homozygous for dpy-20(e1282ts) and therefore are expected to carry two chromosomes that were exposed directly to the RNAi trigger [except in cases of recombination (1.71%) between dpy-20 and oma-1]. The non-dumpy F3 animals would predominantly be of genotype dpy-20(e1282ts)/dpy-20(+) or dpy-20(+)/dpy-20(+). Animals with no progeny or small broods are of ambiguous genotype. To avoid misclassifying the genotype of F3 non-dumpy animals, we excluded from our analysis those animals with zero progeny or with a brood size of <10. Of the animals with broods >10, we separated those with broods of 10–30 animals from those of 31 or more animals. Figure 6B shows the distribution of the silencing frequency of descendants carrying zero, one, or two copies of the chromosomal target locus exposed to dsRNA in the ancestor animal. The silencing efficacy, when measured by brood size, is independent of the origin of the oma-1 allele. This suggests that the silencing determinant is diffuse and not linked to the oma-1 locus.
Figure 6.—
Tests for silencing transmission of oocytes and linkage to the chromosomal locus exposed to dsRNA. (A) Schematic of crosses designed to follow chromosome origin from oocyte transmission. We injected animals that were morphologically dumpy by carrying a homozygous recessive allele of dpy-20(e1282ts). The dpy-20 allele is linked to the oma-1 locus and marks the origin of the chromosome. Injected animals self-fertilized and we used the F1 hermaphrodites to cross with males not exposed to dsRNA and with a wild-type copy of the dpy-20 gene. The cross-progeny were non-dumpy heterozygous (only cross-progeny are non-dumpy). We then allowed heterozygous animals to produce self-fertilized progeny. Dumpy animals are dpy-20(e1282ts) homozygous and non-dumpy animals are either homozygous wild type or heterozygous for dpy-20(e1282ts). F3 animals descended from F2 cross-progeny inherited chromosomes from ancestors exposed or not exposed to dsRNA. We scored the F3 animal's capacity for producing viable progeny. (B) Results of linked heritable silencing assay. We followed the genetic scheme described in A and individually plated F3 animals carrying exposed or nonexposed chromosomes to dsRNA. Homozygous animals carrying the wild-type dpy-20 allele inherited their oma-1 locus from nonexposed animals. To determine if silencing was segregating with the origin of the chromosomes, we used animals having broods of >10 progeny. We used two ranges in brood size (10–30, open bars; >31, shaded bars) as indicators of the efficacy of the silencing achieved. Error bars are 1 SD. The silencing efficacy of F3 animals demonstrates that (1) the transmission through the oocyte is sufficient to transmit silencing capacity and (2) the silencing capacity is unlinked to the origin of the oma-1 locus.
Transmission of the silencing signal through both oocyte and sperm gametic lines:
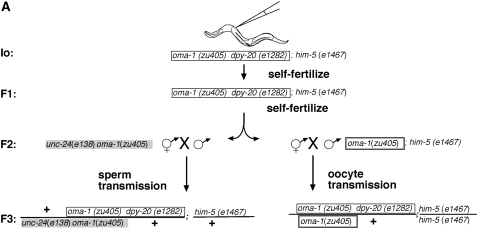
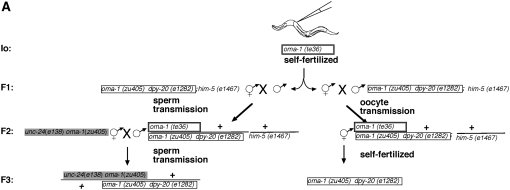
We determined the transmission of silencing through oocyte and sperm gametic lines in the same experiment. We injected hermaphrodites of genotype oma-1(zu405) dpy-20(e1282ts);him-5(e1467), incubated them at 23°, and used both male and hermaphrodite descendants for transmission of the silencing character. Of six injected animals, we picked the one with the largest brood size (>100 progeny) and cloned 50 F1 hermaphrodites. All 50 F1 plates had viable progeny (indicating high efficacy silencing). We chose one F1 plate with a large brood size to select F2 males to assess sperm transmission and their hermaphrodite siblings to assess oocyte transmission (see Figure 7A).
