Abstract
Potato (Solanum tuberosum) has the densest genetic linkage map and one of the earliest established cytogenetic maps among all plant species. However, there has been limited effort to integrate these maps. Here, we report fluorescence in situ hybridization (FISH) mapping of 30 genetic marker-anchored bacterial artificial chromosome (BAC) clones on the pachytene chromosome 6 of potato. The FISH mapping results allowed us to define the genetic positions of the centromere and the pericentromeric heterochromatin and to relate chromatin structure to the distribution of recombination along the chromosome. A drastic reduction of recombination was associated with the pericentromeric heterochromatin that accounts for ∼28% of the physical length of the pachytene chromosome. The pachytene chromosomes 6 of potato and tomato (S. lycopersicum) share a similar morphology. However, distinct differences of heterochromatin distribution were observed between the two chromosomes. FISH mapping of several potato BACs on tomato pachytene chromosome 6 revealed an overall colinearity between the two chromosomes. A chromosome inversion was observed in the euchromatic region of the short arms. These results show that the potato and tomato genomes contain more chromosomal rearrangements than those reported previously on the basis of comparative genetic linkage mapping.
POTATO (Solanum tuberosum, 2n = 4x = 48) is the fourth most important food crop in the world, surpassed only by rice, wheat, and maize. Genetic research of potato has been long hampered by the autotetraploidy and highly heterozygous nature of the potato genome. This challenge, however, has been overcome by the advent of modern molecular marker technology. Several molecular marker-based genetic linkage maps of diploid potato were developed (Bonierbale et al. 1988; Gebhardt et al. 1989, 1991; Tanksley et al. 1992; Jacobs et al. 1995), including a 10,000-marker ultradense map that represents one of the densest genetic maps in any eukaryote (van Os et al. 2006). Other resources for genomics research have also been developed, including expressed sequence tags (ESTs) (Ronning et al. 2003; Flinn et al. 2005), bacterial artificial chromosome (BAC) libraries (Song et al. 2000; Zhang et al. 2003; Chen et al. 2004), and BAC end sequences (Zhu et al. 2008). An international potato genome sequencing consortium has been established recently (http://potatogenome.net) that will make potato one of few major crop species whose genome will be sequenced using a BAC-by-BAC-based approach.
Despite the development of several potato genetic linkage maps, there has been limited effort to integrate the linkage maps with cytogenetic maps (Dong et al. 2000; Song et al. 2000). The chromosomal positions of the molecular makers used in linkage mapping are largely unknown. Furthermore, it is not known if the DNA markers are uniformly distributed along the individual chromosomes. The international potato genome sequencing project will be based on a BAC physical map anchored by the amplified fragment length polymorphism (AFLP) markers used in the development of the ultradense potato linkage map (van Os et al. 2006). The genomewide saturation of these AFLP markers over the entire length of the chromosomes will be a key measure for the success of the sequencing project. The potato chromosomes have been well known to consist of cytologically distinct pericentromeric heterochromatin and distal euchromatin (Yeh and Peloquin 1965). Thus, integration of the genetic linkage map with the cytogenetic map will reveal the euchromatic or heterochromatic locations of the genetic markers.
Fluorescence in situ hybridization (FISH) has become the most common approach to map DNA markers to specific chromosomal domains (Jiang and Gill 1994, 2006). Meiotic pachytene chromosomes are superior to somatic metaphase chromosomes for FISH mapping resolution (Cheng et al. 2002). Integration of genetic linkage maps with pachytene chromosome-based cytogenetic maps has been reported in several plant species (Cheng et al. 2001; Islam-Faridi et al. 2002; Howell et al. 2005; Kim et al. 2005; Kulikova et al. 2001; Walling et al. 2006; Wang et al. 2006; Amarillo and Bass 2007; Chang et al. 2007; Koo et al. 2008; Szinay et al. 2008; Tang et al. 2008, accompanying article, this issue). We report FISH mapping of 30 genetic marker-anchored BACs on the pachytene chromosome 6 of potato. The FISH mapping results allowed us to fully integrate the ultradense potato linkage map with the chromosomal map for chromosome 6. The integration of the two maps revealed a drastic reduction of genetic recombination in the pericentromeric region that consists of almost exclusively cytologically distinct heterochromatin. Comparative cytogenetic mapping of potato and tomato chromosome 6 revealed general similarity/colinearity but with some important differences between these two chromosomes.
MATERIALS AND METHODS
Plant materials and chromosome preparation:
Three potato clones were used in cytological preparations, including a haploid potato clone USW1 (2n = 2x = 24), which was derived from potato cultivar Katahdin, and two diploid potato clones, RH89-039-16 (RH) and SH83-92-488 (SH), which were the parental clones used in developing an ultradense genetic recombination map of potato (van Os et al. 2006). For a comparative analysis between chromosomes 6 of S. tuberosum and S. lycopersicum, tomato varieties Koralik (cherry tomato) and Quinte were included in this study. Immature flower buds were harvested and fixed in 3:1 Carnoy's solution. The procedure for meiotic chromosome preparation was essentially the same as that used for mitotic chromosomes from root tips (Dong et al. 2000) with the following modification: anthers were digested in the enzyme mixture (4% cellulase, 2% pectinase, 0.2% cytohelicase) for 2 hr at 37° (1 hr for tomato). The digested anthers were macerated on glass slides in 3:1 methanol:acetic acid solution with fine-pointed forceps and then “flame dried” over an alcohol flame.
Probes and FISH:
All BAC clones used for FISH mapping were obtained from the RHPOTKEY potato BAC library constructed from the RH clone RH89-039-16 (http://potatogenome.net/FAQPage@DocumentsPane.html#IntegrationStatus), using AFLP markers previously mapped to chromosome 6 (van Os et al. 2006; http://potatogenome.net/FAQPage@DocumentsPane.html#FingerprintingMethod). BAC 39P07, a clone specific to potato chromosome 6 (Dong et al. 2000) previously identified from a S. bulbocastanum BAC library (Song et al. 2000), was used as a reference in initial FISH experiments for chromosome identification for both potato and tomato. BAC RH051A16 hybridized to the pericentromeric heterochromatic regions of all potato chromosomes and was used to visualize the heterochromatin/euchromatin boundaries on potato chromosome 6.
