Abstract
The genus Oenothera has an outstanding scientific tradition. It has been a model for studying aspects of chromosome evolution and speciation, including the impact of plastid nuclear co-evolution. A large collection of strains analyzed during a century of experimental work and unique genetic possibilities allow the exchange of genetically definable plastids, individual or multiple chromosomes, and/or entire haploid genomes (Renner complexes) between species. However, molecular genetic approaches for the genus are largely lacking. In this study, we describe the development of efficient PCR-based marker systems for both the nuclear genome and the plastome. They allow distinguishing individual chromosomes, Renner complexes, plastomes, and subplastomes. We demonstrate their application by monitoring interspecific exchanges of genomes, chromosome pairs, and/or plastids during crossing programs, e.g., to produce plastome–genome incompatible hybrids. Using an appropriate partial permanent translocation heterozygous hybrid, linkage group 7 of the molecular map could be assigned to chromosome 9·8 of the classical Oenothera map. Finally, we provide the first direct molecular evidence that homologous recombination and free segregation of chromosomes in permanent translocation heterozygous strains is suppressed.
THE genus Oenothera (evening primrose) has an outstanding tradition in genetics, summarized in Burnham (1962), Cleland (1972), and Harte (1994). It has been under study for more than a century and is a classical example for hybrid variegation (Kirk and Tilney-Bassett 1978), biparental transmission of plastids (Hagemann 2004), and partial or complete permanent translocation heterozygosity in plants (Holsinger and Ellstrand 1984; Levin 2002; Golczyk et al. 2005). Fundamental findings, namely the rediscovery of the Mendelian rules (De Vries 1900), the independence of plastids as inheritable elements (Renner 1924, 1934, 1936), the first description of polyploidy (Lutz 1907), and the mutation theory (de Vries 1901–1903) were made on Oenothera. There is also commercial interest in the material, since seeds of Oenothera species contain high amounts of γ-linolenic acid. This essential fatty acid is used as a food supply and in medical applications (Barre 2001; Stonemetz 2008). Furthermore, Oenothera tissue culture has been applied for industrial production of pharmaceutically active secondary metabolites (Taniguchi et al. 2002). Consequently, the genus is of interest for commercial breeding and crop improvement by genetic manipulation (De Gyves et al. 2004; Fieldsend 2007).
As no other currently available model, the genus Oenothera allows studying speciation processes, including nuclear organelle co-evolution (Stubbe 1964, 1989; Cleland 1972). Plant genomes are the result of three tightly co-adapted genetic compartments (Herrmann 1997; Herrmann et al. 2003). This becomes obvious, for example, after an exchange of plastids and nuclei. Even between closely related species such an exchange often leads to serious developmental disturbances, so-called interspecific plastome–genome incompatibilities (PGI), characterized by plastid-nuclear disharmony. The impaired cooperation between the two cellular genetic compartments results from the novel combination of foreign plastid and nuclear genes that are not co-adapted (Herrmann et al. 2003; Levin 2003; Schmitz-Linneweber et al. 2005). Therefore, if PGI acts as a hybridization barrier, it appears as an important element for reproductive isolation, which represents a special case of the Dobzhansky–Muller model (Dobzhansky 1937; Turelli and Moyle 2007).
The unique position of Oenothera in addressing the role of compartmental co-evolution in speciation processes basically rests on two aspects: a favorable combination of genetic features and the existence of a well-developed taxonomy, cytogenetics, and formal genetics for the genus (Cleland 1972; Harte 1994; Dietrich et al. 1997). The genetic features include the possibility of wide interspecific crossing, biparental transmission of organelles, fertility of plastome–genome hybrids, and a system of balanced lethal factors, self-incompatibility, selective fertilization, or pollen tube competition in combination with reciprocal translocations of entire chromosome arms, resulting in partial or complete permanent translocation heterozygosity (Burnham 1962; Cleland 1972; Stubbe 1989; Harte 1994; Levin 2002). These features are relatively common throughout the eukaryotic domain, but their combination in Oenothera is unique. Taken together, they allow the exchange of plastids, of individual or multiple chromosome pairs, or even of entire haploid genomes between species, and the production of plastome–genome incompatible plants. In subsection Oenothera, the best studied of all subsections, this possibility led to the identification of five basic, genetically distinguishable plastomes (I–V) that are associated with three haploid nuclear genomes (A, B, and C), occurring in homozygous (AA, BB, CC) or stable heterozygous combinations (AB, BC, AC) (Stubbe 1959). Overall, >300 genetically and taxonomically analyzed as well as morphologically well discernible strains are known from this subsection (Stubbe and Diers 1958; Linder and Jean 1969; Cleland 1972; Jean and Linder 1979; Steiner and Stubbe 1984, 1986; Wasmund and Stubbe 1986; Wasmund 1980, 1990; Schumacher et al. 1992; Schumacher and Steiner 1993), illustrating the enormous potential of the genus for comparative studies of speciation and prespeciation processes (Cleland 1972; Stubbe 1989; Harte 1994; Dietrich et al. 1997).
This communication is part of a more comprehensive project to establish Oenothera as a molecular model, to investigate the impact of plastids in speciation, and to study unique aspects of nuclear genome evolution and epigenetics found in the genus (Cleland 1972; Harte 1994). Application of molecular approaches will also improve commercial breeding programs. So far, we have established various experimental approaches for the genus, notably protoplast and tissue culture, nuclear transformation (Stubbe and Herrmann 1982; Kuchuk et al. 1998; Mehra-Palta et al. 1998), the construction and application of an expressed sequence tag (EST) library (Mráček et al. 2006), and the sequences of the five basic plastome types as well as their analysis (Greiner et al. 2008a,b).
However, molecular markers are rare and not yet available for a large variety of strains and species (Mráček et al. 2006; Larson et al. 2008). Until now, predominantly phenotypic markers for the plastome and the nucleus were employed in Oenothera breeding (Cleland 1972, pp. 109–111; Stubbe 1989). In this report, we describe the development of molecular marker systems for entire haploid genomes, single chromosomes and plastomes, notably amplified fragment length polymorphisms (AFLP), simple sequence length polymorphisms (SSLP), and cleaved amplified polymorphic sequences (CAPS). These molecular markers bypass the limitations of phenotypic markers with regard to resolution and traceability. As documented with various crossing experiments, they enable the use of molecular approaches for the sophisticated Oenothera genetics.
A SHORT INTRODUCTION TO OENOTHERA GENETICS
Although work on Oenothera was pioneering for the development of formal and evolutionary genetics, astonishingly it has received little attention in recent textbooks and in the current mainstream speciation literature, presumably since molecular approaches were largely missing. We therefore provide an outline of some basic aspects of Oenothera genetics that recalls and illustrates the enormous potential of the model and is relevant to understanding the work presented here. Readers familiar with Oenothera genetics may disregard this outline. For details of Oenothera genetics, also see reviews of Burnham (1962), Cleland (1972), Stubbe (1989), Harte (1994), and Levin (2002).
Preservation of structural heterozygosity:
The genetics of Oenothera is a genetics of entire haploid genomes (Renner complexes), which inherit as single units and are entitled with names such as hjohansen, Galbicans, or Gflavens (for terminology, see materials and methods). All loci of a Renner complex are in linkage disequilibrium. How these superlinkage groups assemble is developed below.
Oenothera is a diploid organism with seven chromosome pairs (2n = 14). However, in Oenothera genetics it is useful to not number chromosomes but to number chromosome arms. For example, chromosome I consists of arms 1·2, chromosome II of arms 3·4, and so on, up to chromosome VII with arms 13·14. A reciprocal exchange of chromosome arms leads to an altered, so-called segmental arrangement or chromosome formula of a haploid complex. For example, the arms of chromosomes 1·2 and 3·4 can display three different segmental arrangements, including 1·4 3·2 and 1·3 2·4. Within a Renner complex, all 14 chromosome arms are involved in this process. In general, reciprocal translocations within a given Renner complex are rare, comparable to spontaneous mutations.
A strain harboring two Renner complexes with identical chromosomal arrangements is a homozygous or bivalent forming strain with seven chromosome pairs in meiosis. Oenothera elata subsp. hookeri strain johansen with the Renner complex combination hjohansen·hjohansen can serve as an example. In principle, diakinesis and meiotic segregation in that strain looks identical to that of any other diploid organism (Figure 1, A–C). However, if reciprocal translocations of chromosome arms have occurred in one of the Renner complexes, meiotic pairing patterns are altered. An example illustrates the hybrid hjohansen·Gflavens. The exchange of arm 2 with arm 4 in Gflavens, relative to the hjohansen chromosomes 1·2joh and 3·4joh, leads to the altered segmental arrangement 1·4flavG and 3·2flavG. In diakinesis, chromosomes 1·2joh-2·3flavG-3·4joh-4·1flavG catenate and arrange in a ring of four chromosomes (⊙4). The remaining chromosomes, namely 5·6 7·10 9·8 11·12 and 13·14, still pair as bivalents. The hybrid then possesses the chromosome configuration ⊙4, 5 pairs (Figure 1, D–F). In strains where reciprocal translocations include more chromosomes, larger and/or more rings will appear in meiosis, depending on the translocation pattern, e.g., ⊙6, ⊙4, 2 pairs. If at least one free bivalent is being formed, the situation is designated as partial permanent translocation heterozygosity. However, reciprocal translocations may encompass the entire chromosome complement. In this case, none of the chromosomes can pair as a bivalent or is located in a small ring, and the two Renner complexes assemble in a single ring of 14 (⊙14) incorporating all chromosomes. This pattern is designated terminal or complete permanent translocation heterozygosity. An example is found in Oe. biennis strain suaveolens Grado with the Renner complexes Galbicans and Gflavens (Figure 1, G–I).
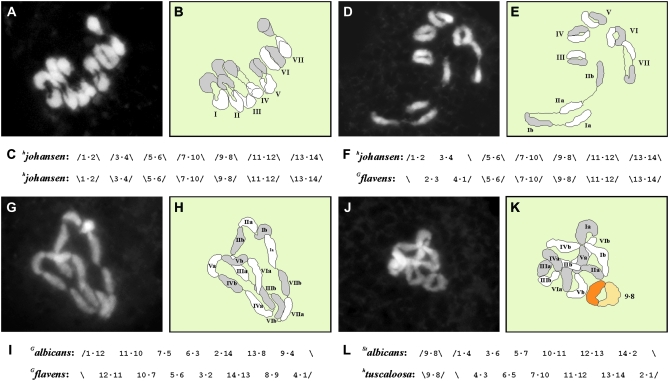
Figure 1.—
Determination of diakinesis/metaphase I configuration in Oenothera strains or hybrids. Chromosome configuration of 7 pairs in strain johansen (hjohansen·hjohansen) (A–C), ⊙4, 5 pairs in hybrid hjohansen·Gflavens (D–F), ⊙14 in strain suaveolens Grado (Galbicans·Gflavens) (G–I), and ⊙12, 1 pair in hybrid Stalbicans·htuscaloosa. Determination in diakinesis via DAPI staining (A, D, G, J) and graphical interpretation (B, E, H, K). The respective chromosome configurations can be predicted by the chromosome formulas of the Renner complexes involved (C, F, I, L). Bivalents are labeled with roman numbers (I–VII), single chromosomes in rings with a combination of roman numbers and letters (Ia, Ib–VIIa, VIIb); so far, chromosome identity cannot be assigned directly in Oenothera. Identity of bivalent 9·8 in K was assigned indirectly since it was deduced from the chromosomal formulas of the Renner complexes involved and thus represents the free pair of Stalbicans·htuscaloosa (L).
The chromosome configuration, i.e., the number and size of rings and bivalents in diakinesis of a strain or hybrid, can be predicted if the segmental arrangement of chromosome arms, the chromosomal formula, for the complexes involved is known (Figure 1).