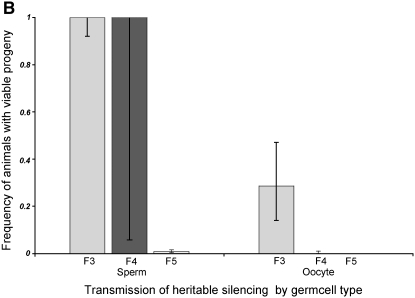
Figure 7.—
Comparison of silencing transmission capacity for oocytes and sperm. (A) Genetic scheme. We first selected one injected animal (I0) to produce F1 and F2 descendants by self-fertilization. We then selected one F2 animal with a large brood size to separately assess sperm and oocyte transmission of silencing. (B) Results of sperm/oocyte comparison. F3 cross-progeny animals were scored individually (7 animals for oocyte transmission and 6 for sperm transmission). F4 descendants were scored in groups of siblings rather than individually since our previous experiments (Figure 2) indicated a consistent F4 bottleneck. In the oocyte experiments, only 2 of 7 F3 animals had progeny: 1 animal had 48 progeny and 1 animal had 1 progeny. None of the F4 animals had F5 progeny. In contrast, in the sperm-transmission experiments, all 6 F3 animals had viable progeny, with brood sizes ranging from 28 to 80. We used groups of F4 animals from sperm transmission and found 26/26 plates with 10 animals to a plate to have viable progeny. To analyze the F5 silencing, we checked the F4 plates for fertile F5 animals. We found 2/26 plates with viable F5 progeny.
F2 male descendants derived from two generations of self-fertilization. The F2 males were crossed to nonexposed hermaphrodites homozygous for oma-1(zu405) and the morphological marker unc-24(e138). Crosses were incubated at the permissive temperature of 16° for essentially a complete generation (6 days), allowing progeny to grow irrespective of the oma-1 rescue status. All six F3 progeny obtained from F2 sperm transmission gave F4 broods. Only two of seven F3 progeny obtained in reciprocal crosses where F2 transmission was through the oocyte lineage gave F4 broods (Figure 7B). In the pedigrees where we had transmitted silencing capacity to the F3 through the F2 male parent, we continued to observe silencing through the F4 and F5 generations. F4 progeny from these experiments were picked in groups of 10 to a plate; of 26 such plates, all had viable progeny. Picking 26 plates of 10 F5 animals, we found that 2 had F6 larvae. By contrast, zero of the 48 F4 animals derived from oocyte transmission animals had viable progeny.
Transmission by germ-cell type starting in a null oma-1 background:
In previous experiments, the neomorphic oma-1 protein derived from the zu405 allele is already present at the time of fertilization, and thus male transmission of silencing factors would not be expected to rescue the oocyte defects with a post-transcriptional silencing mechanism like RNAi. We confirmed this by placing oma-1(zu405) naive hermaphrodites with exposed F1 males at 23°. Of 28 crosses with a single hermaphrodite and a single male incubated at 23°, zero had viable progeny. This made it unavoidable to use the permissive temperature of 16° when the naive strain carrying the oma-1(zu405) supplied the oocyte. Therefore, crosses testing male transmission were done at 16° and those for oocyte transmission at 23°. We were concerned that the difference in transmission between sperm and oocyte was due to the deleterious effects of the zu405 allele on oocytes rather than the germ-cell capacity for effective transmission. The oma-1(zu405) might have secondary effects that make the animals sick after consecutive generations at high temperatures. If this were true, the differences in silencing could be due to an oma-1(zu405) assay.
To address this possibility, we designed an alternative assay. We initiated silencing in animals carrying a loss-of-function allele. Two apparently null alleles were used for loss-of-function studies: (1) zu405te33, a nonsense mutation, and (2) zu405te36, a missense mutation. Each mutant locus also carries zu405. Our assumption, and that of others who have used similar intragenic revertants of gain-of-function alleles (e.g., Greenwald et al. 1983), is that the cis-double mutants behave as a null. The loss-of-function mutations have a high incidence of males (him) of ∼5–15%, which is useful in providing males for crosses.
Initiating silencing in a loss-of-function and viable genetic background allows us to genetically decouple the time line of exposure to the dsRNA innoculum from the deleterious oma-1(zu405) background. The following manipulations (Figure 8A) allowed us to determine and compare the silencing efficacy of animals following (1) one oocyte transmission followed by self-fertilization or (2) two sperm transmissions.