BAC DNA was isolated using the QIAGEN (Valencia, CA) plasmid midikit and labeled with either biotin-16-UTP or digoxigenin-11-dUTP (Roche Diagnostic, Indianapolis) by standard nick translation reaction. The FISH procedure applied to both mitotic and meiotic chromosomes was essentially the same as previously described (Dong et al. 2000). Most BAC probes required sheared potato genomic DNA in the hybridization mixture to reduce the background signals on both potato and tomato chromosomes. To produce more consistent measurements, high-quality pachytene chromosomes were used for repeated probing up to four times, using the procedure described by Cheng et al. (2001). Alternatively, multiprobe FISH cocktails consisting of up to eight different BAC probes were applied. Biotin-labeled and digoxigenin-labeled probes were detected by fluorescein isothiocyanate-conjugated anti-biotin antibody and rhodamine-conjugated anti-digoxigenin antibody (Roche Diagnostic), respectively. Chromosomes were counterstained by 4′,6-diamidino-2-phenylindole (DAPI) in antifade VECTASHIELD solution (Vector Laboratories, Burlingame, CA). Images were captured using a SenSys CCD camera attached to an Olympus BX60 epifluorescence microscope. The CCD camera was controlled using IPLab Spectrum v3.1 software (Signal Analytics, Vienna, VA) on a Macintosh computer. Final image adjustments were done with Adobe Photoshop software.
Cytological measurements and construction of a FISH-based physical map of potato chromosome 6:
The total length of midpachytene chromosome 6, the chromosome arm ratio (r = length of the long arm/length of the short arm), the percentage of heterochromatin (on the total length and for each arm), and the position of the interstitial knob were measured in potato genotypes USW1 and RH and in tomato genotype Koralik. To estimate the chromatin condensation pattern along potato chromosome 6, measurements were also done at different pachytene stages.
To allow a direct comparison between genetic and physical positions of the genetic markers, measurements of BAC positions along the chromosome were taken according to a methodology described by Cheng et al. (2001). The length of the linkage map of potato chromosome 6 of clone RH is 57.6 cM (https://cbsgdbase.wur.nl/UHD/chromdrawmapframe.php?parent=RH&ch=6). Thus, the length of pachytene chromosome 6 was divided into 57.6 fractional lengths (FL). The position of each marker-anchored BAC clone on a pachytene chromosome 6 was calculated using
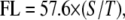 |
where S is the distance (in micrometers) between the FISH hybridization site and the end of the short arm, and T is total length of the chromosome (in micrometers). FL data were collected from pachytene chromosomes 6 of average length of 41.1 ± 5.2 μm. All measurements were made on digital images using IPLab software.
RESULTS
Morphology of pachytene chromosome 6 of potato and tomato:
Potato pachytene chromosomes were prepared from a haploid potato clone USW1 (2n = 2x = 24) and the diploid clone RH that was chosen for sequencing by the Potato Genome Sequencing Consortium (http://potatogenome.net/). The length of potato chromosome 6 (in USW1) at different pachytene stages varied from ∼23 to ∼67 μm (supplemental Table 1). The morphological features of midpachytene chromosomes 6 in both USW1 and RH genotypes are summarized in Table 1 and shown in Figure 1. In general, the morphology of the midpachytene chromosomes 6 from USW1 (Figure 1b) and RH (Figure 1c) is highly similar. On average, the total length of a midpachytene chromosome 6 of USW1 was 43 ± 4 μm (n = 16), with a long arm of 34 ± 3 μm and a short arm of 9 ± 1 μm. The subtelomeric chromosome 6 had an arm ratio of ∼4:1 (Table 1).
TABLE 1.
Length, arm ratio, percentage of heterochromatin, knob, and centromere position of potato (USW1 and RH) and tomato chromosome 6
| Genotypea | n | Total chr. length (μm) | LAa (μm) | SAa (μm) | Arm ratio | Het. totalb (%) | Het. LA (%) | Het. SA (%) | Knob positionc (%) | Cen positionc (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| S. tuberosum (RH) | 11 | 47.2 ± 2.9 | 37.9 ± 2.5 | 9.3 ± 0.9 | 4.1:1 | 27.0 ± 4.1 | 20.5 ± 3.9 | 53.3 ± 5.8 | 49.7 ± 2.4 | 19.7 ± 1.5 |
| S. tuberosum (USW1) | 16 | 42.8 ± 3.8 | 34.1 ± 3.4 | 8.8 ± 0.7 | 3.9:1 | 27.7 ± 2.9 | 21.3 ± 3.5 | 52.4 ± 4.9 | 53.6 ± 4.3 | 20.5 ± 1.5 |
| S. lycopersicum (Koralik) | 11 | 45.5 ± 3.9 | 37.2 ± 3.7 | 8.7 ± 0.7 | 4.3:1 | 26.8 ± 2.6 | 22.5 ± 3.1 | 44.9 ± 2.9 | NA | 19.2 ± 2.4 |
LA, long arm; SA, short arm.
[Total heterochromatin (μm)/total chromosome length (μm)] × 100.
Knob (centromere) position (%) is (S/T) × 100, where S = distance of knob (centromere) (in micrometers) from the end of the short arm, and T = length of chromosome 6 (in micrometers). NA, the knob was not observed in tomato.
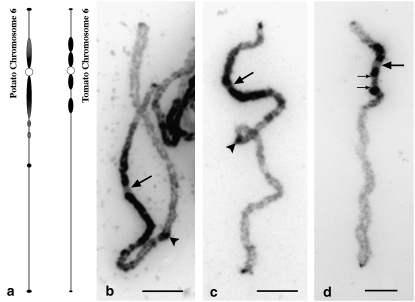
Figure 1.—
Morphology and heterochromatin distribution on pachytene chromosome 6 of potato and tomato. (a) Idiograms of potato and tomato pachytene chromosome 6. Heterochromatic regions are represented by solid/shaded thickenings. Shaded thickenings indicate regions that were less stained by DAPI than the regions marked by solid thickenings. Hatched thickenings indicate regions that are separated into multiple small knobs on early pachytene chromosomes. (b) A midpachytene chromosome 6 of USW1. All large arrows point to the centromeres. Arrowheads point to the major knob on the long arm of potato chromosome 6. Several small knobs are visible between the proximal heterochromatin and the major knob (arrowhead) on the long arm. (c) A midpachytene chromosome 6 of RH. (d) A midpachytene chromosome 6 of tomato. Identification of this chromosome was confirmed by FISH mapping of a potato chromosome BAC RH034P18 (data not shown). Two small arrows point to the ends of the euchromatic domain within the pericentromeric heterochromatin on the long arm. Bars, 5 μm.
Potato chromosome 6 showed a distinct heterochromatin and euchromatin distribution pattern on the basis of DAPI staining. Heterochromatin accounts for ∼28% of the length of the chromosome. Most of the heterochromatin was confined to the pericentromeric region, spanning ∼21% of the length of the long arm and ∼52% of the length of the short arm (Table 1). Small heterochromatic domains were observed at both ends of the chromosome (Figure 1, b and c). In addition, an interstitial heterochromatic knob was consistently detected on the long arms of both USW1 and RH (Figure 1, a–c). This knob, however, was not observed in potato lines described by Tang et al. (2008), which shows intraspecific variation of the heterochromatin distribution in the potato genome.