Rings of chromosomes in diakinesis have consequences for inheritance, since they change linkage equilibrium. Each ring or free chromosome pair constitutes a single linkage group. For example, a hybrid with the chromosome configuration ⊙6, ⊙4, 2 pairs displays four linkage groups. Two linkage groups are represented by the free, bivalent-forming chromosomes and two by the ring of six and ring of four chromosomes, respectively. A hybrid with the chromosome configuration of 7 pairs, in turn, displays seven linkage groups, one for each chromosome. If chromosomes assemble in a ring of 14 chromosomes, only a single linkage group is found, involving the entire chromosome complement of both haploid genomes.
Exchange of plastids between species:
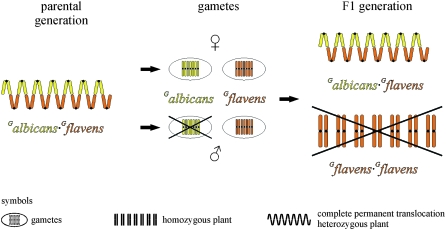
The occurrence of just one superlinkage group results in the genetics of complete permanent translocation heterozygosity. Figure 2 illustrates the maintenance of the complete permanent translocation heterozygote Oe. biennis strain suaveolens Grado. Its meiotic ring, which contains the Renner complexes Galbicans and Gflavens with the chromosome configuration ⊙14, mentioned above, is considered to prevent free segregation of chromosomes and practically homologous recombination. Consequently, the Galbicans and Gflavens complexes are not mixed and are inherited as units of a single linkage group. Male and female gametes exclusively contain chromosomes of the haploid sets of either Galbicans or Gflavens. Gametophytic lethal factors ensure that the complex Galbicans is inherited strictly maternally by the egg cell (♀). The Galbicans pollen is abortive and eliminated. Galbicans is therefore designated the α-complex or egg cell complex. In the case considered, the β-complex Gflavens is inherited biparentally via egg and pollen (♂♀) and is designated the pollen complex. However, sporophytic lethal factors prevent the occurrence of Gflavens·Gflavens homozygotes in the offspring of Oe. biennis strain suaveolens Grado. Consequently, the F1 generation is identical to the parental generation without segregation of traits, since chromosomes separate as a set without intermixing. Comparable to apomixis, a clone is produced. Species obeying this pattern are designated as true breeding species.
Figure 2.—
Maintenance of the permanent translocation heterozygote Oe. biennis strain suaveolens Grado. Free segregation of chromosomes and homologous recombination is suppressed in the complete permanent translocation heterozygote between the haploid Renner complexes Galbicans (yellow) and Gflavens (orange). Gametophytic lethal factors repress germination of Galbicans pollen, and sporophytic lethal factors eliminate homozygous Gflavens·Gflavens offspring in F1. Referring to the parental generation, identical offspring are produced in F1. For a detailed explanation, see text.
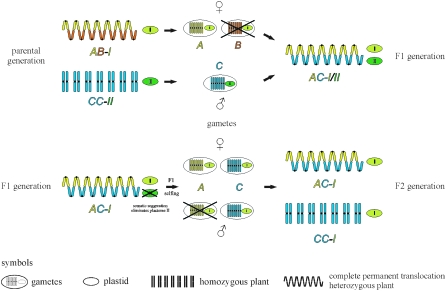
Knowledge about chromosomal formulas and lethal factors along with biparental transmission of plastids offer the possibility of exchanging plastids and/or single pairs or sets of chromosomes between Oenothera species. Figure 3 illustrates such an exchange of plastids and haploid genomes between species via sexual crosses in just two generations. In the chosen example, a complete permanent translocation heterozygous hybrid AB associated with plastome I as seed parent is crossed with the homozygote CC-II. [We have chosen this example for simplicity, although the combinations AC-I, CC-I, and CC-II are incompatible and barley viable. In the experiment they have to be nurtured with additional compatible plastomes, e.g., by AC-I/IV, CC-I/VI, and CC-II/IV (Stubbe 1960).] The homozygous Renner complex C lacks gametophytic or sporophytic lethal factors and self-incompatibility alleles. It forms seven bivalents in meiosis. In a cross between AB and CC, only the hybrid AC occurs in F1, since complex B cannot be inherited paternally as its pollen is abortive. The chromosomal formulas of the A and C complexes were chosen in a way that hybrid AC assembles a meiotic ring of 14 chromosomes. This meiotic configuration thus allows ready exchange of plastids. Due to biparental plastid inheritance, the F1 generation carries two plastome types, namely plastomes I and II. Somatic segregation of the two plastome types leads to sorting out of plastids (Figure 4A), resulting in flowers on a plant carrying exclusively plastome I (or plastome II, respectively). Selfing of appropriate flowers ensures that only plastome I is inherited to the next generation. However, in F2 the nuclear genome splits into the progenies CC-I and AC-I. In the chosen example, complex A is exclusively inherited by the egg cell, which prevents the generation of AA homozygotes in the offspring, but, as described above, complex C lacks gametophytic or sporophytic lethal factors. Since the ring of 14 chromosomes should inhibit free segregation of individual chromosomes and practically homologous recombination between haploid genomes in the hybrid AC, the coding potential of the A and C genomes is not mixed and a seemingly unchanged, homozygous CC genotype, now associated with plastome I, occurs in F2. With this breeding strategy, plastids as well as entire haploid genomes were exchanged. Starting in the parental generation with the heterozygous hybrid AB-I and the homozygous combination CC-II, crossing end products in F2 are the new combinations AC-I and CC-I (Figure 3).
Figure 3.—
Exchange of plastids and genome rearrangement between Oenothera strains. Repression of homologous recombination in a complete permanent translocation heterozygote and somatic segregation leading to sorting out of the two plastid types in F1 (I and II) allow the exchange of plastomes as well as haploid genomes (A, B, and C) in F2. For detailed explanation, see text.
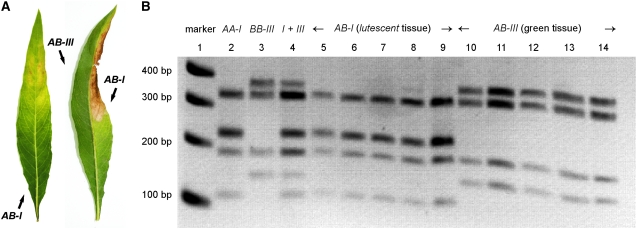
Figure 4.—
Lutescent phenotype and somatic segregation of two plastome types in the F1 generation of a cross between Oe. elata subsp. hookeri strain johansen (AA-I) and Oe. grandiflora strain tuscaloosa (BB-III), resulting in so-called hybrid variegation (A). Variegated tissue is generated if plastomes are inherited by both sexes and only one of the plastomes is incompatible with the nuclear genome. Separation of the two tissue types differing in their plastid genome results from the statistical process of sorting out, i.e., the random segregation of the two plastid types during cell divisions (Kirk and Tilney-Bassett 1978; Birky 2001). Incompatible and green tissue was correlated with a plastome type via a BamHI CAPS marker (Table 3) (B). Marker alleles rrn16-trnIGAU I3 and -II/III8 were amplified from plastomes Ijoh and IIItusca and digested with BamHI (lanes 2 and 3). Lane 4 shows a mixture of cleaved Ijoh and IIItusca PCR products. Applying that marker to tissues of F1 individuals, plastome Ijoh exclusively correlates with the lutescent phenotype (lanes 5–9) and plastome IIItusca with green tissue (lanes 10–14).
MATERIALS AND METHODS
Plant material:
Genetic constitution and corresponding references for Oenothera strains used in this work are listed in supplemental Table 1. For a detailed taxonomy, see Dietrich et al. (1997) and Dietrich (1977).
Terminology of Renner complexes:
Several superscripts are used in this work to specify the source or genetic behavior of distinct Renner complexes. Throughout the literature lethal-factor-free Renner complexes are commonly designated haplo-complexes or haploid-complexes, characterized by the superscript “h,” e.g., hjohansen. Additional superscripts are employed to further specify the source of a given Renner complex. Following the terminology suggested by Renner (1917, 1956), Oenothera strains and species can be described by their Renner complexes, e.g., Oe. biennis (albicans·rubens) or Oe. suaveolens (albicans·flavens). Refined genetic analysis made it necessary to distinguish Renner complexes of the same kind from different sources, e.g., albicans Grado and albicans Standard (Stubbe 1953). Since additions such as “Grado” or “Standard” interfere with the notation, they were abbreviated with “G” or “St” and used as superscripts—Galbicans or Stalbicans in this example. This designation, originally developed by Stubbe (1960), was adapted to all Renner complexes with Latin names, since different haplotypes can be recognized in nature for all of them and molecular analysis allows their discrimination. Abbreviations describing sources of different Renner complexes used in the text are St, Standard; G, Grado; S, Sweden; and Th, Thorn.
Field growth and crossing experiments:
To grow Oenothera in the experimental field, seeds were sown in January, choked at the two-leaf to four-leaf state in trays of 54 seedlings, and kept in a greenhouse until end of April. After 1 or 2 weeks of hardening, rosettes were planted outside. Flowering of evening primroses started during summer. A few strains such as pupurata, douthat 1, or biennis München behave biennially, depending on seasonal variations.
Strains of Oe. grandiflora were short-day treated to induce early flowering in the season. This treatment, performed in a greenhouse after plants had broken rosette stage at a height of 40 cm, ensured that the material flowered and set seeds simultaneously with the other strains under study. Without this treatment Oe. grandiflora starts blooming later in autumn and does not complete its generation cycle before winter in the climate of Munich. Alternatively, seeds of Oe. grandiflora were sown in autumn in a greenhouse to achieve shoot formation before winter. During winter the material grew under a natural short-day period. Some of these plants were not transferred to the field later in spring and were kept in the greenhouse until flowering, since seasonal cold interference may inhibit flower induction.
For crossing experiments, immature anthers were removed the day before maturation and flowering. The emasculated flower was guarded and pollinated with the desired pollen of the male crossing mate during the following day and guarded again. Pollination was repeated the next day. Seeds were harvested ∼6 weeks after pollination and dried at room temperature. No dormancy is needed for propagating the next generation. Dry seeds can be stored at −20° for decades. Without freezing, seeds lose their ability to germinate after ∼4 years.
To determine the plastome type of a flower bud, leaf material of three successive bracts on a stem was pooled and used for DNA isolation. Oenothera bracts are arranged in orthostiches with an angle of 120° around the stem. Pooling material ensures the recognition of stems carrying two different plastome types.
Determination of chromosome configurations:
Inflorescences of appropriate age were collected from several plants of each genotype, fixed with acetic acid/ethanol (3: 1), and stored at −20°. The anther loculi were gently squeezed in a drop of 45% acetic acid under a cover slip. After freezing in dry ice, cover glasses were removed. The preparations were air dried and mounted in a drop of 4′,6-diamidino-2-phenyloindole (DAPI) staining solution (2 mg/ml; dissolved in a mixture of glycerol:McIlvaine buffer, 1:2). Images were taken with a Zeiss epifluorescence microscope equipped with a cooled CCD monochrome camera. For digital arrangement of meiotic configurations in one focal plane, five to seven frames were captured. The frames were stacked and combined applying the Combine ZM software (http://www.hadleyweb.pwp.blueyonder.co.uk).
DNA isolation:
Total DNA was isolated from green leaf material using the DNeasy plant mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol with minor modifications: Approximately 50–100 mg of plant material (fresh weight) was homogenized. To 400 μl AP1 buffer, supplied by the manufacturer, 4.0 μl of 10% polyvinylpyrrolidone 10,000 and 0.4 μl 1 m sodium ascorbate solution were added. To increase yield, DNA was eluted twice from the DNeasy Mini Spin Column with 50 μl provided by AE buffer. Yields were in a range of 100 ng DNA/μl but decreased with age of the material.
PCR reaction and product sequencing:
PCR was performed using standard protocols. When Oenothera DNA was used as a template, the initial denaturation time was prolonged to 5 min. Single PCR products, derived from homozygous Oenothera strains, were directly sequenced after cleaning with QIAquick PCR Purification kit (Qiagen). Nucleotide sequences were determined with an ABI 377 robot (Applied Biosystems, Foster City, CA). Cycling reactions were performed using DYE ET Terminator Cycle Sequencing kit (Amersham Biosciences, Uppsala, Sweden) and cycling conditions recommended by the supplier.