F1 hermaphrodite progeny of injected oma-1(zu405te36)IV parents were crossed with oma-1(zu405)dpy-20(e1282ts)IV;him-5(e1467)V males at 23°. Cross-progeny are F2 (from injection) and heterozygous oma-1(zu405)/oma-1(zu405te36). These F2 animals are allowed to self and we identified the oma-1(zu405) homozygote animals by the linked marker dpy-20(e1282ts).
In a reciprocal set of experiments, F1 male progeny of an injected oma-1(zu405te36) hermaphrodite parent were crossed with dpy-20(e1282ts)oma-1(zu405)IV hermaphrodites at 16° to yield cross-progeny F2, which are likewise heterozygous oma-1(zu405)/oma-1(zu405te36) but which result from F1 transmission through the male. The silencing character is then transmitted one further generation through the male lineage by mating males from the cross with unc-24(e138)oma-1(zu405) hermaphrodites.
Figure 8.—
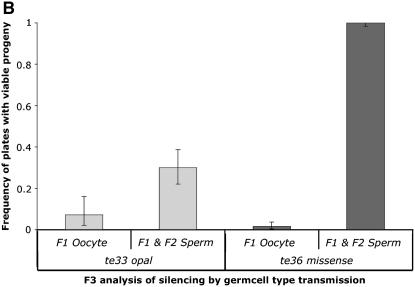
Transmission as a function of the oma-1 genetic background. (A) We examined the transmission of the silencing character in two loss-of-function backgrounds of oma-1. The genetic scheme shows the missense mutation zu405te36 as the starting point for the experiment. The same scheme was followed to examine the silencing efficacy, starting with the loss-of-function nonsense mutation zu405te33. Loss-of-function strains (zu405te36 and zu405te33) of oma-1 were injected with dsRNA trigger A2 and crossed to gain-of-function zu405 strains. Sperm transmission experiments were done at 16° and oocyte transmission at 23°. (B) Calculated frequencies of silencing were measured by the frequency of homozygous zu405 F3 animals with viable F4 progeny.
We repeated the genetic manipulations described in Figure 8A, starting with allele zu405te33. Figure 8B shows the silencing frequency in F3 animals (three generations removed from initial dsRNA injection and first-generation homozygous zu405). We observed transmission of the silencing activity through sperm with both oma-1 null alleles, with transmission appearing more effective in animals carrying missense allele zu405te36 as compared to nonsense allele zu405te33. In both cases, transmission through the oocyte lineage was less efficient than through the male. These results are consistent with silencing activity mediated by a silencing mechanism not linked to the oma-1 locus, since the tested allele is introduced in the F2 generation.
Sperm vs. seminal fluid transmission assay:
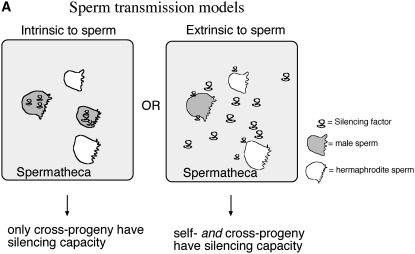
We envisioned two possible vehicles for sperm to transmit silencing activity: (1) the silencing factor was within the sperm in the limited cytoplasm (1% of the oocyte cytoplasm volume) or in the nuclear contents or (2) the silencing factor was carried outside of the sperm in the seminal fluid that accompanies sperm as it travels into the hermaphrodite during copulation (Figure 9A). The use of extracellular transmission of cytoplasmic signals by sperm has a precedent in C. elegans. The major sperm protein oocyte maturation protein is provided by sperm to oocytes in extracellular vesicles carried in the seminal fluid (Kosinski et al. 2005). This molecular mechanism allows sperm, with its limited cytoplasm, to release signals into the oocyte environment.