While the intensity of DAPI staining of euchromatin vs. heterochromatin was significantly different, the transition between the heterochromatic and the euchromatic regions was often not sharp and thus difficult to locate on some chromosomes, especially the transition zone on the long arm. Several small interspersed heterochromatic and euchromatic domains were observed in the transition zone on the long arm of chromosome 6 of both USW1 and RH (Figure 1, b and c), especially on well-extended early pachytene chromosomes. The DAPI staining intensity associated with the pericentromeric heterochromatin was not uniform on early pachytene chromosomes. The proximal heterochromatin was more brightly stained than the distal heterochromatin, especially on the short arm (Figure 1, b and c).
In general, the pachytene chromosome 6 of tomato cv. Koralik shares a similar total chromosome length, arm ratio, and heterochromatin distribution pattern to potato pachytene chromosome 6 but with some distinct differences (Table 1, Figure 1d). The transitions between the pericentromeric heterochromatin and euchromatin were generally less ambiguous in tomato than in potato. The pericentromeric heterochromatin on the long arm of tomato chromosome 6 from both genotypes Koralik and Quinte was clearly separated by a euchromatic domain (Figure 1d). On average, ∼23% of the long arm of tomato chromosome 6 is heterochromatic, which is similar to potato. However, this estimate is comprehensive of the euchromatic domain within the pericentromeric heterochromatin on the long arm. In addition, the distinct interstitial knob in the long arm of potato chromosome 6 was not observed on the tomato chromosome 6 in both varieties analyzed (Figure 1d).
The condensation pattern of potato pachytene chromosome 6:
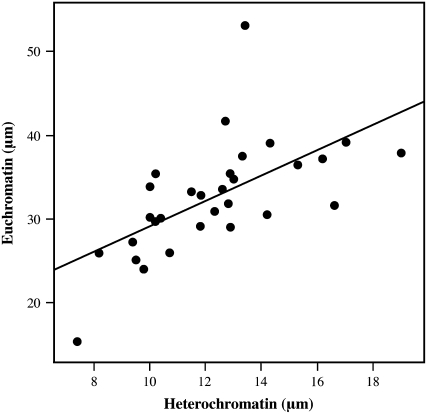
Length variation and chromatin condensation pattern of pachytene chromosomes are very important for FISH mapping purposes. Although FISH mapping data are usually expressed as relative distances, a differential condensation pattern of euchromatin vs. heterochromatin of the chromosome could dramatically affect the resulting cytogenetic map. This, in turn, would mean that the relative distances on the cytogenetic map refer only to pachytene chromosomes at a particular stage. To investigate whether chromatin condensation pattern (euchromatin vs. heterochromatin) was uniform along potato chromosome 6, we measured the euchromatic and heterochromatic regions of 31 pachytene chromosomes. The total chromosome length varied from ∼23 μm (with total euchromatic region of 15 μm) to ∼67 μm (with total euchromatic region of 53 μm), corresponding to a 2.9-fold variation (supplemental Table 1). Most chromosomes observed fell into the 34- to 56-μm range, which corresponds to a length variation of 1.6-fold, and had an average ratio between total euchromatin and heterochromatin of 2.7. We plotted the total length of the euchromatic regions against the total length of the heterochromatic regions for each chromosome. Correlation between total euchromatin and heterochromatin (micrometers) was highly significant (r = 0.616, P = 0.0002; Figure 2). In addition, a highly significant correlation was found between the length of the short arm and the length of long arm (r = 0.748, P < 0.0001). These results indicated an overall uniform chromatin condensation pattern along chromosome 6, at least within the length range reported here.
Figure 2.—
Correlation of condensation between the euchromatin and the heterochromatin of potato chromosome 6. Total length of euchromatin and heterochromatin regions was compared for 31 pachytene chromosomes 6 from potato clone USW1.
The genetic position of the centromere of potato chromosome 6:
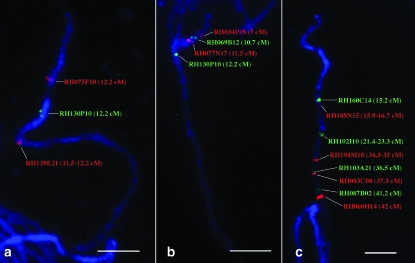
An ultradense genetic recombination map of potato with >10,000 AFLP loci was constructed using two diploid heterozygous potato clones, RH and SH (van Os et al. 2006). The genetic positions of the centromere of chromosome 6 were tentatively mapped to 12.2 cM (bin 17) and to 2.9 cM (bin 5) in the linkage maps of the RH parent and the SH parent, respectively. The centromeric positions were predicted on the basis of the significant density of markers in these regions on the linkage maps (van Os et al. 2006) and, in the case of the RH parent, it was confirmed by half-tetrad analysis (Park et al. 2007).
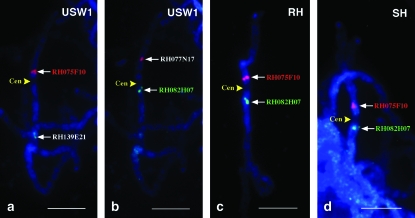
Cytologically, the centromere of potato chromosome 6 can be unambiguously identified on the basis of its characteristic light DAPI staining at the pachytene stage (Figure 1). The lightly stained centromere is flanked by brightly stained heterochromatin on both sides. FISH mapping of several BACs anchored by AFLP markers mapped between 11.5 and 12.2 cM (bin range 16–17, Table 2) enabled us to integrate this cytological feature into the genetic map (Figure 3). The centromere was localized at 11.8 FL, between BAC RH075F10 located on the short arm and BAC RH082H07 on the long arm. BAC RH075F10 was anchored by AFLP marker 7786 at 12.2 cM of the RH map (bin 17), whereas BAC RH082H07 was anchored by AFLP marker 12,647 at 11.5–12.2 cM of the RH map (bin range 16–17) and at 1.5–3.6 cM of the SH map (bin range 3–6), respectively. The physical locations of these two marker-anchored BACs relative to the centromere were consistent among potato genotypes USW1, RH, and SH (Figure 3).
TABLE 2.