If a genomic locus is amplified in a permanent translocation heterozygote, usually two bands, originating in the α- and β-complexes, respectively, are derived from the same primer pair. Both bands were cloned individually with the pGEM-T Easy Vector system (Promega, Mannheim, Germany) according to the supplier's manual. For sequencing analysis the primer pairs T7for (5′-TAATACGACTCACTATAGGG-3′) and SP6rev (5′-ATTTAGGTGACACTA TAGAAT-3′) were used. Several independent clones of the same PCR product were sequenced.
Design and analysis of SSLP and CAPS markers:
Distinct ESTs were chosen to generate PCR-based markers as described previously (Mráček et al. 2006). CAPS markers were designed using the software SNP2CAPS (Thiel et al. 2004). The PCR products used for genotyping were digested with appropriate restriction endonucleases supplied by Fermentas International (Burlington, Ontario, Canada) or New England Biolabs (Ipswich, MA) according to the manufacturer's protocol and analyzed on 2–3% agarose gels.
Design and application of AFLP markers:
AFLP genotyping is a PCR-based approach that involves restriction of genomic DNA, followed by ligation of adapters to the fragments generated and selective PCR amplification of a subset of these fragments (Vos et al. 1995). AFLP genotyping was optimized by selection of appropriate enzymes, primer lengths, and PCR reagents, which are crucial for generating banding patterns with detectable polymorphisms (Ridout and Donini 1999). The restriction enzymes SacI and MseI and appropriate primer combinations, which gave a high number of polymorphic bands, were chosen for analyzing genetic diversity. AFLP analysis was performed essentially as described (Peters et al. 2001), except that a fluorescence-based approach was used (Huang and Sun 1999). In brief, in the first reaction, restriction and ligation to MseI and SacI adapters took place simultaneously. The assays contained 100–300 ng DNA, 20 nmol ATP, 2.5 units MseI, 5 units SacI with 1× buffer, 15 μg bovine serum albumin, 1.2 Weiss units T4 DNA ligase (Qiagen), 50 pmol of MseI adapter, and 5 pmol of SacI adapter in a total volume of 20 μl. They were incubated at 37° for 3 hr (AFLP reaction I). For the first PCR amplification cycle (AFLP reaction II) 4 μl of a 10-fold diluted AFLP reaction I with 10 mm Tris–HCl and 0.1 mm EDTA, pH 8.0 (0.1 TE buffer) was used. AFLP reaction II mixtures were amplified in 20 μl using standard PCR conditions and SacI and MseI primers, each containing one additional selective base (SacI + 1, MseI + 1). AFLP reaction II's were conducted as follows: 20 sec at 94°, followed by 20 cycles of 30 sec at 56° and 2 min at 72°. They were terminated by incubation at 60° for 30 min. The products obtained were diluted 10-fold with 0.1 TE buffer. In a second amplification cycle (AFLP reactions III) AFLP fragments were labeled using the selective primers SacI + 2 labeled with the fluorescent dyes 6-FAM (6-carboxy-fluorescein) or Joe (2,7-dimethoxy-4,5-dichloro-6-carboxy-fluorescein). For PCR, 4 μl of diluted AFLP reaction II, 0.1 μm SacI + 2 primer, and 0.25 μm MseI + 3 primer were used. Selective primers are listed in supplemental Table 2. The reactions were as follows: incubation at 94° for 2 min and then 10 cycles of 20 sec at 94° and 30 sec at 66°. The annealing temperature was decreased by 1°/cycle. The reactions were completed with an incubation at 72° for 2 min. In a subsequent step, repeated 20 times, the assays were incubated for 20 sec at 94°, 30 sec at 56°, and 2 min at 72°, followed by a final incubation at 60° for 30 min. All primer combinations used for AFLP analyses are listed in supplemental Table 3. AFLP products were mixed with an equal volume (1 μl) formamide dye (80% formamide, 10 mg/ml dextran blue, 5 mm EDTA) and 0.15 μl of GENESCAN-500 ROX (carboxy-X-rhodamine) internal lane standard (Applied Biosystems). Mixtures were heated at 80°–90° for 2 min and subsequently kept at 4° prior to loading. To visualize the DNA fingerprints, fragments were separated in a 5% denaturing polyacrylamide gel on an ABI Prism 377 DNA automated sequencer (Applied Biosystems). The gel was prepared using Long Ranger gel solution (50% stock solution) (Cambrex, Rockland, ME). The MATRIX file was generated with the dyes 6-FAM, ROX, NED, and Joe (MWG Biotech, Munich, Germany). Gel images were captured by GeneScan software (Applied Biosystems). The amplified fragments were recognized by fluorescence laser scanning. Nomenclature used for primer combinations and markers was similar to Peters et al. (2001). The first two letters represent restriction enzymes, the next three numbers the distinct IDs for primer combinations, and the final number the size of a given AFLP maker.
Calculation of genetic linkage:
Logarithm of odds (LOD) scores and genetic linkage were calculated with the Kosambi function, using the Joinmap program (van Ooijen and Voorrips 2001).
RESULTS
Genomewide suppression of homologous recombination:
To date, rearrangements of Renner complexes, plastid exchanges as well as repression of both homologous recombination and free segregation of chromosomes due to a meiotic ring formation in Oenothera strains, have been characterized indirectly and exclusively by cytological analysis in combination with formal genetics on the basis of only a few phenotypic markers (Cleland 1972; Stubbe 1989; Harte 1994; Stubbe and Steiner 1999). Molecular proof for these findings was lacking.
To verify the theory and to estimate the quality of an interspecific plastid exchange between Oenothera species, we compared the natural strain Oe. elata subsp. hookeri strain johansen, which is associated with plastome I (johansen Ijoh; genotype: hjohansen·hjohansen Ijoh), with the artificial strain johansen IIIlam (genotype: hjohansen·hjohansen IIIlam), a combination of the nucleus of Oe. elata subsp. hookeri strain johansen and the plastome of Oe. glazioviana strain rr-lamarckiana Sweden (IIIlam). The strain johansen Ijoh was collected in 1927 (Cleland 1935) and inbred since then. The artificial hybrid strain johansen IIIlam was established in 1983 and propagated by continuous selfing. It was kindly provided by Wilfried Stubbe. The strain johansen IIIlam was not produced with the minimum number of crosses necessary to establish such a hybrid. Instead, its Renner complex hjohansen was combined four times during the crossing program in a ring of 14 chromosomes with Renner complex Galbicans of Oe. biennis strain suaveolens Grado, resulting in the hybrid Galbicans·hjohansen (supplemental Figure 1). According to formal Oenothera genetics, in this hybrid homologous recombination and free segregation of chromosomes should not have happened between the two haploid genomes.
AFLP fingerprinting involving 10 different primer combinations gave rise to 711 bands in the strains johansen Ijoh and johansen IIIlam. Although their hjohansen complex was combined four times during the crossing program with a different Renner complex, only a single band was specific for strain johansen IIIlam; all others were identical (Table 1). Both hjohansen complexes differed by only 0.14%. In general, genetic variation between strains of Oenothera subspecies is in the range of 10%; variation between species or basic nuclear genotypes is substantially higher (Rauwolf 2008).
TABLE 1.
AFLP genotyping of the strains johansen Ijoh and johansen IIIlam and of the Renner complex Galbicans
| Primer combinationa | No. of bands johansen Ijoh | No. of band johansen IIIlam | No. of polymorphic bands between johansen Ijoh and johansen IIIlam | No. of Galbicans-specific markers detected between Galbicans and hjohansen |
|---|---|---|---|---|
| sm261 | 75 | 75 | 0 | 7 |
| sm263 | 78 | 78 | 0 | 2 |
| sm267 | 108 | 108 | 0 | 3 |
| sm276 | 49 | 49 | 0 | 4 |
| sm279 | 47 | 48 | 1 | 8 |
| sm280 | 80 | 80 | 0 | 4 |
| sm281 | 54 | 54 | 0 | 2 |
| sm285 | 65 | 65 | 0 | 4 |
| sm290 | 66 | 66 | 0 | 5 |
| sm299 | 88 | 88 | 0 | 6 |
| Total | 710 | 711 | 1 | 45 |
Of 711 bands generated, only a single polymorphism was detected between the strains johansen Ijoh and johansen IIIlam. The polymorphic primer combination is indicated by italics. The combination Galbicans·Galbicans could not be investigated directly, since it is not realizable genetically. Galibcans was therefore analyzed in a Galbicans·hjohansen background.
For primer combination and sequences, see supplemental Tables 2 and 3.
AFLP fingerprinting of plants containing the Renner complexes Galbicans·hjohansen, using the same primer combinations as before, revealed a total of 45 markers specific for the Galbicans complex (Table 1). The additional band of strain johansen IIIlam compared johansen Ijoh originated in the Galbicans complex and was also confirmed there (sm279_291.7), suggesting the transfer of that sequence interval from Galbicans to hjohansen by homologous recombination during the breeding program to generate the artificial hjohansen·hjohansen IIIlam combination. Thus, only 1 of the 45 possible markers detected (or 2.2%) was transferred from Galbicans to hjohansen. AFLP makers for both hjohansen and Galbicans are available upon request.
Since polymorphisms between the strains johansen Ijoh and johansen IIIlam are negligible, but common between hjohansen and Galbicans, our data illustrate the lack of homologous recombination (disregarding telomeric parts of chromosomes) and exclude free segregation of chromosomes between the haploid genomes Galbicans and hjohansen in a ring of 14 chromosomes during the crossing program.
The data corroborate and extend Oenothera genetics, postulating that in ring-forming hybrids individual Renner complexes, i.e., entire haploid genomes, behave as a single coupling group. Free segregation of chromosomes is suppressed by the meiotic ring and if recombination occurs as a rare event, it is restricted to telomeric ends (Cleland 1972, pp. 113–122). Direct molecular evidence for this finding will be presented below. Furthermore, the genomewide sheltering from exchange of genetic material in the nucleus provides an advantageous genetic situation for exchanges of plastids between Oenothera species.
Genotyping Renner complexes:
If Renner complexes behave as single linkage groups in complete permanent translocation heterozygous combinations, in principle only a single marker should be needed to follow up the fate of a particular complex in crossing programs. The SSLP marker M40 was considered to be a suitable choice for genotyping a large variety of Renner complexes. It was derived from the EST cluster C_1231-11-B04 of Oe. elata subsp. hookeri strain hookeri de Vries (Mráček et al. 2006) and encodes a chloroplast-located sedoheptulose–bisphosphatase, and its gene is interrupted by two variant intervening sequences. Genomic M40 alleles known so far are intron spanning and highly polymorphic between the A genome hjohansen and the B genome htuscaloosa, suggesting that the region is suitable for detecting further polymorphisms among different Renner complexes.
PCR products of M40 were amplified in 20 homozygous and 5 permanent translocation heterozygous strains. Homozygous strains harbor two identical haploid Renner complexes, so-called haplo-complexes, and are free of lethal factors. For example, strain tuscaloosa of Oe. grandiflora carries the haplo-complex htuscaloosa twice. Consequently, the primer pair M40for (5′-ACCGTCTCCTCCAAGCACTGC-3′) and M40rev (5′-TCAGCCCTTTGTCCG AAGTCG-3′) amplifies only a single band of 500 bp (Figure 5, lane 1, and Table 2). Strain bauri Standard, a complete permanent translocation heterozygote, contains two different Renner complexes (Stlaxans and Stundans). Their M40 alleles differ, since the primer pair amplified two bands of 474 and 579 bp (Figure 5, lane 2, and Table 2). The bauri Standard bands could originate in either Stlaxans or Stundans and were assigned by crosses with htuscaloosa (Figure 5). Tuscaloosa as seed parent results in the F1 hybrid htuscaloosa·Stundans. Due to gametophytic lethal factors, the combination htuscaloosa·Stlaxans was not produced in this crossing direction. The Stlaxans complex is inherited strictly maternally. Taking bauri Standard as the pollen donor, the only realizable hybrid is htuscaloosa·Stundans; the Stundans complex is inherited strictly paternally. Investigating htuscaloosa·Stundans with marker M40 displays two PCR products of 500 and 579 bp (Figure 5, lane 3). Since the former originates in the htuscaloosa complex, the 579-bp band must be linked to Stundans. The 474-bp PCR product, detected in the strain bauri Standard, in turn can serve as a marker for Stlaxans. A comparable strategy was performed to assign M40 marker alleles to the Renner complexes of further permanent translocation heterozygous strains such as ammophila Standard (Strigens·Stpercurvans), rr-lamarckiana Sweden (r-Svelans·r-Sgaudens), suaveolens Grado (Galbicans·Gflavens), and suaveolens Standard (Stalbicans·Stflavens) (Table 2).