Figure 9.—
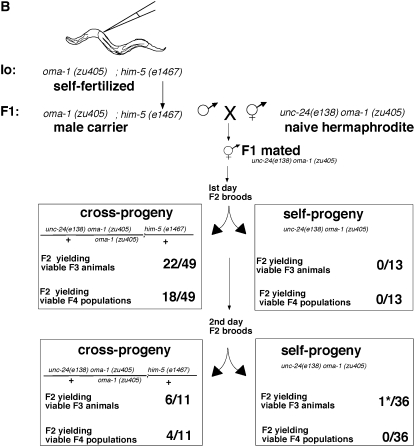
Explicit test of male transmission of silencing. (A) Males during copulation transmit both sperm and seminal fluid. The male-derived silencing efficacy can be explained by at least two models: (1) The silencing factor is inside the sperm and (2) the silencing factor is transmitted through the male in the seminal fluid. (B) Schematic of crosses designed to test the male transmission of silencing through the sperm or the seminal fluid. It was critical for this experiment that we identify self-progeny animals that had been fertilized after their parent hermaphrodite had received male sperm and seminal fluid. To ensure this, we transferred the parent hermaphrodites each day and scored only self-progeny that derive from mothers that had previously produced cross-progeny. Operationally, this was carried out by mating individual F1 male silencing carriers with five naive hermaphrodites for 6–12 hr, transferring the hermaphrodites to individual fresh plates to allow egg laying for 1 day (first brood), and transferring hermaphrodite mothers to a second plate for an additional day (second brood). Of 50 mated hermaphrodites, 6 met the criterion that they had some cross-progeny on the first day of transfer and some self-progeny on the second day of transfer. The self-progeny broods on these six plates from the second transfer consist of self- and cross-progeny that were fertilized subsequent to the transfer of sperm and seminal fluid from males to the mother hermaphrodite. We then compare silencing transmission to self-progeny and cross-progeny from these broods. The boxes summarize the viability of F3 and F4 cross- and self-progeny from first and second transfers at 25°. The data show that carrier males transfer the silencing trait to cross-progeny and not to self-progeny. This is consistent with a signal intrinsic to sperm and not one carried in the seminal fluid. The asterisk indicates that a single viable F3 larva was produced from 1 of the 36 F2 animals in this experiment; this animal yielded no F4 progeny and may have represented a rare “spontaneous rescue” affecting ∼1 in 104 progeny of oma-1(zu405) mothers.
To determine if the male capacity for silencing is extracellular (carried in the seminal fluid) or intracellular (within the sperm itself), we scored the silencing efficacy in cross-progeny and self-progeny of individual hermaphrodites mated to F1 male carriers of heritable silencing. It was important for this analysis to ensure that mating had occurred prior to self-fertilization. Thus, an essential part of the experiment was to score self-progeny of hermaphrodites that had already mated and produced cross-progeny. As above, these experiments began by injecting dsRNA for oma-1 into oma-1(zu405)IV;him-5(e1467)V animals, placing the injectees in a 25° incubator and 3 days later selecting plates with the largest brood sizes as our source of F1 male carriers (9B). We crossed F1 males with oma-1(zu405) unc-24(e138) hermaphrodites at 16°. We scored the silencing capacity of cross-progeny and self-progeny siblings of mated hermaphrodites by looking at the viability of F3 and F4 animals. We found that only cross-progeny had a viable F3 from which populations of F4 progeny emerged. This is consistent with a signal intrinsic to sperm.
Effects of selection scheme on generational silencing profile:
As discussed above, choices of which animals to propagate in each generation to assess silencing heritability would be expected to influence quantitative and qualitative aspects of the observed populations. All preceding data were obtained with a “maximum efficacy” selection in which we chose animals to transfer at each generation on the basis of maximal silencing of the oma-1 locus (i.e., maximum viability) in the immediate families of candidate animals. An alternative “intermediate efficacy” selection protocol (Figure 10A) differed from the “highest efficacy silencing” group by two criteria: (1) Plates genetically related by having the same parent (sibling group) were chosen for further pedigree analysis only if they included at least one plate with no viable progeny (i.e., only animals with at least one nonrescued sibling were followed) and (2) from the candidate groups fulfilling the first criterion, we selected for animals with moderate (30–80) brood sizes.