Genetic and physical position of AFLP marker-anchored potato BAC clones
| BAC clone | Marker ID | Chromosome arm | cM | FLa | Relative physical locationb | n |
|---|---|---|---|---|---|---|
| RH177C17 | 20,054 | Short arm | 0.0 | 0.7 ± 0.2 | 1.2 ± 0.4 | 10 |
| RH160K03 | 7,679 | 0.8 | d | d | ||
| RH083L10 | 20,669 | 0.0–1.6 | 1.8 ± 0.3 | 3.1 ± 0.5 | 11 | |
| RH081K17 | 20,298 | 0.8–1.6 | d | d | ||
| RH097M07 | 7,688 | 1.6 | d | d | ||
| RH136O23 | 7,689 | 1.6 | d | d | ||
| 39P07 | GP79 | c | 2.4 ± 0.4 | 4.2 ± 0.7 | 7 | |
| RH084L03 | 12,631 | 3.1 | 3.2 ± 0.5 | 5.6 ± 0.9 | 7 | |
| RH034P18 | 7,717 | 7.0 | 4.7 ± 0.5 | 8.1 ± 0.9 | 14 | |
| RH125D13 | 7,717 | 7.0 | d | d | ||
| Hetero-euchromatin boundary | 5.6 ± 0.8 | 9.8 ± 1.4 | 16 | |||
| RH069B12 | 7,751 | 10.7 | 6.7 ± 0.6 | 11.6 ± 1.0 | 8 | |
| RH077N17 | 7,752 | 11.5 | 8.1 ± 1.0 | 14.0 ± 2.0 | 5 | |
| RH018J03 | 12,623 | 7.0–12.2 | 8.7 ± 1.0 | 15.1 ± 1.7 | 6 | |
| RH143A24 | 12,659 | 11.5–12.2 | d | d | ||
| RH075F10 | 7,786 | 12.2 | 10.8 ± 1.6 | 18.8 ± 2.9 | 10 | |
| RH022P07 | 7,786 | 12.2 | d | d | ||
| Centromere | 11.8 ± 0.9 | 20.5 ± 1.5 | 16 | |||
| RH082H07 | 12,647 | Long arm | 11.5–12.2 | 14.2 ± 1.8 | 24.7 ± 3.1 | 3 |
| RH130P10 | 7,801 | 12.2 | 15.2 ± 1.0 | 26.5 ± 1.7 | 10 | |
| RH060M13 | 20,173 | 12.2–14.4 | 18.0 ± 1.3 | 31.3 ± 2.2 | 5 | |
| Hetero-euchromatin boundary | 21.6 ± 1.8 | 37.5 ± 3.2 | 16 | |||
| RH139E21 | 7,753 | 11.5–12.2 | 22.1 ± 2.3 | 38.4 ± 4.0 | 12 | |
| RH057H05 | 7,891 | 13.7 | 23.3 ± 1.3 | 40.5 ± 2.2 | 5 | |
| RH160C14 | 7,905 | 15.2 | 25.2 ± 1.1 | 43.7 ± 1.9 | 12 | |
| RH188N15 | 20,129 | 15.9–16.7 | 26.3 ± 1.1 | 45.7 ± 1.9 | 12 | |
| RH094G20 | 7,933 | 20.3 | 30.2 ± 0.9 | 52.5 ± 1.6 | 4 | |
| Knob | 30.9 ± 2.4 | 53.6 ± 4.3 | 16 | |||
| RH102I10 | 12,834 | 21.4–23.3 | 33.2 ± 0.8 | 57.7 ± 1.4 | 11 | |
| RH051B02 | 20,052 | 24.9 | 33.8 ± 1.0 | 58.6 ± 1.7 | 4 | |
| RH194M18 | 7,989 | 34.3–35.0 | 38.7 ± 0.4 | 67.2 ± 0.8 | 7 | |
| RH103A21 | 8,016 | 36.5 | 40.8 ± 1.3 | 70.9 ± 2.2 | 8 | |
| RH083C08 | 8,040 | 37.3 | 41.4 ± 0.6 | 71.8 ± 1.0 | 7 | |
| RH087B02 | 8,060 | 41.2 | 44.5 ± 0.9 | 77.3 ± 1.6 | 8 | |
| RH060H14 | 8,076 | 42.0 | 46.3 ± 0.6 | 80.5 ± 1.0 | 7 | |
| RH200K19 | 12,982 | 53.7 | 49.8 ± 0.6 | 86.4 ± 1.0 | 6 | |
| RH204G08 | 12,998 | 56.8–57.6 | 52.7 ± 0.6 | 91.5 ± 1.0 | 6 |
Fraction length (FL) = (S/T) × 57.6, where S is the distance (micrometers) from the FISH site to the end of the short arm, T is the total length of the chromosome (micrometers), and 57.6 is the length (in centimorgans) of the linkage map of chromosome 6.
Relative physical location = (S/T) × 100, where S is the distance (in micrometers) from the FISH site to the end of the short arm of the chromosome, and T is the total length of the chromosome (in micrometers). All measurements were made on pachytene chromosomes 6 of average length of 41.1 ± 5.2 μm.
BAC 39P07 is anchored by marker GP79 that was mapped to 4.4 cM on the potato linkage group 6 by Tanksley et al. (1992).
The location of BAC RH177C17 on the pachytene chromosome was completely overlapped with RH160K03; RH083L10 overlapped with RH081K17, RH097M07, and RH136O23; RH034P18 overlapped with RH125D13; RH143A24 overlapped with RH018J03; and RH022P07 overlapped with RH075F10.
Figure 3.—
Determination of the genetic position of the centromere of potato chromosome 6 by FISH mapping of marker-anchored BAC clones. (a) FISH mapping of BACs RH075F10 and RH139E21 on pachytene chromosome 6 of USW1. (b) FISH mapping of BACs RH077N17 and RH082H07 on pachytene chromosome 6 of USW1. (c) FISH mapping of BACs RH075F10 and RH082H07 on pachytene chromosome 6 of RH. (d) FISH mapping of BACs RH075F10 and RH082H07 on pachytene chromosome 6 of SH. The positions of the centromeres (Cen) are shown by yellow arrowheads. Bars, 5 μm.
The genetic positions of the euchromatin–heterochromatin boundaries on potato chromosome 6:
To determine the genetic positions of the euchromatin–heterochromatin boundaries we mapped several BACs, anchored by AFLP markers between 10.7 and 14.4 cM, to the pachytene chromosome positions relative to the brightly DAPI-stained pericentromeric heterochromatin (Table 2, Figure 4). On the basis of the DAPI staining pattern, the transition between euchromatin and heterochromatin on the short arm and the long arm was estimated at ∼10% (5.6 FL) and ∼37% (21.6 FL) of the total chromosome length from the end of the short arm, respectively. BAC RH069B12 was anchored by AFLP marker 7751 mapped to 10.7 cM (bin 15), and it was mapped very close to the euchromatin/heterochromatin junction on the short arm (Figure 4a). The FISH signal generated by this BAC was located ∼12% from the end of the short arm, at 6.7 FL (Table 2). On the long arm, the boundary was located between BAC RH060M13 (identified by AFLP marker 20,173 at 12.2–14.4 cM, bin range 17–20) and BAC RH139E21 (identified by marker 7753 at 11.5–12.2 cM, bin range 16–17) (Figure 4a). FISH signals derived from RH060M13 and RH139E21 were detected at ∼31% (18.0 FL) and ∼38% (22.1 FL) away from the end of the short arm, respectively.