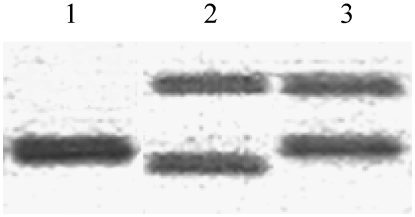
Figure 5.—
Assignment of M40 alleles to the Renner complexes Stlaxans·Stundans in the strain bauri Standard: In tuscaloosa, a single band of 500 bp was amplified (lane 1); in bauri Standard, two bands of 474 and 579 bp were detected (lane 2). The two bands of 500 and 579 bp detectable in the F1 hybrid htuscaloosa·Stundans assign the latter to Stundans (lane 3).
TABLE 2.
SSLP and selected restriction endonuclease patterns of the M40 microsatellite regions in various Renner complexes
| M40 allele | Renner complex | SSLP (bp) | CAPS (bp) (MboII) | CAPS (bp) (MspI) | CAPS (bp) (SpeI) | GenBank accession no. |
|---|---|---|---|---|---|---|
| A1 | Galbicans, Stalbicans, hblandina de Vries, hfranciscana de Vries, hfranciscana E. & S., hhookeri de Vries, Stlaxans, hpurpurata, Strigens | 474 | 210, 170, 67, 27 | 319, 155 | 249, 126, 99 | EU432376, EU432377, EU432378, EU432379, EU432380, EU432381, EU432382, EU432383, EU432384 |
| A2 | hchapultepec, hcholula, hpuebla, htoluca, Stundans | 579 | 285, 200, 67, 27 | 431, 148 | 352, 135, 92 | EU432385, EU432386, EU432387, EU432388, EU432389 |
| A3 | hjohansen, r-Svelans | 583 | 349, 207, 27 | 428, 155 | 352, 132, 99 | EU432390, EU432391 |
| B1 | hdecipiens de Vries, hdeserens de Vries, r-Sgaudens | 470 | 207, 169, 67, 27 | 315, 155 | 371, 99 | EU432392, EU432393, EU432394 |
| B2 | hbellamy A, hBA-castleberry A-4, hBA-chastang 7, Gflavens, hstockton 1 | 499 | 226, 179, 67, 27 | 325, 174 | 364, 135 | EU432395, EU432396, EU432397, EU432398, EU432399 |
| B3 | Stflavens | 500 | 227, 179, 67, 27 | 325, 175 | 365, 135 | EU432400 |
| B4 | htuscaloosa | 500 | 227, 179, 67, 27 | 325, 175 | 365, 135 | EU432401 |
| C1 | hdouthat 1, hwilliamsville 2, hwilson creek 1 | 496 | 243, 159, 67, 27 | 496 | 364, 132 | EU432402, EU432403, EU432404 |
The corresponding Oenothera strains and species are listed in supplemental Table 1.
All marker regions mentioned were analyzed by sequencing. The sequences are deposited in GenBank (Table 2). In all Renner complexes investigated, the marker allele M40 displays two introns. In general, they contain length polymorphisms resulting in eight SSLPs among the 29 Renner complexes investigated. At least one specific SSLP was found for each of the three basic nuclear genome types A, B, and C. PCR products converted into CAPS markers allow discrimination of single genotypes on 2–3% agarose gels (Table 2).
Genotyping of basic plastome types and their subplastomes:
To verify interspecific plastome exchanges, plastome-specific markers are required. For this purpose, the plastid rrn16-trnIGAU spacer region was investigated from 42 Oenothera strains. A high degree of polymorphism in this region was reported for a limited number of strains (Hornung et al. 1996; Sears et al. 1996). Amplification with the primer pairs 16S SEQ (+) (5′-CTTGTACAC ACCGCCCGTCACACT-3′) and trnI PCR (+) (5′-CCAGGCACAACGACGCAATTATCA-3′) and sequence analysis of the PCR products derived uncovered a BamHI restriction polymorphism in the rrn16-trnIGAU spacer. Within subsection Oenothera, 17 different BamHI restriction patterns could be detected, of which 13 could readily be distinguished on 2% agarose gels (supplemental Figure 2 and Table 3). The marker alleles rrn16-trnIGAU I1 and rrn16-trnIGAU I2 of strains chapultepec, cholula, puebla, or toluca cannot be discerned via a BamHI, but rather by BsmBI digestion (rrn16-trnIGAU I1: 619 bp, 261 bp and rrn16-trnIGAU I2: 870 bp). For all five basic plastomes at least one specific allele was detected (Table 3). The patterns of subplastome variation obtained reflect both phylogenetic relationships between the chosen strains and the basic plastome types (see discussion).
TABLE 3.
BamHI restriction and SSLP pattern of the rrn16-trnIGAU spacer region in Oenothera plastomes and subplastomes used in this study
| rrn16-trnIGAU allele | Strain | Species | Plastome type | SSLP (bp) | CAPS (bp) (BamHI) | GeneBank accession no. |
|---|---|---|---|---|---|---|
| I1 | chapultepec | Oe. elata subsp. elata | I | 880 | 322, 229, 220, 109 | EU262892 |
| I2 | cholula, puebla, toluca | Oe. elata subsp. elata | I | 870 | 322, 220, 218, 110 | EU262893, EU282392, EU282393 |
| I3 | franciscana de Vries, franciscana E.& S., johansen | Oe. elata subsp. hookeri | I | 1058 | 322, 224, 220, 182, 110 | EU282394, EU282395, EU262894, |
| I4 | hookeri de Vries | Oe. elata subsp. hookeri | I | 876 | 322, 247, 197, 110 | EU262895 |
| I5 | bauri Standard | Oe. villosa subsp. villosa | I | 891 | 322, 262, 197, 110 | EU262896 |
| II/III1 | biennis München, castleberry B-8, conferta Standard, purpurata, suaveolens Fünfkirchen, suaveolens Grado, suaveolens Standard | Oe. biennis, Oe. grandiflora, Oe. biennis × Oe. glazioviana | II or III | 977 | 322, 257, 216, 182 | EU282396, EU282397, EU282398, EU282399, EU282400, EU262897, EU282401 |
| II/III2 | coronifera Standard, nuda Standard | Oe. biennis, Oe. glazioviana | II | 981 | 322, 257, 220, 182 | EU282402, EU262898 |
| II/III3 | bellamy A, biennis de Vries, chastang 7, chicaginensis Colmar, horsesheads 2, marienville 3, stockton 1 | Oe. biennis, Oe. grandiflora, Oe. nutans | II or III | 963 | 322, 318, 182, 141 | EU282404, EU262899, EU282405, EU282403, EU282406, EU282407, EU282408 |
| II/III4 | mitchell | Oe. nutans | III | 980 | 335, 322, 182, 141 | EU262900 |
| II/III5 | castleberry A-4 | Oe. grandiflora | III | 845 | 341, 322, 182 | EU262901 |
| II/III6 | elkins 2 | Oe. nutans | III | 940 | 322, 295, 182, 141 | EU262902 |
| II/III7 | lawrenceville 3 | Oe. biennis | II | 781 | 322, 318, 141 | EU262903 |
| II/III8 | tuscaloosa | Oe. grandiflora | III | 1009 | 364, 322, 182, 141 | EU262904 |
| II/III9 | blandina de Vries, decipiens de Vries, deserens de Vries, rr-lamarckiana Sweden | Oe. glazioviana | III | 617 | 322, 295 | EU282409, EU282410, EU282411, EU262905 |
| IV1 | ammophila Standard, atrovirens Standard, silesiaca Standard, st. stephen | Oe. oakesiana, Oe. parviflora | IV | 961 | 322, 236, 221, 182 | EU282415, EU262906, EU282412, EU282413 |
| V1 | douthat 1 | Oe. argillicola | V | 1101 | 322, 236, 221, 182, 140 | EU262907 |
| V2 | williamsville 2, wilson creek 1 | Oe. argillicola | V | 1102 | 322, 236, 221, 182, 141 | EU282414, EU262908 |
| ber | berteriana Schwemmlea | Oe. villaricae | NA | 685 | 328, 357 | EU262909 |
Supplemental Table 1 lists references describing species, strain, and plastome type for each single accession.
The strain berteriana Schwemmle of Oe. villaricae belongs to subsection Munzia, from which no plastome types have been defined.
New combinations of genetic compartments:
To demonstrate potential and applicability of the marker systems described above, two incompatible plastome–genome combinations, AB-I and BB-II, as well as a genetically compatible one (AB-III) were established in crossing experiments.
With appropriate parental lines, the production of heterozygous interspecific plastome–genome incompatible hybrids, such as AB-I, may be rather easy. Many of them arise already in the F1 generation. The homozygotes Oe. elata subsp. hookeri strain johansen (AA-I; hjohansen·hjohansen Ijoh) and Oe. grandiflora strain tuscaloosa (BB-III; htuscaloosa·htuscaloosa IIItusca) were used to generate both the incompatible AB-I and the compatible AB-III hybrids. Since plastids are inherited biparentally in Oenothera and segregate somatically in the F1 generation, the AB-I and AB-III hybrids, hjohansen·htuscaloosa Ijoh and hjohansen·htuscaloosa IIItusca, respectively, can be derived directly from a cross of both parental species. Compatible and incompatible tissue usually segregates on the same individual. The segregation of plastomes was checked by the PCR polymorphism described above and found to be consistent with the expected compatible or incompatible phenotypes, respectively (Figure 4).
A different strategy was chosen to produce the incompatible combination BB-II. In contrast to AB-I, which requires only a single cross, generation of BB-II demands a relatively complex crossing program because a foreign plastid has to be incorporated into a homozygous nuclear background, as will be explained subsequently. The possibility of such plastid exchanges is unique to Oenothera genetics.
The first crossing mate, strain suaveolens Grado of Oe. biennis (AB-II), is a complete permanent translocation heterozygote and its Renner complexes Galbicans and Gflavens form a ring of 14 chromosomes in diakinesis (supplemental Table 4, first row). The strain produces two different egg cells. The α-complex (Galbicans) is exclusively inherited maternally. The β-complex (Gflavens) is predominantly inherited paternally and found facultatively in egg cells. The second crossing partner, Oe. grandiflora strain tuscaloosa (BB-III), displays a different breeding behavior. It forms seven bivalents in meiosis, with twice the htuscaloosa complex (supplemental Table 4, second row). The complex is inherited biparentally and lacks gametophytic or sporophytic lethal factors.