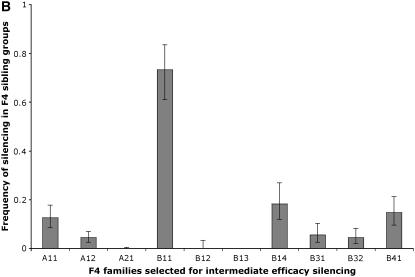
Figure 10.—
Relaxed stringency of early selection allows some persistence of silencing in the F4 generation. (A) Schematic of assay. We designed a selection process to evaluate the relationship between the strength of the silencing response measure by the silencing frequency in a particular pedigree, to the persistence of the silencing across generations. Degrees of silencing efficacy were determined by the silencing frequency and brood size of the selected animals. We used the frequency of silencing to classify pedigrees as transmitting at highest, intermediate, or low silencing efficacy. When then used brood size as a second criterion to guide the selection of individuals to analyze the silencing frequency of the next generation. In the “highest silencing efficacy” group, we selected from plates with the largest brood sizes (>90). In the intermediate silencing efficacy group, we selected individuals from plates with broods between 30 and 80. Animals where most siblings have no viable progeny represent low-silencing-efficacy groups and were not used. (B) Intermediate silencing efficacy populations overcome the F4 bottleneck. We followed the less stringent selection scheme of intermediate silencing efficacy and found that 7/10 F4 sibling groups had at least some viable F5 progeny. This is in contrast to the F4 bottleneck that we observed when we used the “highest silencing efficacy” selection (data in Figure 3 and data not shown). Error bars represent 1 SD. As the manner in which animals are chosen to carry forward the silencing trait is critical in determining the behavior of descendant populations, we describe the selection process for the intermediate silencing efficacy group in some detail as follows: The F4 animals, classified as descendants of continuous intermediate silencing efficacy selection, were derived from one of five injected animals. Of the original five injected animals, we picked all viable progeny and arbitrarily assigned each a color (purple, red, green, orange, and blue). Three days later, all injected animals had viable progeny. We individually plated the F1 animals and scored the frequency of viable F2 progeny. Only the orange F1 family had no viable progeny (n = 15). All F1 plates from blue (n = 40), red (n = 54), purple (n = 91), and green (n = 20) had viable progeny. We selected F2 animals from eight F1 families: two blue, one green, two purple, and three red. Each F1 family gave rise to an F2 sibling group (designated by two letters). From the blue family, the BD group had 100% plates with viable progeny while the BE group had only 7.8%. From the green family, GF had 80%; from the purple families, both PH and PJ had 100% transmission; and from the red families, RA had 94.7%, RB had 80.7%, and RC had 100%. The RA and RB lineages fulfilled the criteria for selection of intermediate silencing efficacy. To extract the populations with smaller brood sizes, we removed the F2 animals at day 2. On day 3, we scored the F2 plates. This allowed us to better assign a generation to animals by increasing the age difference between F3 adults and young F4 larvae. Two days after removing the F2 adults, we surveyed all plates of F2 animals with F3 broods. Plates with large brood sizes had depleted the bacterial lawns. Plates with “smaller” F3 broods were not depleted of bacteria (fewer worms on plate, more food per worm) and their growth was uninterrupted. We used F4 animals from small broods to represent the intermediate silencing efficacy groups.
We note that results from the “highest” and “intermediate” efficacy selections are quite similar in broad outline (Figure 10B). We observed a silencing for several generations followed by a dramatic drop in silencing. Nevertheless, we observed a distinct F4 generation profile. Particularly striking is the retention of partial silencing in the F4 generation following “intermediate efficacy selection” as compared to a complete loss following earlier “highest efficacy selection.”
DISCUSSION
We found that a single dose of dsRNA targeting the germline-active gene oma-1 can lead to silencing that lasts multiple generations. Heritable silencing was initiated by the RNAi response, with a biphasic time course involving nearly complete but temporary suppression of the target locus lasting three to four generations, followed by a much lower frequency of long-term silencing. Heritable silencing frequency and persistence was dependent on the dose of induction trigger. When the trigger was limited, the silencing efficacy was preferentially distributed in early born progeny. Both sperm and oocytes were capable of transmitting the silencing signal to descendant populations. Surprisingly, the frequency of silencing after sperm transmission appeared to be higher than for oocyte transmission. The silencing achieved by sperm transmission involved a signal intrinsic to sperm.