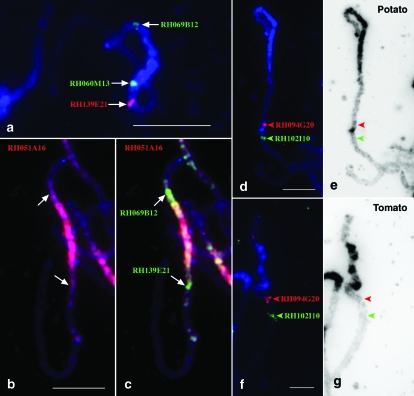
Figure 4.—
The genetic positions of the heterochromatic domains on potato chromosome 6. (a) BAC RH069B12 is located in the euchromatin–heterochromatin boundary on the short arm. The boundary on the long arm is not sharp but can be assigned to between BAC RH060M13 and BAC RH139E21. (b) A potato (USW1) pachytene chromosome 6 was probed with BAC RH051A16. The FISH signals (red) cover the entire pericentromeric region as well as the interstitial knob on the long arm. (c) Two BAC clones (green), RH069B12 and RH139E21, were comapped with RH051A16 (red). The most prominent green signals (arrows) were mapped within the euchromatin–heterochromatin boundaries. (d) FISH mapping of two BAC clones, RH094G20 (red) and RH102I10 (green), which span the knob on the long arm of potato chromosome 6. (e) The DAPI-stained pachytene chromosome was converted into a black-and-white image to enhance the contrast of the knob region. The arrowheads point to the location of the two BAC clones. (f) FISH mapping of two BACs, RH094G20 (red) and RH102I10 (green) on tomato pachytene chromosome 6. (g) The DAPI-stained pachytene chromosome was converted into a black-and-white image. The arrowheads point to the location of the two BAC clones. No knob is observed between the two arrowheads. Bars, 5 μm.
As mentioned above, the transition between euchromatin and heterochromatin was often not sufficiently sharp for unambiguous localization based on DAPI staining. We used the FISH signals derived from a highly repetitive BAC clone RH051A16 to assist the identification of the heterochromatic region. The FISH signals generated by BAC RH051A16 covered almost all of the DAPI-bright regions (supplemental Figure 1). Co-FISH mapping with BAC RH051A16 confirmed that the pericentromeric heterochromatin of chromosome 6 was confined between AFLP marker 7751 on the short arm and markers 7753/20,173 on the long arm (Figure 4, b and c). This region corresponds to <2 cM (bins 15–17) on the genetic map of the RH parent and to ∼16 FL on the cytogenetic map (Table 2).
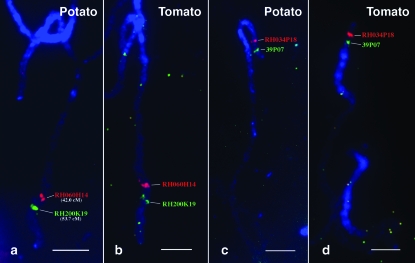
FISH also allowed the integration in the genetic map of the heterochromatic knob located on the long arm. This knob was located at 30.9 FL, between BACs RH094G20 (anchored by AFLP 7933 marker at 20.3 cM) and RH102I10 (identified by AFLP marker 12,834 at 21.4–23.3 cM) in both USW1 and RH (Figure 4, d and e). These two BACs were mapped to the similar positions on tomato chromosome 6 (Figure 4, f and g).
Correlation between genetic and physical distances on potato chromosome 6:
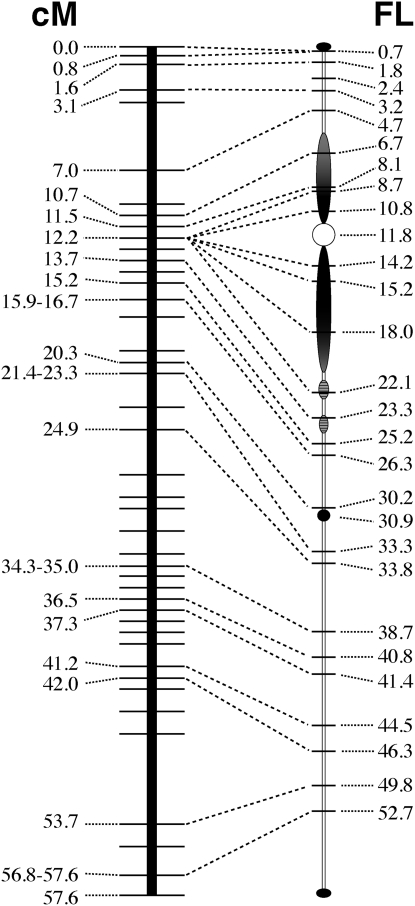
To investigate the relationship between genetic and physical distances of the AFLP markers mapped on potato chromosome 6, the chromosomal positions of 30 AFLP maker-anchored BACs were mapped on the pachytene chromosome 6 (Table 2, Figures 5 and 6). The AFLP markers anchoring the BACs are spaced at an average of ∼3 cM intervals from 0.0 cM (bin 1) to the end of the linkage map of RH (56.8–57.6 cM, bins 73–74). We mapped a total of 13 BACs on the short arm and 17 on the long arm, including 9 BACs in the pericentromeric heterochromatin region. In general, the order of individual BACs along the chromosome was concordant with the order of the AFLP markers along the linkage map (Table 2). However, the FISH results allowed us to resolve the order of the tightly linked markers located in the pericentromeric region. These markers were mapped to the same centimorgan/bin (or centimorgan/bin range) on the linkage map. For example, the markers used to anchor RH075F10 and RH130P10 were both mapped to 12.2 cM on the RH genetic map (Table 2). Physically, RH075F10 was located on the short arm at 10.8 FL, and RH130P10 was mapped on the long arm at 15.2 FL (Figure 5a).
Figure 5.—
FISH mapping of AFLP marker-anchored BAC clones on potato pachytene chromosome 6. (a) FISH mapping of three BACs anchored by markers mapped at 12.2 cM. These three BACs span the centromere and most of the heterochromatin on the long arm. (b) FISH mapping of four BACs by anchored markers mapped at between 7 and 12.2 cM. (c) FISH mapping of eight BACs located in the euchromatic region on the long arm. Bars, 5 μm.