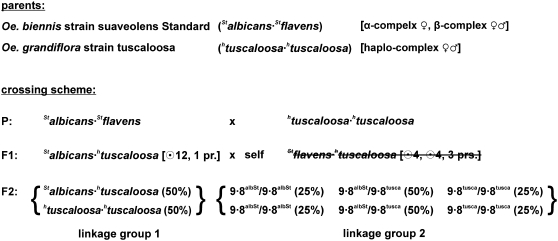
Figure 6 presents the crossing scheme to exchange plastomes between these two species. With Oe. biennis strain suaveolens Grado as female parent and Oe. grandiflora strain tuscaloosa as pollen donor, the F1 generation obtained is not uniform. Two egg cells, differing in their genetic constitution, Galbicans or Gflavens, are produced by suaveolens Grado. Each of them can be combined with htuscaloosa, giving rise to a non-Mendelian nuclear genome splitting in F1. The two populations are designated twin hybrids, with the genotypes Galbicans·htuscaloosa (AB) and Gflavens·htusclaoosa (BB). Due to biparental plastid transmission in Oenothera, both plastome types are inherited (plastome IIsuavG from suaveolens Grado and plastome IIItusca from tuscaloosa). Therefore, all F1 offspring are chimeric for its plastome (IIsuavG/IIItusca). One of the twin hybrids, Gflavens·htuscaloosa, was discarded. Its diakinesis configuration displays a small ring of four and five bivalents (⊙4, 5 pairs) (supplemental Table 4, third row), which results in six linkage groups so that the hybrid is not constant in successive generations. The chromosome formula of the second twin hybrid, Galbicans·htuscaloosa, allows a ring of 14 chromosomes (⊙14) (supplemental Table 4, fourth row). Therefore, the complexes Galbicans and htuscaloosa should not be mixed because of repression of recombination and of free chromosome segregation in the ring. If Galbicans·htuscaloosa is now selfed, the progeny splits into two populations in F2 with the genetic constitution Galbicans·htuscaloosa and htuscaloosa·htuscaloosa, a consequence of the maternal inheritance of Galbicans and the biparental inheritance of the freely segregating lethal factor-free complex htuscaloosa. The interspecific exchange of plastids again takes place by somatic segregation and sorting out of the two plastomes IIsuavG and IIItusca in the F1 hybrid Galbicans·htuscaloosa IIsuavG/IIItusca. Selfing of flower buds carrying exclusively plastome IIsuavG led to the desired offspring htuscaloosa·htuscaloosa IIsuavG (BB-II) (Figure 7C).
Figure 6.—
Crossing scheme to exchange plastome III of Oe. grandiflora strain tuscaloosa with plastome II of Oe. biennis strain suaveolens Grado. For a detailed description, see text.
Figure 7.—
Phenotypic and molecular discrimination of crossing intermediates and end products in the BB-II crossing program: Galbicans·htuscaloosa with plastomes IIsuavG or IIItusca (AB-II/III) (A). Native, compatible combination htuscaloosa·htuscaloosa or hybrid Gflavens·htuscaloosa with plastome IIItusca (BB-III) (B). Incompatible combination htuscaloosa·htuscaloosa IIsuavG (BB-II) (C). Molecular discrimination of twin hybrids and plastomes (D): Galbicans·htuscaloosa (AB), heterozygous for M40 alleles A1 and B4, digested with SpeI (lane 1); htuscaloosa·htuscaloosa (BB) homozygous for M40 allele B4, digested with SpeI (lane 2); plastome IIsuavG rrn16-trnIGAU allele II/III1, digested with BamHI (lane 3); and plastome IIItusca rrn16-trnIGAU allele II/III8 digested with BamHI (lane 4); the first lane (M) shows a 100-bp ladder (New England BioLabs, Ipswich, MA). Detailed explanations are given in the text. The weaker signals characteristic of the Galbicans complex in lane 1 are primer specific and independent of the complex with which Galbicans is associated.
In the outlined crossing program, two basic genome combinations (AB and BB) and two basic plastome types (II and III) have to be distinguished. The nuclear genotypes can be discerned and separated phenotypically. Figure 7A shows the leaf shape of the hybrid Galbicans·htuscaloosa (AB). It is easily distinguishable from a BB leaf of strain tuscaloosa (htuscaloosa·htuscaloosa) or the hybrid htuscaloosa·Gflavens (Figure 7, B and C). The molecular marker is consistent with this assignment. Lane 1 in Figure 7D shows that Galbicans·htuscaloosa is heterozygous for the M40 alleles A1 (Galbicans) and B4 (htuscaloosa) as checked by SpeI digestion. Lane 2 shows the SpeI restriction pattern derived from homozygous M40-B4 alleles (htuscaloosa·htuscaloosa; BB). The BB combination Gflavens·htuscaloosa gives the same pattern. M40 alleles are listed in Table 2. Thus, the progeny in the splitting generations Galbicans·htuscaloosa and Gflavens·htuscaloosa in F1, as well as Galbicans·htuscaloosa and htuscaloosa·htuscaloosa in F2, can be reliably and readily monitored with phenotypic and molecular markers.
The crucial step during the crossing program is the selfing of the F1 hybrid Galbicans·htuscaloosa (Figure 6). Selection has to happen on flower buds carrying exclusively plastome IIsuavG. In this case phenotypic discrimination is not possible, since plastids with plastome types II or III are both green in an AB nuclear background (Stubbe 1989). With the plastidic CAPS marker, plastome identity of a flower bud can be checked as illustrated in Figure 7D, lines 3 (BamHI digest of the rrn16-trnIGAU allele II/III1 for plastome IIsuavG) and 4 (BamHI digest of the rrn16-trnIGAU allele II/III8 for plastome IIItusca).
The marker system is also suitable for verifying the genetic identity of the incompatible combination BB-II, which differs phenotypically from its compatible counterpart BB-III by a yellow-green leaf phenotype (lutescent) (Figure 7, B and C).
Generation and assignment of nuclear co-dominant markers to the molecular Oenothera map:
PCR-based, co-dominant markers for the Renner complexes hjohansen hhookeri de Vries, htuscaloosa, and hbellamy A were generated using clones of the Oenothera EST library as templates (Mráček 2005; Mráček et al. 2006; supplemental Table 4). In this study their number was increased and many of them were assigned to the molecular Oenothera map generated for the haplo-complexes hjohansen·htuscaloosa (Table 4). Both Renner complexes share the same chromosomal formula. Their hybrid displays seven bivalents in diakinesis and all chromosomes are lethal free. Therefore, the chromosomes are not in linkage disequilibrium to each other and the seven linkage groups found in hjohansen·htuscaloosa represent the seven chromosomes of the classical Oenothera map, namely 1·2 3·4 5·6 7·10 9·8 11·12 13·14. The molecular Oenothera map, predominantly based on AFLP markers, will be published in a subsequent communication. However, classical and molecular maps remain to be integrated.
TABLE 4.
Assignment of PCR-based codominant markers to the seven linkage groups of the molecular Oenothera map of the hybrid hjohansen·htuscaloosa representing chromosomes 1·2 3·4 5·6 7·10 9·8 11·12 13·14
| Marker allele | GenBank accession no. | Renner complex | Linkage group | Marker type (enzyme) | Predicted PCR products in (bp)a | Predicted restriction fragments (bp)a | Primer | Primer sequence (5′–3′) | EST/cluster accessionb |
|---|---|---|---|---|---|---|---|---|---|
| M02-A1 | EU483117 | hjohansen | LG 4 | CAPS (ApeKI) | 288 | 250, 38 | M02for | tggccatggcgacacaagcctc | C_4044-89-F11 |
| EU483118c | hhookeri de Vries | ||||||||
| M02-B1 | EU483119 | htuscaloosa | 288 | 186, 64, 38 | M02rev | cctcaacctgagccttacggag | |||
| EU483120c | hbellamy A | ||||||||
| M03-A1 | EU483121 | hjohansen | NA | SNP | 466 | NA | M03for | atatcacctggtactgctagct | C_4496-95-C09 |
| EU483122c | hhookeri de Vries | ||||||||
| M03-B1 | EU483123 | htuscaloosa | 466 | M03rev | aactccctccaatctgaagggt | ||||
| EU483124c | hbellamy A | ||||||||
| M07-A1 | EU483125 | hjohansen | LG 3 | CAPS (PstI) | 356 | 356 | M07for | accatacccatatacccagtgc | S_4170-90-H07 |
| EU483126c | hhookeri de Vries | ||||||||
| M07-B1 | EU483127 | htuscaloosa | 356 | 299, 57 | M07rev | tcaagcggcttcggtgcatctc | |||
| EU483128c | hbellamy A | ||||||||
| M08-A1 | EU483129 | hjohansen | LG 6 | CAPS (BsuRI) | 282 | 247, 35 | M08for | ctcagccaggaggacctcaagc | S_3501-84-D07 |
| EU483130c | hhookeri de Vries | ||||||||
| M08-B1 | EU483131 | htuscaloosa | 282 | 173, 74, 35 | M08rev | gaggtgggtatcgacctcgtcg | |||
| M13-A1 | EU447201 | hjohansen | NA | CAPS (NciI) | 328 | 241, 86, 1 | M13for | atcgatcatccatggcca | C_1649-15-A03 |
| M13-B1 | EU447202 | htuscaloosa | 328 | 123, 118, 86, 1 | M13rev | cgagaatggatcacctcca | |||
| M15-A1 | EU447203 | hjohansen | NA | CAPS (StyI) | 399 | 162, 149, 88 | M15for | ttggaggagttgcagttacaga | C_179-81-E03 |
| M15-B1 | EU447204 | htuscaloosa | 391 | 242, 149 | M15rev | cgtccaagtaggtcaggctct | |||
| M17-A1 | EU447205 | hjohansen | NA | CAPS (TaqI) | 512 | 427, 85 | M17for | gagatcacagtagtaatggcttcca | C_1955-18-D05 |
| M17-B1 | EU447206 | htuscaloosa | 503 | 403, 85, 15 | M17rev | catctgcagtggtagatctctga | |||
| M19-A1 | EU447207 | hjohansen | LG 1 | CAPS (PflMI) | 396 | 208, 188 | M19for | aatcctaatggctgcctctaca | C_2501-25-D11 |
| M19-B1 | EU447208 | htuscaloosa | 396 | 396 | M19rev | cacactgcctcaccgaact | |||
| M23-A1 | EU447209 | hjohansen | NA | CAPS (StyI) | 364 | 364 | M23for | ccacgcgaactctttaacact | C_46-4-A11 |
| M23-B1 | EU447210 | htuscaloosa | 362 | 231, 131 | M23rev | ggagtggatgacctcgagct | |||
| M28-A1 | EU447211 | hjohansen | LG 2 | CAPS (BsuRI) | 278 | 182, 39, 30, 27 | M28for | ggctccgacatccttgtggag | S_56-4-B10 |
| M28-B1 | EU447212 | htuscaloosa | 278 | 212, 39, 27 | M28rev | gcgactaaggggacgctatcg | |||
| M33-A1 | EU447213 | hjohansen | NA | SSLP | 309 | Not specifiable on agarose gels | M33for | tcaggcctcaagagctcagcc | C_943-8-C09 |
| M33-B1 | EU447214 | htuscaloosa | 310 | M33rev | acctcaagtggggagtccttg | ||||
| M38-A1 | EU447215 | hjohansen | LG 2 | CAPS (BsuRI) | 213 | 156, 44, 13 | M38for M38rev | ggcaaagctatggccactctc | S_1191-10-F12 |
| M38-B1 | EU447216 | htuscaloosa | 213 | 200, 13 | gtccgaccaagcagcgacgtt | ||||
| M39-A1 | EU447217 | hjohansen | LG 5 | CAPS (BclI) | 680 | 374, 262, 44 | M39for | ccaaagtggtatcgcggtgtc | S_1214-10-H11 |
| M39-B1 | EU447218 | htuscaloosa | 680 | 374, 306 | M39rev | ggaaccagtacgtagtacgttgc | |||
| M40 | See Table 2 | LG 2 | See Table 2 | M40for | accgtctcctccaagcactgc | C_1231-11-B04 | |||
| M40rev | tcagccctttgtccgaagtcg | ||||||||
| M41-A1 | EU447219 | hjohansen | LG 6 | CAPS (EarI) | 267 | 161, 106 | M41for | acaccctcttatcaccaatggc | C_1234-11-B07 |