Multigenerational silencing after RNAi treatment:
Earlier analysis of heritable silencing in C. elegans had been carried out with a number of loci. Following microinjection of pos-1 dsRNA, one study (Grishok et al. 2000) demonstrated clear inheritance of pos-1 silencing through at least one generation of either the hermaphrodite or male germline and showed that this inheritance could occur in the absence of activity for the target locus. Particularly striking was the ability to transmit pos-1 silencing for one generation through males in the absence of the target locus. The oma-1 analysis allows generational silencing pedigrees to be extended to subsequent generations with marking of the initial targeted locus and continuous and functional measurement of target gene activity. The oma-1 assays provided a sensitive means for tracking the silencing bottleneck that occurs three to four generations following injection.
A recent study (Vastenhouw et al. 2006) used injected dsRNA and transient bacteria-mediated dsRNA feeding to follow longer-term inheritance for silencing of a GFP transgene and for a variety of endogenous loci. Much of the analysis of Vastenhouw et al. was carried out many generations subsequent to the initial delivery of dsRNA and following extended periods of strong selection for phenotypic effects consistent with gene silencing. The requirements defined in that work are likely to be relevant to processes in a “late” phase of silencing, when the initial pleiotropy resulting from the RNAi response has been restricted. The stabilized or reinforced “late” phase phenotypes are conceivably a consequence of prolonged and stringent selection for unusual epigenetic or genetic characteristics.
We found that the high-efficiency initial phases of inherited silencing reflect transmission of a silenced character that is unlinked to the target chromosomal locus. Such inheritance would suggest a diffusible molecule not coupled to the chromosome. Given the specificity for the locus, an attractive hypothesis is that the critical inherited signal at these stages would be a silencing RNA. One hypothesis for the transmission of such an RNA signal would be a passive diffusion of silencing RNA from the original trigger. Alternatively, there may be a trigger-initiated amplification process by which larger populations of silencing RNA are generated following the initial microinjection. Relevant to any proposed amplification is the observation that silencing efficiency dramatically decreases three to four generations following the injection. This decrease would be inconsistent with a simple self-renewing trigger population. Thus we expect that there may be specific mechanisms that limit long-term amplification. One such mechanism would require a small number of molecules derived from the original foreign RNA innoculum (or its initial amplification products) to silence effectively at each generation. A mechanism with a limited capacity to engage the initial dsRNA innoculum could also explain the consistency of the bottleneck over a 125-fold range of dsRNA concentrations. Supplemental Figure S1 shows a series of simulations in which the initial trigger population is “diluted” through several generations of inheritance. Such models depend on some means by which the animal would preferentially deliver the trigger population to the germ lineage and to subsequent progeny; certainly precedents for such mechanisms are evident from the germline-associated P granules that can be observed in C. elegans embryos, larvae, and adults. The simulations certainly show consistency between the size of the initial injected RNA pool and the generational persistence of the silencing effect. We note, however, that small numbers of molecules predicted from such models would likely be insufficient to directly silence the target RNA population (oma-1 mRNA); instead, we might expect that the injected dsRNA innoculum would continue to function through the characterized RdRP-based amplification mechanism present in C. elegans.
The ability to inherit oma-1 silencing in the absence of the originally exposed oma-1(zu405) chromosome indicates some degree of mobility of the silenced character. Certainly one type of model for such mobility would invoke an RNA trigger population, which might act in the cytoplasm with no reference to the chromosome or to the nucleus. Although such models might be favored at present by parsimony, we certainly cannot rule out nuclear activities in heritable RNAi. In particular, one intriguing group of models would involve the chromosome as a repository of an epigenetic signal that could then transfer to homologous chromosomes in mitotic or meiotic cells. Paramutation is a phenomenon in which the silencing is triggered by allelic conversion, leading to an inherited epigenetic signal without affecting the genomic sequence. There are examples of paramutation in maize and in mice (Chandler 2007). The mechanism(s) by which these occur are unknown. In maize, paramutation is extremely stable, with 100% penetrance. Analysis of paramutation in the b1 locus in maize indicates the involvement of an RNA-directed RNA polymerase and suggests that transfer of silencing information to chromosomal loci is directed by RNA (Alleman et al. 2006). In one study of mice, it was proposed that the silencing of the gene Kit can be initiated by either a paramutagenic allele (Kit*) or by injecting RNA directly into one-cell embryos (Rassoulzadegan et al. 2006). Although these studies have alternative models for the initiation of paramutation, both sets of researchers have proposed an RNA-mediated process in the maintenance of silencing heritability.