Figure 6.—
Integration of genetic linkage map of potato chromosome 6 with potato pachytene chromosome 6. The genetic linkage map of potato chromosome 6 is according to van Os et al. (2006).
The relationship between the genetic and physical distances along chromosome 6 is illustrated in Figure 6. Overall, the average recombination frequency along the short arm (12.2 cM/10.8 FL) was similar to that along the long arm (45.4 cM/43.4 FL). However, recombination was not evenly distributed along the physical length of chromosome 6. A significant disproportion between genetic and physical distances was found in the region of the chromosome spanning 10.7–12.2 cM on the genetic map and 6.7–22.1 FL on the cytogenetic map, which largely corresponds to the pericentromeric heterochromatin. Within this region, suppression of recombination was found between the markers anchoring the BACs RH075F10, RH082H07, RH130P10, RH060M13, and RH139E21, which all genetically mapped to 12.2 cM. Indeed, BACs RH075F10 and RH139E21 are physically separated by ∼11 FL (Table 2). In addition, suppression of recombination seemed to be more severe in the heterochromatic region on the long arm, effecting ∼2 FL on the short arm and ∼11 FL on the long arm.
Several other regions also show disproportion between genetic and physical distances. In the euchromatic region of the short arm, BACs RH084L03 (3.1 cM) and RH034P18 (7.0 cM) are separated by ∼4 cM but by only 1.5 FL (Figure 6). On the long arm, RH102I10 (21.4–23.3 cM) and RH051B02 (24.9 cM) are separated genetically by 1.6–3.5 cM but physically by ∼0.5 FL, a three- to sixfold difference (Figure 6). Similarly, RH060H14 (42.0 cM) and RH200K19 (53.7 cM) are separated by 11.7 cM, but physically by 3.5 FL, an approximately threefold difference (Figures 6 and 7a). These disparities between the genetic and physical maps, it should be noted, indicate a genetic map longer than the physical map, which is the opposite of the trend in the pericentromeric region.
Figure 7.—
Comparative FISH mapping of potato and tomato pachytene chromosome 6. (a) FISH mapping of RH060H14 (42.0 cM) and RH200K19 (53.7 cM) on potato pachytene chromosome 6. The region spanned by these two BACs shows an increased recombination rate. (b) BACs RH060H14 and RH200K19 were located at similar positions on tomato pachytene chromosome 6. (c) FISH mapping of BACs 39P07 (green) and RH034P18 (red) on potato pachytene chromosome 6. (d) FISH mapping of BACs 39P07 (green) and RH034P18 (red) on tomato pachytene chromosome 6. These two BACs showed a reverse order as compared with that on potato chromosome 6. Bars, 5 μm.
Comparative FISH mapping on tomato pachytene chromosome 6:
Comparative linkage mapping based on a common set of markers revealed five chromosomal inversions differentiating the potato and tomato genomes, but otherwise demonstrated an overall genetic colinearity between the two species (Tanksley et al. 1992). To compare potato and tomato chromosome 6 from a cytological standpoint, a set of the potato BACs used in this study was FISH mapped on tomato pachytene chromosome (Table 3 and Figures 7 and 8). BAC 39P07, a clone specific to potato chromosome 6 (Dong et al. 2000), was also included for chromosome identification, as this clone was identified by RFLP marker GP79 previously mapped on linkage group 6 of both tomato and potato (Tanksley et al. 1992).
TABLE 3.
Comparative FISH mapping of potato BAC clones in potato and tomato
| Relative physical location (%)a
|
|||||
|---|---|---|---|---|---|
| BAC clone | Chromosome arm/region | Potato USW1 | n | Tomato cv. Koralik | n |
| RH160K03, RH177C17 | Short arm | 1.2 ± 0.4 | 10 | 10.1 ± 0.7 | 6 |
| RH081K17, RH136O23 | 3.1 ± 0.5 | 11 | 8.6 ± 0.8 | 6 | |
| 39P07 | 4.2 ± 0.7 | 7 | 6.9 ± 1.1 | 5 | |
| RH084L03 | 5.6 ± 0.9 | 7 | 5.7 ± 0.8 | 6 | |
| RH034P18, RH125D13 | 8.1 ± 0.9 | 14 | 4.0 ± 0.5 | 10 | |
| Hetero-euchromatin boundary | 9.8 ± 1.4 | 16 | 10.3 ± 1.6 | 11 | |
| Centromere | 20.5 ± 1.5 | 16 | 19.2 ± 2.4 | 11 | |
| Hetero-euchromatin boundary | 37.5 ± 1.2 | 16 | 37.4 ± 3.6 | 11 | |
| RH094G20 | Long arm | 52.5 ± 1.6 | 4 | 45.2 ± 2.7 | 6 |
| Knob | 53.6 ± 4.3 | 16 | b | b | |
| RH102I10 | 57.7 ± 1.4 | 11 | 51.2 ± 4.2 | 6 | |
| RH194M18 | 67.2 ± 0.8 | 7 | NA | NA | |
| RH103A21 | 70.9 ± 2.2 | 8 | NA | NA | |
| RH060H14 | 80.5 ± 1.0 | 7 | 77.4 ± 2.1 | 5 | |
| RH200K19 | 86.4 ± 1.0 | 6 | 84.7 ± 2.7 | 7 | |
| RH204G20 | 91.5 ± 1.0 | 6 | 92.0 ± 2.8 | 3 | |
NA, measurement data not available. However, the relative order of these BACs in tomato was the same as in potato.
Relative physical location is determined as specified in Table 1. BAC clones are listed following the order in potato. All measurements for tomato were made on pachytene chromosomes 6 of average length of 45.2 ± 5.1 μm.
The knob is not present in tomato.