| M41-B1 | EU447220 | htuscaloosa | 267 | 267 | M41rev | tctccacgagagtgtccgtgg | |||
| M43-A1 | EU447221 | hjohansen | Chromosome 9·8 | CAPS (BsuRI) | 275 | 206, 47, 22 | M43for | accacattcctcaaagctccg | S_1221-11-A06 |
| M43-B1 | EU447222 | htuscaloosa | 275 | 118, 88, 47, 22 | M43rev | cggaagcaagaagctctttgg | |||
| M44-A1 | EU447223 | hjohansen | NA | SSLP | 305 | Not specifiable on agarose gels | M44for | tcaacaatggctgccgcagtg | C_1431-12-H01 |
| M44-B1 | EU447224 | htuscaloosa | 311 | M44rev | agtgcttcaccttcgccggag | ||||
| M46-A1 | EU447225 | hjohansen | LG 5 | CAPS (XhoI) | 194 | 108, 86 | M46for | aaatggcgtccatggcgctta | S_1491-13-D02 |
| M46-B1 | EU447226 | htuscaloosa | 194 | 194 | M46rev | cttgggactcaagctcggcag | |||
| M47-A1 | EU447227 | hjohansen | LG 4 | CAPS (TaqI) | 257 | 257 | M47for | tgggtgggattgccctacgtg | S_1494-13-D05 |
| M47-B1 | EU447228 | htuscaloosa | 257 | 175, 82 | M47rev | gcgacaaccttaaccatgtcg | |||
| M48-A1 | EU447229 | hjohansen | NA | CAPS (BseRI) | 247 | 97, 95, 52, 3 | M48for | tcctcctagccactccactgc | C_1483-13-C06 |
| M48-B1 | EU447230 | htuscaloosa | 244 | 189, 52, 3 | M48rev | agcttctggtggagcttggct | |||
| M50-A1 | EU447231 | hjohansen | LG 6 | CAPS (HhaI) | 225 | 225 | M50for | ctgctccaccacaatggctgc | C_1598-14-D12 |
| M50-B1 | EU447232 | htuscaloosa | 225 | 148, 77 | M50rev | accaacgaaccgtctagccag | |||
| M52-A1 | EU447233 | hjohansen | NA | CAPS (HpyCH4III) | 235 | 120, 115 | M52for | aagcagccatggcgacatctc | S_1607-14-E09 |
| M52-B1 | EU447234 | htuscaloosa | 235 | 235 | M52rev | tccatgtagggcatcgagtcc | |||
| M57-A1 | EU447235 | hjohansen | NA | CAPS (BsrI) | 193 | 126, 67 | M57for | ctgatgttccttcccaagatg | C_2295-22-H03 |
| M57-B1 | EU447236 | htuscaloosa | 193 | 193 | M57rev | agaatgacccacggaatgtcc | |||
| M58-A1 | EU447237 | hjohansen | Chromosome 9·8 | CAPS (BsrI) | 533 | 424, 109 | M58for | gatccggaggatggaagtcct | S_2302-22-H10 |
| M58-B1 | EU447238 | htuscaloosa | 544 | 280, 155, 109 | M58rev | ctgaactgccacggctgttgg | |||
| M59-A1 | EU447239 | hjohansen | Chromosome 9·8 | CAPS (RsaI) | 703 | 322, 199, 178, 4 | M59for | tgctctccgccacaatgtccg | C_2346-23-D11 |
| M59-B1 | EU447240 | htuscaloosa | 699 | 517, 178, 4 | M59rev | caaaccctctggtggccacac | |||
| M60-A1 | EU447241 | hjohansen | NA | CAPS (AluI) | 231 | Not specifiable on agarose gels | M60for | gcaaccaacaatggcggtctg | C_2590-26-F11 |
| M60-B1 | EU447242 | htuscaloosa | 231 | M60rev | ctcttaccgcagccggaatcc | ||||
| M74-A1 | EU447243 | hjohansen | LG 3 | CAPS (HhaI) | 250 | 208, 40, 2 | M74for | aatggcggctctccagcagac | C_3913-88-C06 |
| M74-B1 | EU447244 | htuscaloosa | 250 | 104, 54, 50, 40, 2 | M74rev | tggtttcgagagtaccgttgg | |||
| M75-A1 | EU447245 | hjohansen | Chromosome 9·8 | CAPS (AluI) | 149 | 120, 29 | M75for | gtctgttatatcgagtgctgggac | C_4066-89-H09 |
| M75-B1 | EU447246 | htuscaloosa | 149 | 64, 56, 29 | M75rev | cctgatcagccatgcatctgag | |||
| M82-A1 | EU447247 | hjohansen | NA | CAPS (NaeI) | 223 | 223 | M82for | agcaccatggtgagcacctcc | C_5307-94-E09 |
| M82-B1 | EU447248 | htuscaloosa | 223 | 137, 86 | M82rev | agtagggcaaatcgattccctc | |||
| M86-A1 | EU447249 | hjohansen | LG 3 | CAPS (DdeI) | 208 | 84, 66, 45, 13 | M86for | tccctcatttctctacctccagag | C_4643-96-G12 |
| M86-B1 | EU447250 | htuscaloosa | 208 | 150, 58 | M86rev | accagccatagcaacgacgcc | |||
| M88-A1 | EU447251 | hjohansen | LG 3 | CAPS (DdeI) | 176 | 176 | M88for | accacagtctccgcagtaact | C_4753-98-B06 |
| M88-B1 | EU447252 | htuscaloosa | 176 | 152, 24 | N88rev | tgttgagcccaatccgaggtc | |||
| M95-A1 | EU447253 | hjohansen | Chromosome 9·8 | CAPS (HhaI) | 315 | 299, 16 | M95for | tcggactcagcaatggcgctc | C_5102-112-A10 |
| M95-B1 | EU447254 | htuscaloosa | 315 | 252, 47, 16 | M95rev | tggtggctgtctgtgctcgaa | |||
| M97-A1 | EU483132 | hjohansen | LG 6 | CAPS (ApeKI) | 357 | 246, 71, 29, 11 | M97for | atgaaagcacaaggagtcctc | S_1348-12-C09 |
| EU483133c | hhookeri de Vries | ||||||||
| M97-B1 | EU483134 | htuscaloosa | 375 | 346, 29 | M97rev | cgagaatgaagctgcctaaga | |||
| EU483135c | hbellamy A | ||||||||
| M98-A1 | EU483136 | hjohansen | LG 2 | CAPS (BbvI) | 459 | 223, 221, 15 | M98for | aagccgagatcatcctgcaatgg | C_1202-10-G11 |
| M98-A2 | EU483137c | hhookeri de Vries | 459 | 236, 223 | M98rev | aggcaaaataaaacggggatacagc | |||
| M98-B1 | EU483138 | htuscaloosa | 470 | 470 | |||||
| EU483139c | hbellamy A | ||||||||
Not each PCR product was fully sequenced; lengths in base pairs were derived from the position of the primer in the EST sequence used as template.
EST sequence, gene function, and corresponding Arabidopsis ortholog can be obtained from GenBank using the Oenothera EST cluster accession.
Allele first published without sequence in Mráček (2005).
Correlation of the classical and molecular Oenothera maps:
Classical Oenothera maps are based on the segmental arrangements of chromosome arms and their relative locations to each other (Figure 1). Chromosomal arrangements or formulas are available for >300 Oenothera strains (Stubbe and Diers 1958; Linder and Jean 1969; Cleland 1972; Jean and Linder 1979; Steiner and Stubbe 1984, 1986; Wasmund and Stubbe 1986; Wasmund 1980, 1990; Schumacher et al. 1992; Schumacher and Steiner 1993) and phenotypic characters were assigned to individual chromosome arms. The extension of the classical map with molecular markers is clearly one of the most important future steps in Oenothera research and breeding programs. It allows immediate and comprehensive genetic identity of a variety of strains.
To address this question, in a case study an attempt was made to combine methods of molecular and classical Oenothera genetics. Since chromosomes of the classical map can be identified only by their genetic behavior, an appropriate cross was chosen to distinguish chromosome 9·8 from the rest of the genome by segregation analysis.
A cross of strains suaveolens Standard (Stalbicans·Stflavens) and tuscaloosa (htuscaloosa·htuscaloosa) resulted in the F1 hybrid Stalbicans·htuscaloosa. The hybrid closely resembles hybrid Galbicans·htuscaloosa (⊙14) used above for interspecific plastid exchange (Figure 6). However, the crucial difference is that Stalbicans·htuscaloosa displays the chromosome configuration of a ring of 12 chromosomes and one free bivalent (⊙12, 1 pair). The latter is formed by chromosome 9·8, as illustrated in Figure 1, J–L.
A diakinesis configuration of ⊙12, 1 pair, represents two independent linkage groups. The 12 chromosomes that constitute a single linkage group do not segregate in a Mendelian manner. Like Galbicans in Figure 6, Stalbicans is exclusively inherited by the egg cell. The selfed F1 hybrid Stalbicans·htuscaloosa, therefore, splits into Stalbicans·htuscaloosa and htuscaloosa·htuscalosoa in a theoretical ratio of 1:1. The lethal combination of a homozygous Stalbicans complex (Stalbicans·Stalbicans) is not found. However, segregation of the Renner complexes Stalbicans and htuscaloosa involves six chromosomes each, which are part of the ring. The seventh, chromosome 9·8, is free and does not cosegregate with the torso complexes Stalbicans and htuscaloosa. Since chromosome 9·8 lacks lethal factors (Renner 1942; Stubbe 1953), it can segregate independently in a Mendelian manner (9·8albSt 9·8albSt/9·8albSt 9·8tusca/9·8tusca 9·8tusca; ratio 1:2:1) (Figure 8). [The statement of Cleland (1972, p. 110) that the phenotypic marker pil, located on chromosome arm 8 of Stalbicans, is homozygous lethal obviously arose from a misinterpretation of Renner's sometimes complex German (Renner 1942).]
Figure 8.—
Assembly and segregation behavior of the hybrid Stalbicans·htuscaloosa. The F2 population was used to identify chromosome 9·8 (linkage group 2), which segregates independently from the large, strongly linked coupling groups of the torso complexes Stalbicans or htuscaloosa (linkage group 1). Details are given in the text.
The presence of two linkage groups can be monitored with molecular markers. The large linkage group, containing lethal factors and involving 12 chromosomes, can readily be distinguished from the small one including a single bivalent and lacking lethal factors. Since the seven Oenothera linkage groups in the AFLP map were identified with the htuscaloosa complex (see above), the freely segregating bivalent of Stalbicans·htuscaloosa must be identical with one of the seven coupling groups in the map of hjohansen·htuscaloosa.
Eight co-dominant markers, representing all seven Oenothera chromosomes as individual linkage groups, were checked for heterozygosity in the hybrid Stalbicans·htuscaloosa (AB). The strategy was to digest derived PCR products with restriction endonucleases that are appropriate for distinguishing the Renner complexes hjohansen (A) and htuscaloosa (B) (Table 4). All markers described resembled the restriction pattern of the A genome hjohansen in the A genome Stalbicans (supplemental Table 5).
Analysis of 38 Stalbicans·htuscaloosa F2 plants uncovered a clear linkage of markers M19, M40, M41, M46, M47, M50, and M74 with phenotypic markers for Stalbicans·htuscaloosa and htuscaloosa·htuscaloosa, respectively (LOD = 11.13). In the large coupling group, only Stalbicans·htuscaloosa or htuscaloosa·htuscaloosa genotypes could be detected in the expected ratio of ∼50% (Figure 8). A second, freely segregating coupling group detected with marker M58 was confirmed with a LOD >10. All allelic combinations (M58-A1/M58-A1, M58-A1/M58-B1, and M58-B1/M58-B1) were found in this linkage group. Since M58 is part of linkage group 7, these data therefore assign linkage group 7 to chromosome 9·8 (Table 4 and supplemental Table 5). This finding opens the intriguing possibility in Oenothera genetics of identifying all individual chromosomes (see discussion). Plants genotyped in F2 as htuscaloosa·htuscaloosa M58-A1/M58-A1 represent an interspecific chromosome exchange. Furthermore, the strong linkage of M19, M40, M41, M46, M47, M50, and M74, representing six chromosomes in a ring of 12, provides the first direct molecular evidence for the suppression of homologous recombination and free segregation through meiotic rings in Oenothera.