Whatever form in which the RNA would be inherited must allow sperm and oocyte transmission. We were surprised to observe the most efficient transmission through spermatogenesis. If germ-cell silencing is dependent on the induction-trigger concentration, we might expect the silencing efficacy to be dependent on germ-cell type. Sperm, which have <1% of the volume of oocytes (Kimble and White 1981), might be expected to exhibit <1/100 of the silencing capacity of an oocyte. C. elegans sperm carry RNA in an observed perinuclear “halo” that has yet to be characterized (Ward et al. 1981).
It is conceivable that this RNA population carries the signals for gene silencing to the next generation. It is of interest to note that transmission through sperm occurs even in the absence of a large pool of oma-1 mRNA expressed during spermatogenesis (Detwiler et al. 2001; Shimada et al. 2002; Reinke et al. 2004). Although oma-1 is classified as oocyte specific at both the transcript and protein levels, low levels of expression of oma-1 mRNAs or of alternative transcripts from this genomic region during spermatogenesis have not been ruled out.
Working with biases inherent in a pedigree-based analysis:
Nondestructive pedigree-based assays are a powerful tool for analyzing the phenomena of heritable silencing. Viability as an indicator for silencing affords flexibility in our selection of animals used to analyze each generation. Additionally, each silenced injected animal gives rise to a population of F1 animals and each silenced F1 can initiate an F2 population. How the selection of few individuals to represent a population affects the outcome is unknown a priori. In a multigenerational experiment, even subtle effects could have cumulative consequences that could substantially bias results.
Several potential selections are an inherent part of any oma-1(zu405) silencing assay. First, even with an apparently normal morphology and growth of the non-rescued parent population [(oma-1(zu405)] at 16°, there may be some selection from growth of animals. Second, there is certainly a strong selection for loss of oma-1(zu405) activity at growth temperatures >21°. Third, there may be deleterious effects of multigenerational silencing of oma-1, despite the presence of the compensating oma-2 gene. The rigors of multigenerational selection may introduce biases in population or long-term studies. It should be stressed that, in these experiments, any selective biases would likely have negligible effects in experiments that last only one to two generations. Long-term silencing, however, where successive populations are under selective pressure and undergoing cycles of reproduction, introduces the potential for more subtle bias and requires more careful interpretation. To definitively investigate the heritable silencing effects induced by dsRNA and account for unintentional biases that may become fixed through recurrent selections, we designed several methodical selection protocols with distinct selection criteria. Although we selected individuals from high-transmission broods, the enrichment for the trait (silencing) is derived from the analysis of one-sixth of the population (14 animals from a brood of 90–110) at each generation. We note that, although selections for three to six consecutive generations have a limited capacity to enrich for rare genetic mutations in an isogenic strain, it could certainly skew an epigenetic character of the population.
Phenotypic diversity allows selection for RNA-based epigenetics in populations:
To tease out the characteristics of heritable silencing intrinsic to the induction of RNAi, we chose different subpopulations from which to analyze the frequency and persistence of silencing. We found that selecting animals with high efficacy of silencing leads to a severe drop at the F4 generation. When we relaxed the stringency of the initial selection to permit intermediate efficacy of silencing in early generations, we found the silencing in later generations was characterized by greater variability within and between families in pedigrees. The variability extended to the F4 generations, where we found wide variation in silencing frequency among families.
These transmission data are consistent with the engagement of at least two different silencing processes in the injected populations. A high-efficiency but short-term process presumably accounts for the bulk of rescued animals in the first few generations following injection and for the majority of results in this study. Our present data are also consistent with the coexistence of a longer-term silencing process (as described previously), which we have not characterized in detail. We note, however, the challenges in late generations of discerning processes specifically initiated by the original trigger (through the RNAi mechanism or other processes) from those that arise from strong or continued selection for phenotypic character.