Figure 8.—
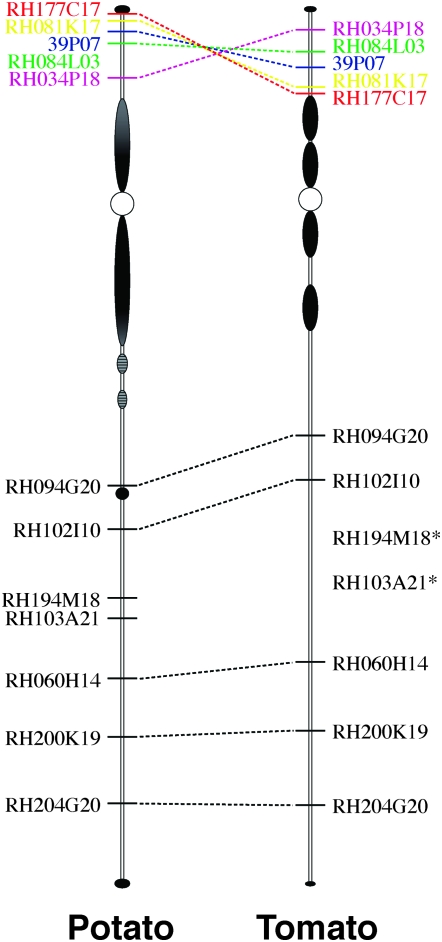
The comparative chromosomal positions of 12 potato BACs on potato and tomato pachytene chromosome 6. The exact chromosomal positions of BACs RH194M18 and RH103A21 in tomato were not determined, but relative order of these two clones in tomato is the same as that in potato.
Cytologically, the centromere and the boundaries of the pericentromeric heterochromatin were estimated to be at similar physical positions in the two species. The potato BAC clones on the long arm were mapped to similar positions on the pachytene chromosome 6 of tomato (Table 3, Figures 7, a and b, and 8). However, on the short arm the relative order between BAC RH177C17 (0.0 cM) and RH034P18 (7.0 cM) was inverted on tomato compared to potato (Figures 7, c and d, and 8). The chromosomal region spanned by these two clones almost includes the entire euchromatic portion of the short arm in both species (Figure 8; supplemental Figure 2). In potato, RH177C17 (0.0 cM) was detected very close to the telomere of the short arm. RH034P18 (7.0 cM) was located near the proximal heterochromatin (Figure 8). In tomato, however, RH177C17 was detected at the euchromatin/heterochromatin boundary, whereas RH034P18 was close to the telomere (Figure 8). These results suggest that a potato/tomato inversion likely spans the entire euchromatic portion of the short arms of the chromosome 6. A similar pachytene chromosome-based FISH mapping using tomato BACs revealed the same potato/tomato inversion (Tang et al. 2008).
DISCUSSION
Severe repression of genetic recombination in the pericentromeric heterochromatin:
Suppression of recombination in the pericentromeric regions is a common genetic phenomenon in all eukaryotes. However, the sizes of the recombination-suppressed pericentromeric domains vary significantly in different plant species. In rice, these domains, which encompass the functional centromeres, contain only 1.5–5.5 megabases of DNA, which on average account for ∼10% of the rice chromosomes (Yan et al. 2005, 2006; H. H. Yan and J. Jiang, unpublished data). However, in several grass species with large genomes, such as barley, recombination is almost completely suppressed in the proximal halves of the chromosome arms (Künzel et al. 2000). Suppression of recombination in the pericentromeric regions in tomato was indicated by both classical and molecular marker-based genetic and cytogenetic mapping (Khush and Rick 1968; Tanksley et al. 1992; Koo et al. 2008; Szinay et al. 2008). The recombination-suppressed domains in tomato appeared to be correlated with the pericentromeric heterochromatin on the basis of mapping of recombination nodules (RNs) on synaptonemal complexes (SCs) (Sherman and Stack 1995). RNs were rarely observed in the pericentromeric regions in all tomato chromosomes.
Suppression of recombination in the pericentromeric domains of individual potato chromosomes was clearly illustrated by a single region with significant marker clustering on the majority of the linkage groups of the ultradense genetic linkage map (van Os et al. 2006). However, the marker clustering data do not reveal the physical span of the recombination-suppressed regions. Potato BACs anchored by AFLP markers mapped at 10.7–12.2 cM were localized to the heterochromatin–euchromatin transition regions on both arms (Figure 6). Thus, the integration of genetic and physical maps revealed that the entire pericentromeric domain of potato chromosome 6 is severely suppressed in recombination. Cytologically distinct heterochromatin was observed in the pericentromeric regions of all potato chromosomes (Yeh and Peloquin 1965) (supplemental Figure 1). Thus, recombination suppression is likely to be associated with the pericentromeric heterochromatin in other potato chromosomes. It is also interesting to note that the recombination in the euchromatin is enhanced relative to heterochromatin, but the recombination rate in the euchromatic region varies widely. The chromosomal domain at FL 46.3–49.8 shows a significantly higher recombination rate than the rest of the chromosome (Figure 6).
Potato–tomato synteny:
Potato and tomato have diverged for ∼12 million years (Desa and Drouin 1996). The pachytene chromosomes from these two species share highly similar morphology (Barton 1950; Yeh and Peloquin 1965). The genetic colinearity between potato and tomato chromosomes was well demonstrated by comparative genetic linkage mapping, using a common set of molecular markers (Bonierbale et al. 1988; Tanksley et al. 1992). Five chromosomal inversions of marker order were found to be associated with potato/tomato chromosomes 5, 9, 10, 11, and 12. These inversions appear to be paracentric and involve the entire chromosome arms (Tanksley et al. 1992). FISH mapping of potato BACs on tomato chromosome 6 revealed a similar inversion involving part of the short arm (Figures 7 and 8). Although the order of two DNA markers (GP164 and GP79) suggested an inversion associated with tomato chromosome 6 (van Wordragen et al. 1994), this inversion was not revealed by the previous comparative linkage mapping possibly due to the relatively few markers mapped on the short arms of potato/tomato chromosome 6 (Gebhardt et al. 1991; Tanksley et al. 1992). Since the short arms of potato and tomato chromosome 6 share a similar heterochromatin distribution pattern, the proximal breakpoint of this inversion is likely located within the euchromatin. The chromosomal position of BACs RH177C17 and RH034P18 in potato and tomato demonstrates that this inversion possibly involves the entire euchromatic portion of the short arms, which has also been revealed by Tang et al. (2008) using a different set of tomato and potato BAC clones. Our results also suggest that the five other inversions associated with potato and tomato chromosomes (Tanksley et al. 1992) may also span only the euchromatic portions of the respective arms, rather than the complete arms.
The discovery of the inversion associated with chromosome 6 suggests that the potato and tomato genomes may contain significantly more structural rearrangements than those revealed by the previous comparative linkage mapping. Rearrangements located in the pericentromeric heterochromatin are difficult to assess by linkage mapping due to the cosegregation of the markers in the mapping population. It is interesting to note that the pericentromeric heterochromatin in the long arm of tomato chromosome 6 is separated by a euchromatic domain (Figure 1d) that is not observed in the corresponding region in potato. An inversion in the tomato chromosome, with one breakpoint in the euchromatin and one breakpoint in the heterochromatin, can result in the difference between the potato and the tomato chromosome 6. Such an inversion involving the pericentromeric heterochromatin and euchromatin was demonstrated in Arabidopsis thaliana (Fransz et al. 2000). Complete sequencing of potato and tomato chromosome 6 will reveal the mechanism that caused the difference in heterochromatin distribution in these two chromosomes.