DISCUSSION
Benefit of co-dominant markers to Oenothera breeding:
Molecular co-dominant markers are of intrinsic interest for breeding programs; however, in Oenothera research they were largely missing (Mráček et al. 2006; Larson et al. 2008). They are important for various lines of research, such as in investigations of plastome–genome incompatible hybrids, genome divergence and patterns, and mapping of speciation relevant loci for commercial interests or breeding programs. Until now, neither basic nor subplastome types could be distinguished without performing extensive restriction fragment length polymorphism (RFLP) analyses (Herrmann et al. 1980; Gordon et al. 1981, 1982; Chapman et al. 1999), and in interspecific plastome exchanges different plastome types were marked with bleached plastome mutants (Stubbe 1960, 1989; supplemental Figure 1). Time-consuming crossing steps were required to establish or remove these mutant plastomes (Stubbe 1960, 1989). The rrn16-trnIGAU marker allele (Table 3) allows an easy monitoring of all basic plastome types and a large number of subplastomes. It renders the use of plastome mutants dispensable and reduces the number of crosses significantly.
Substantial progress was also made in identifying nuclear genomes, e.g., with the marker allele M40 (Table 2). Crosses involving ring-forming hybrids can now be monitored for all basic Oenothera genotypes and a large number of Renner complexes. A major advantage here is the possibility to screen splitting generations for different Renner complex combinations already at the seedling stage and by a single PCR. To date, only mature plants could be checked using phenotypic markers. The advantages of molecular approaches are obvious, especially for annual herbs, such as Oenothera.
Co-dominant markers listed in Table 4, when applied to Oenothera genomes, indicate that already minor nucleotide changes are appropriate for generating CAPS markers from almost all genes studied (∼60% at an average of only 300 bp). Of 34 markers studied, 22 could be mapped to the seven coupling groups of the hybrid hjohansen·htuscaloosa, representing chromosomes 1·2 3·4 5·6 7·10 9·8 11·12 13·14 of the classical Oenothera map illustrating that the Oenothera EST library (Mráček et al. 2006) is a rich source for PCR-based markers. Furthermore, with appropriate genetic lines and the nuclear marker M58 in a pilot study it was possible to assign chromosome 9·8 to linkage group 7 of the molecular linkage map. For the first time this offers the possibility of combining data from a century of classical genetic research on Oenothera genomes with modern molecular approaches and of integrating classical and molecular Oenothera maps completely.
The classical maps allow predicting the segregation pattern in superlinkage groups involving single or multiple chromosomes (Cleland 1972, pp. 43–64). They allow a directed exchange of single or multiple chromosome pairs between genomes at the diploid level. Single chromosome pairs can be specifically placed in a breed without disturbing the combination of characters located in the remaining genome (Figure 8). This strategy can be applied, for example, to study the impact of individual linkage groups on the genome and/or plastome evolution in Oenothera but also to introduce single chromosomes with desired traits into commercial cultivars. This approach has been neglected in professional Oenothera breeding programs (Fieldsend 2007) since monitoring of such crosses is realizable only with phenotypic markers, which is inherently difficult in general. Furthermore, some chromosomes in various strains lack phenotypic markers, rendering an analysis of their segregation without molecular approaches impossible (Cleland 1972, pp. 109–111). Therefore, molecular markers assigned to the classical Oenothera map provide immediate access to (i) various phenotypic markers or other loci located on that map and (ii) to 300 strains for which the chromosomal formulas are known (Stubbe and Diers 1958; Linder and Jean 1969; Cleland 1972; Jean and Linder 1979; Steiner and Stubbe 1984, 1986; Wasmund and Stubbe 1986; Wasmund 1980, 1990; Schumacher et al. 1992; Schumacher and Steiner 1993).
In summary, the markers described in this work represent significant progress in Oenothera genetics. They allow a precise and easy molecular identification of plastomes, Renner complexes, and individual nuclear chromosomes or chromosome arms (see below) in crossing programs. Presumably, they will also have an impact on commercial Oenothera breeding.
Alignment of classical and molecular Oenothera maps:
None of the nuclear molecular markers described so far had been assigned to the classical Oenothera map. Assigning linkage group 7 to chromosome 9·8 is therefore a milestone for future breeding. The same strategy can be applied to all other chromosome pairs. The prerequisite for such an approach—hybrids of hjohansen or htuscaloosa with various Renner complexes having one of the seven chromosomes as a free, individual pair—is available for all seven chromosomes.
For example, the hybrid Stlaxans·htuscaloosa possesses the chromosome configuration ⊙4, ⊙4, ⊙4, 1 pair. In this case, chromosome 1·2 is the only bivalent (1 pair). Segregation analysis with molecular markers in the F2 generation should display four coupling groups, three consisting of four chromosomes each (⊙4), and one with a single chromosome, identical to chromosome 1·2. The same principle allows assignment of all further chromosomes using, e.g., the hybrids Stpingens·hjohansen (chromosome 3·4), hjohansen·r-Sgaudens (chromosome 5·6), hjohansen·Stpercurvans (chromosome 7·10), htuscaloosa·Stundans (chromosome 11·12), and Thtingens·htuscaloosa (chromosome 13·14).
After all seven coupling groups of hjohansen·htuscaloosa have been associated with chromosomes, chromosome arms can be identified in a next step. A possible hybrid with which to work is the hybrid Stalbicans·hcholula with the freely segregating pair 1·4. Since chromosome 1·2 was already identified from Stlaxans·htuscaloosa, as outlined above, all markers assigned to chromosome 1·2 in hjohansen·Stlaxans, which also map to chromosome 1·4 of Stalbicans·hcholula, must be part of arm 1. Consequently, the remaining markers on 1·2 of Stlaxans·htuscaloosa, which are not part of coupling group 1·4 in Stalbicans·hcholula, must reside on arm 2. Conversely, identification of arm 1 in chromosome 1·4 will also characterize arm 4. Characterization of arm 4, in turn, allows assignment of arm 3 on chromosome 3·4.
The strategy can readily be applied to all other chromosome arms. For example, with the hybrid hcholula·Stpercurvans, having 6·8 as a free pair, arms 5, 6, 8, and 9 can be identified. The rich source of Oenothera strains analyzed allows determining all remaining chromosome arms.
Phylogenetic pattern of the plastid rrn16-trnIGAU marker allele:
The rrn16-trnIGAU spacer region used as a marker allele for the identification of basic and subplastome types in crossing experiments can also serve to monitor gene flow and recent hybridizing events within subsection Oenothera. The marker may be an indicator of how much subplastome variation exists within the subsection, without performing laborious RFLP analysis of the entire plastid chromosome (Herrmann et al. 1980). Knowledge of the degree of variation is indispensable to correlate differences between five sequenced reference plastomes (Greiner et al. 2008a) with speciation or prespeciation events. Although the phylogenetic significance of the rrn16-trnIGAU spacer region may be limited, since short direct repeats, synapomorphies, and parallel changes are common in this region (Hornung et al. 1996; Sears et al. 1996), some inferences may be drawn: In both species with plastome IV, Oe. oakesiana and Oe. parviflora, only a single marker allele (rrn16-trnIGAU IV1) was detected in four strains. Also, alleles rrn16-trnIGAU V1 and V2 present in three strains of Oe. argillicola were identical, disregarding a single-base-pair polymorphism. For plastome I, the closely related strains of Oe. elata subsp. elata (chapultepec, cholula, puebla, and toluca) possess two highly similar alleles, rrn16-trnIGAU I1 and -I2, which differ in only a small deletion. In Oe. elata subsp. hookeri, in turn, two alleles were found in four different strains. Strain bauri Standard of Oe. villosa subsp. villosa displayed a distinct allele as well. Taken together, the alleles described are specific for the basic plastomes I, IV, or V and appear to represent unequivocal genetic markers for these plastome types (Table 3).
The data also confirm tendencies of subplastome evolution. The single allele of plastome IV in Oe. oakesiana and Oe. parviflora indicates that plastome IV of these two species is monophyletic and probably arose from a hypothetical ancestor with the genomic constitution CC-IV (Dietrich et al. 1997). In the AA-I clade, Oe. elata subsp. elata, Oe. elata subsp. hookeri, and Oe. villosa subsp. villosa were proposed to represent different evolutionary lineages (Dietrich et al. 1997), a fact supported by distinct rrn16-trnIGAU marker alleles. It would be particularly interesting to see whether subplastome patterns reflect solely mechanistic imprecision of DNA replication or repair with no phylogenetic relevance or correlate in part with prespeciation processes, e.g., geographic trends and genetic drifts of subpopulations in species with floating chromosome formulas, which in several instances are known in continental dimension (Cleland 1972, pp. 227–298).
The situation is quite different for plastomes II and III, found in Oe. biennis, Oe. glazioviana, Oe. grandiflora, and Oe. nutans. No specific alleles that are related to the genetic behavior of their plastomes were detected (Table 3). Allele rrn16-trnIGAU II/III3 was found in plastomes II or III of Oe. biennis (AB-II or BA-III), Oe. grandiflora (BB-III), and Oe. nutans (BB-III). Allele rrn16-trnIGAU II/III1 is not characteristic for plastome type II since it was also found in the strain castleberry B-8 of Oe. grandiflora (BB-III). These data indicate gene flow between the four species. Variation among plastomes II and III appears to be more widely spread than between other plastome types, since nine different plastome II/III alleles were found. Alleles rrn16-trnIGAU II/III1 and -II/III3 probably reflect two major patterns for both plastome II and plastome III. So far, alleles rrn16-trnIGAU II/III4–II/III8 were found to be specific for single strains (Table 3).
Acknowledgments
We thank Elisabeth Gerick for excellent technical assistance and work in our crossing programs. We thank also Ulrich Wissnet for nursing the plant material for years in Munich. We are especially grateful to Wilfried Stubbe and Werner Dietrich, University of Düsseldorf, who established the comprehensive Oenothera collection, of which a small fraction was used in this work. We thank Dario Leister for critical reading and Volker Mohler for calculations using the JoinMap program. The work was supported by the Deutsche Forschungsgemeinschaft (SFB-TR1 to R.G.H. and J.M.) and the Hanns-Seidel-Stiftung supported by the Bundesministerium für Bildung und Forschung to S.G.
References
- Barre, D. E., 2001. Potential of evening primrose, borage, black currant, and fungal oils in human health. Ann. Nutr. Metab. 45 47–57. [DOI] [PubMed] [Google Scholar]
- Birky, C. W., Jr., 2001. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 35 125–148. [DOI] [PubMed] [Google Scholar]
- Burnham, C. R., 1962. Discussions in Cytogenetics. Burgess, Minneapolis.
- Chapman, M. J., D. L. Mulcahy and D. B. Stein, 1999. Identification of plastid genotypes in Oenothera subsect. Munzia by restriction fragment length polymorphisms (RFLPs). Theor. Appl. Genet. 98 47–53. [Google Scholar]
- Cleland, R. E., 1935. Cyto-taxonomic studies on certain Oenotheras from California. Proc. Am. Philos. Soc. 75 339–429. [Google Scholar]
- Cleland, R. E., 1972. Oenothera cytogenetics and evolution, pp. 1–370 in Experimental Botany, Vol. 5, edited by J. F. Sutcliffe and P. Mahlberg. Academic Press, San Diego/New York/London.
- de Gyves, E. M., C. A. Sparks, O. Sayanova, P. Lazzeri, J. A. Napier et al., 2004. Genetic manipulation of gamma-linolenic acid (GLA) synthesis in a commercial variety of evening primrose (Oenothera sp.). Plant Biotechnol. J. 2 351–357. [DOI] [PubMed] [Google Scholar]
- de Vries, H., 1900. Das Spaltungsgesetz der Bastarde. Ber. Dtsch. Bot. Ges. 18 83–90. [Google Scholar]
- de Vries, H., 1901–1903. Die Mutationstheorie. von Veit, Leipzig, Germany.
- Dietrich, W., 1977. The South American species of Oenothera sect. Oenothera (Raimannia, Renneria; Onagraceae). Ann. Mo. Bot. Gard. 64 425–626. [Google Scholar]
- Dietrich, W., W. L. Wagner and P. H. Raven, 1997. Systematics of Oenothera section Oenothera subsection Oenothera (Onagraceae), pp. 1–234 in Systematic Botany Monographs, Vol. 50, edited by C. Anderson. The American Society of Plant Taxonomists. Laramie, WY.