Acknowledgments
We thank Karen Beemon, Victor Corces, Cecilia Mello, Fredrick Tan, Sam Gu, Jonathan Gent, Judith Yanowitz, Anne Villeneuve, Poornima Parameswaran, Julia Pak, Steve Johnson, Ayelet Lamm, Jamie Saynuk, Susan Parrish, Chaya Krishna, and members of the Fire lab for help and support. Some of the strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources. This work was supported by National Institutes of Health (NIH) grants R01GM37706 (A.Z.F.) and R01HD37933 (R.L.) and NIH training grant 2T32GM007231 (R.M.A.).
References
- Alleman, M., L. Sidorenko, K. McGinnis, V. Seshadri, J. E. Dorweiler et al., 2006. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442 295–298. [DOI] [PubMed] [Google Scholar]
- Aoki, K., H. Moriguchi, T. Yoshioka, K. Okawa and H. Tabara, 2007. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26 5007–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, E., A. A. Caudy, S. M. Hammond and G. J. Hannon, 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409 363–366. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V. L., 2007. Paramutation: from maize to mice. Cell 128 641–645. [DOI] [PubMed] [Google Scholar]
- Detwiler, M. R., M. Reuben, X. Li, E. Rogers and R. Lin, 2001. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1 187–199. [DOI] [PubMed] [Google Scholar]
- Greenwald, I., P. W. Sternberg and H. R. Horvitz, 1983. The lin-12 locus specifies cell fate in Caenorhabditis elegans. Cell 34 435–444. [DOI] [PubMed] [Google Scholar]
- Grishok, A., 2005. RNAi mechanisms in Caenorhabditis elegans. FEBS Lett. 579 5932–5939. [DOI] [PubMed] [Google Scholar]
- Grishok, A., H. Tabara and C. C. Mello, 2000. Genetic requirements for inheritance of RNAi in C. elegans. Science 287 2494–2497. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., H. R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble, J. E., and J. G. White, 1981. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81 208–219. [DOI] [PubMed] [Google Scholar]
- Kosinski, M., K. McDonald, J. Schwartz, I. Yamamoto and D. Greenstein, 2005. C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132 3357–3369. [DOI] [PubMed] [Google Scholar]
- Lin, R., 2003. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev. Biol. 258 226–239. [DOI] [PubMed] [Google Scholar]
- Maine, E. M., J. Hauth, T. Ratliff, V. E. Vought, X. She et al., 2005. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired DNA during C. elegans meiosis. Curr. Biol. 15 1972–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, G., and T. Tuschl, 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431 343–349. [DOI] [PubMed] [Google Scholar]
- Mello, C., and A. Fire, 1995. DNA transformation. Methods Cell Biol. 48 451–482. [PubMed] [Google Scholar]
- Pak, J., and A. Fire, 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315 241–244. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan, M., V. Grandjean, P. Gounon, S. Vincent, I. Gillot et al., 2006. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature 441 469–474. [DOI] [PubMed] [Google Scholar]
- Reinke, V., I. S. Gil, S. Ward and K. Kazmer, 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131 311–323. [DOI] [PubMed] [Google Scholar]
- Shimada, M., H. Kawahara and H. Doi, 2002. Novel family of CCCH-type zinc-finger proteins, MOE-1, -2 and -3, participates in C. elegans oocyte maturation. Genes Cells 7 933–947. [DOI] [PubMed] [Google Scholar]
- Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish et al., 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107 465–476. [DOI] [PubMed] [Google Scholar]
- Sijen, T., F. A. Steiner, K. L. Thijssen and R. H. Plasterk, 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315 244–247. [DOI] [PubMed] [Google Scholar]
- Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok et al., 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99 123–132. [DOI] [PubMed] [Google Scholar]
- Vastenhouw, N. L., K. Brunschwig, K. L. Okihara, F. Muller, M. Tijsterman et al., 2006. Gene expression: long-term gene silencing by RNAi. Nature 442 882. [DOI] [PubMed] [Google Scholar]
- Ward, S., Y. Argon and G. A. Nelson, 1981. Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J. Cell Biol. 91 26–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C., and C. Moffitt, 2001. A critique of methods of sampling and reporting pathogens in populations of fish. J. Aquat. Anim. Health 13 300–309. [Google Scholar]
- Williams, C., and C. Moffitt, 2005. Estimation of pathogen prevalence in pooled samples using maximum likelihood methods and open-source software. J. Aquat. Anim. Health 17 386–391. [Google Scholar]