- Dobzhansky, T., 1937. Genetics and the Origin of Species. Columbia University Press, New York.
- Fieldsend, A. F., 2007. The impact of plant breeding on seed oil content and quality in evening primrose crops, pp. 29–36 in Proceedings of the Joint International Conference on Long-Term Experiments. Agricultural Research and Natural Resources, May 31–June 1, Debrecen-Nyírlugos, Hungary.
- Golczyk, H., R. Hasterok and A. J. Joachimiak, 2005. FISH-aimed karyotyping and characterization of Renner complexes in permanent heterozygote Rhoeo spathacea. Genome 48 145–153. [DOI] [PubMed] [Google Scholar]
- Gordon, K. H. J., E. J. Crouse, H. J. Bohnert and R. G. Herrmann, 1981. Restriction endonuclease cleavage site map of chloroplast DNA from Oenothera parviflora (Euoenothera plastome IV). Theor. Appl. Genet. 59 281–296. [DOI] [PubMed] [Google Scholar]
- Gordon, K. H. J., E. J. Crouse, H. J. Bohnert and R. G. Herrmann, 1982. Physical mapping of differences in chloroplast DNA of the five wild-type plastomes in Oenothera subsection Euoenothera. Theor. Appl. Genet. 61 373–384. [DOI] [PubMed] [Google Scholar]
- Greiner, S., X. Wang, U. Rauwolf, M. V. Silber, K. Mayer et al., 2008. a The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Res. 36 2366–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner, S., X. Wang, R. G. Herrmann, U. Rauwolf, K. Mayer et al., 2008. b The complete nucleotide sequences of the 5 genetically distinct plastid genomes of Oenothera, subsection Oenothera: II. A microevolutionary view using bioinformatics and formal genetic data. Mol. Biol. Evol. 25 2019–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann, R., 2004. The sexual inheritance of plant organelles, pp. 93–114 in Molecular Biology and Biotechnology of Plant Organelles: Chloroplasts and Mitochondria, edited by H. Daniell and C. D. Chase. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Harte, C., 1994. Oenothera: contributions of a plant to biology, pp. 1–261 in Monographs on Theoretical Applied Genetics, Vol. 20, edited by R. Frankel, M. Grossman, H. F. Linskens, P. Maliga and R. Riley. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Herrmann, R. G., 1997. Eukaryotism: towards a new interpretation, pp. 73–118 in Eukaryotism and Symbiosis, edited by H. E. A. Schenk, R. G. Herrmann, K. W. Jeon, N. E. Müller and W. Schwemmler. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Herrmann, R. G., P. Seyer, R. Schedel, K. Gordon, C. Bisanz et al., 1980. The plastid chromosomes of several dicotyledons, pp. 97–112 in Biological Chemistry of Organelle Formation, edited by T. Bücher, W. Sebald and H. Weis. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Herrmann, R. G., R. M. Maier and C. Schmitz-Linneweber, 2003. Eukaryotic genome evolution: rearrangement and coevolution of compartmentalized genetic information. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger, K. E., and N. C. Ellstrand, 1984. The evolution and ecology of permanent transloction heterozygotes. Am. Nat. 124 48–71. [Google Scholar]
- Hornung, S., H. Fulgosi, P. Dörfel and R. G. Herrmann, 1996. Sequence variation in the putative replication origins of the five genetically distinct basic Euoenothera plastid chromosomes (plastomes). Mol. Gen. Genet. 251 609–612. [DOI] [PubMed] [Google Scholar]
- Huang, J., and M. Sun, 1999. A modified AFLP with fluorescence-labelled primers and automated DNA sequencer detection for efficient fingerprinting analysis in plants. Biotechnol. Tech. 13 277–278. [Google Scholar]
- Jean, R., and R. Linder, 1979. Oenothera suzukiana species nova. Analyse floristique et cytogénétique. Cytologia. 44 757–780. [Google Scholar]
- Kirk, J. T. O., and R. A. E. Tilney-Bassett, 1978. The Plastids: Their Chemistry, Structure, Growth and Inheritance. Elsevier, Amsterdam.
- Kuchuk, N., R. G. Herrmann and H.-U. Koop, 1998. Plant regeneration from leaf protoplasts of evening primrose (Oenothera hookeri). Plant Cell Rep. 17 601–604. [DOI] [PubMed] [Google Scholar]
- Larson, E. L., S. M. Bogdanowicz, A. A. Agrawal, M. T. J. Johnson and R. G. Harrison, 2008. Isolation and characterization of polymorphic microsatellite loci in common evening primrose (Oenothera biennis). Mol. Ecol. Res. 8 434–436. [DOI] [PubMed] [Google Scholar]
- Levin, D. A., 2002. The Role of Chromosomal Change in Plant Evolution. Oxford University Press, New York.
- Levin, D. A., 2003. The cytoplasmic factor in plant speciation. Syst. Bot. 28 5–11. [Google Scholar]
- Linder, R., and R. Jean, 1969. Oenothera ersteinensis, espèce nouvelle. Bull. Soc. Bot. France 116 523–529. [Google Scholar]
- Lutz, A. M., 1907. A preliminary note on the chromosomes of Oenothera lamarckiana and one of its mutants, Oenothera gigas. Science 27 151–152. [DOI] [PubMed] [Google Scholar]
- Mehra-Palta, A., H.-U. Koop, S. Goes, E.-M. Troidl, G. Nagy et al., 1998. Tissue culture of wild-type, interspecific genome/plastome hybrids and plastome mutants of evening primrose (Oenothera): controlled morphogenesis and transformation. Plant Cell Rep. 17 605–611. [DOI] [PubMed] [Google Scholar]
- Mráček, J., 2005. Investigation of genome-plastome incompatibility in Oenothera and Passiflora. Ph.D. Thesis, Ludwig-Maximilians-Universität, Munich, Germany.
- Mráček, J., S. Greiner, W. K. Cho, U. Rauwolf, M. Braun et al., 2006. Construction, database integration, and application of an Oenothera EST library. Genomics 88 372–380. [DOI] [PubMed] [Google Scholar]
- Peters, J. L., H. Constandt, P. Neyt, G. Cnops, J. Zethof et al., 2001. A physical amplified fragment-length polymorphism map of Arabidopsis. Plant Physiol. 127 1579–1589. [PMC free article] [PubMed] [Google Scholar]
- Rauwolf, U., 2008. Mapping of genomes and plastomes of subsection Oenothera with molecular marker technologies. Ph.D. Thesis, Ludwig-Maximilians-Universität, Munich, Germany.
- Renner, O., 1917. Versuche über die gametische Konstitution der Oenotheren. Z. Indukt. Abstamm. Vererbungsl. 18 121–294. [Google Scholar]
- Renner, O., 1924. Die Scheckung der Oenotherenbastarde. Biol. Zentralbl. 44 309–336. [Google Scholar]
- Renner, O., 1934. Die pflanzlichen Plastiden als selbständige Elemente der genetischen Konstitution. Ber. Math.-Physik. Kl. Sächs. Akad. Wiss. Leipzig 86 241–266. [Google Scholar]
- Renner, O., 1936. Zur Kenntnis der nichtmendelnden Buntheit der Laubblätter. Flora 30 218–290. [Google Scholar]
- Renner, O., 1942. Über das Crossing-over bei Oenothera. Flora 136 117–214. [Google Scholar]
- Renner, O., 1956. Europäische Wildarten von Oenothera III. Planta 47 219–254. [Google Scholar]
- Ridout, C. J., and P. Donini, 1999. Use of AFLP in cereals research. Trends Plant Sci. 4 76–79. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., S. Kushnir, E. Babiychuk, P. Poltnigg, R. G. Herrmann et al., 2005. Pigment deficiency in nightshade/tobacco cybrids is caused by the failure to edit the plastid ATPase alpha-subunit mRNA. Plant Cell 17 1815–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, E., and E. E. Steiner, 1993. Cytological analysis of complex-heterozygotes in populations of Oenothera grandiflora (Onagraceae) in Alabama. Plant Syst. Evol. 184 77–87. [Google Scholar]
- Schumacher, E., E. E. Steiner and W. Stubbe, 1992. The complex-heterozygotes of Oenothera grandiflora L'Her. Bot. Acta 105 375–381. [Google Scholar]
- Sears, B. B., L. L. Stoike and W. L. Chiu, 1996. Proliferation of direct repeats near the Oenothera chloroplast DNA origin of replication. Mol. Biol. Evol. 13 850–863. [DOI] [PubMed] [Google Scholar]
- Steiner, E. E., and W. Stubbe, 1984. A contribution to the population biology of Oenothera grandiflora L'Her. Am. J. Bot. 71 1293–1301. [Google Scholar]
- Steiner, E. E., and W. Stubbe, 1986. Oenothera grandiflora revisited: a new view of its population structure. Bull. Torrey Bot. Club 113 406–412. [Google Scholar]
- Stonemetz, D., 2008. A review of the clinical efficacy of evening primrose. Holist. Nurs. Pract. 22 171–174. [DOI] [PubMed] [Google Scholar]
- Stubbe, W., 1953. Genetische und zytologische Untersuchungen an verschiedenen Sippen von Oenothera suaveolens. Z. Indukt. Abstamm. Vererbungsl. 85 180–209. [PubMed] [Google Scholar]
- Stubbe, W., 1959. Genetische Analyse des Zusammenwirkens von Genom und Plastom bei Oenothera. Z. Vererbungsl. 90 288–298. [Google Scholar]
- Stubbe, W., 1960. Untersuchungen zur genetischen Anaylse des Plastoms von Oenothera. Z. Bot. 48 191–218. [Google Scholar]
- Stubbe, W., 1964. The role of the plastome in evolution of the genus Oenothera. Genetica 35 28–33. [Google Scholar]
- Stubbe, W., 1989. Oenothera:– an ideal system for studying the interaction of genome and plastome. Plant Mol. Biol. Rep. 7 245–257. [Google Scholar]
- Stubbe, W., and L. Diers, 1958. Die Chromosomenformel des albicans-Komplexes der Sippe Grado von Oenothera suaveolens. Z. Vererbungsl. 89 320–322. [Google Scholar]
- Stubbe, W., and R. G. Herrmann, 1982. Selection and maintenance of plastome mutants and interspecific genome/plastome hybrids from Oenothera, pp. 149–165 in Methods in Chloroplast Molecular Biology, edited by M. Edelman, R. B. Hallick and N.-H. Chua. Elesevier, Amsterdam.
- Stubbe, W., and E. Steiner, 1999. Inactivation of pollen and other effects of genome-plastome incompatibility in Oenothera. Plant Syst. Evol. 217 259–277. [Google Scholar]
- Taniguchi, S., Y. Imayoshi, T. Hatano, K. Yazaki and T. Yoshida, 2002. Hydrolysable tannin production in Oenothera tetraptera shoot tissue culture. Plant Biotechnol. 19 357–363. [Google Scholar]
- Thiel, T., R. Kota, I. Grosse, N. Stein and A. Graner, 2004. SNP2CAPS: a SNP and INDEL analysis tool for CAPS marker development. Nucleic Acids Res. 32 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and L. C. Moyle, 2007. Asymmetric postmating isolation: Darwin's corollary to Haldane's rule. Genetics 176 1059–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen, J. W., and R. E. Voorrips, 2001. JoinMap-Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands.
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee et al., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmund, O., 1980. Cytologenetische Untersuchungen zur Systematik einiger Sippen der Subsektion Euoenothera der Gattung Oenothera (Onagraceae). State Examination thesis, Heinrich-Heine-Universität, Düsseldorf, Germany.
- Wasmund, O., 1990. Cytogenetic investigation on Oenothera nutans (Onagraceae). Plant Syst. Evol. 169 69–80. [Google Scholar]
- Wasmund, O., and W. Stubbe, 1986. Cytogenetic investigations on Oenothera wolfii (Onagraceae). Plant Syst. Evol. 154 79–88. [Google Scholar]