Acknowledgments
We thank Brian Yandell for his help on statistical analysis of the pachytene chromosome condensation pattern. This work was supported by grant DBI-0604907 from the National Science Foundation. The RHPOTKEY library was provided by the Laboratory of Plant Breeding, Wageningen University (Wageningen, The Netherlands), Applied Science Foundation STW (Utrecht, The Netherlands), and Keygene N.V. (Wageningen, The Netherlands).
References
- Amarillo, F. I. E., and H. W. Bass, 2007. A transgenomic cytogenetic sorghum (Sorghum propinquum) bacterial artificial chromosome fluorescence in situ hybridization map of maize (Zea mays L.) pachytene chromosome 9: evidence for regions of genome hyperexpansion. Genetics 177 1509–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, D. W., 1950. Pachytene morphology of the tomato chromosome complement. Am. J. Bot. 37 639–643. [Google Scholar]
- Bonierbale, M. W., R. L. Plaisted and S. D. Tanksley, 1988. RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S. B., L. K. Anderson, J. D. Sherman, S. M. Royer and S. M. Stack, 2007. Predicting and testing physical locations of genetically mapped loci on tomato pachytene chromosome 1. Genetics 176 2131–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q., S. Sun, Q. Ye, S. McCuine, E. Huff et al., 2004. Construction of two BAC libraries from the wild Mexican diploid potato, Solanum pinnatisectum, and the identification of clones near the late blight and Colorado potato beetle resistance loci. Theor. Appl. Genet. 108 1002–1009. [DOI] [PubMed] [Google Scholar]
- Cheng, Z., G. G. Presting, C. R. Buell, R. A. Wing and J. Jiang, 2001. High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157 1749–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z. K., C. R. Buell, R. A. Wing and J. Jiang, 2002. Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes and extended DNA fibers. Chromosome Res. 10 379–387. [DOI] [PubMed] [Google Scholar]
- deSa, M. M., and G. Drouin, 1996. Phylogeny and substitution rates of angiosperm actin genes. Mol. Biol. Evol. 13 1198–1212. [DOI] [PubMed] [Google Scholar]
- Dong, F., J. Song, S. K. Naess, J. P. Helgeson, C. Gebhardt et al., 2000. Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor. Appl. Genet. 101 1001–1007. [Google Scholar]
- Flinn, B., C. Rothwell, R. Griffiths, M. Lague, D. DeKoeyer et al., 2005. Potato expressed sequence tag generation and analysis using standard and unique cDNA libraries. Plant Mol. Biol. 59 407–433. [DOI] [PubMed] [Google Scholar]
- Fransz, P. F., S. Armstrong, J. H. de Jong, L. D. Parnell, G. van Drunen et al., 2000. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100 367–376. [DOI] [PubMed] [Google Scholar]
- Gebhardt, C., E. Ritter, T. Debener, U. Schachtschabel, B. Walkemeier et al., 1989. RFLP analysis and linkage mapping in Solanum tuberosum. Theor. Appl. Genet. 78 65–75. [DOI] [PubMed] [Google Scholar]
- Gebhardt, C., E. Ritter, A. Barone, T. Debener, B. Walkemeier et al., 1991. RFLP maps of potato and their alignment with the homoeologous tomato genome. Theor. Appl. Genet. 83 49–57. [DOI] [PubMed] [Google Scholar]
- Howell, E. C., S. J. Armstrong, G. C. Barker, G. H. Jones, G. J. King et al., 2005. Physical organization of the major duplication on Brassica oleracea chromosome O6 revealed through fluorescence in situ hybridization with Arabidopsis and Brassica BAC probes. Genome 48 1093–1103. [DOI] [PubMed] [Google Scholar]
- Islam-Faridi, M. N., K. L. Childs, P. E. Klein, G. Hodnett, M. A. Menz et al., 2002. A molecular cytogenetic map of sorghum chromosome 1: fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 161 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, J. M. E., H. J. Vaneck, P. Arens, B. Verkerkbakker, B. T. L. Hekkert et al., 1995. A genetic map of potato (Solanum tuberosum) integrating molecular markers, including transposons, and classical markers. Theor. Appl. Genet. 91 289–300. [DOI] [PubMed] [Google Scholar]
- Jiang, J. M., and B. S. Gill, 1994. Nonisotopic in situ hybridization and plant genome mapping: the first 10 years. Genome 37 717–725. [DOI] [PubMed] [Google Scholar]
- Jiang, J. M., and B. S. Gill, 2006. Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49 1057–1068. [DOI] [PubMed] [Google Scholar]
- Khush, G. S., and C. M. Rick, 1968. Cytogenetic analysis of the tomato genome by means of induced deficiencies. Chromosoma 23 452–484. [Google Scholar]
- Kim, J. S., M. N. Islam-Faridi, P. E. Klein, D. M. Stelly, H. J. Price et al., 2005. Comprehensive molecular cytogenetic analysis of sorghum genome architecture: distribution of euchromatin, heterochromatin, genes and recombination in comparison to rice. Genetics 171 1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, D.-H., S.-H. Jo, J.-W. Bang, H.-M. Park, S. Lee et al., 2008. Integration of cytogenetic and genetic linkage maps unveils the physical architecture of tomato chromosome 2. Genetics 179 1211–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikova, O., G. Gualtieri, R. Geurts, D. J. Kim, D. Cook et al., 2001. Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J. 27 49–58. [DOI] [PubMed] [Google Scholar]
- Künzel, G., L. Korzun and A. Meister, 2000. Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, T. H., J. B. Kim, R. C. B. Hutten, H. J. van Eck, E. Jacobsen et al., 2007. Genetic positioning of centromeres using half-tetrad analysis in a 4x-2x cross population of potato. Genetics 176 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronning, C. M., S. S. Stegalkina, R. A. Ascenzi, O. Bougri, A. L. Hart et al., 2003. Comparative analyses of potato expressed sequence tag libraries. Plant Physiol. 131 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, J. D., and S. M. Stack, 1995. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J., F. Dong and J. Jiang, 2000. Construction of a bacterial artificial chromosome (BAC) library for potato molecular cytogenetics research. Genome 43 199–204. [PubMed] [Google Scholar]
- Szinay, D., S.-B. Chang, L. Khrustaleva, S. Peters, E. Schijlen et al., 2008. High-resolution chromosome mapping of BACs using multi-colour FISH and pooled-BAC FISH as a backbone for sequencing tomato chromosome 6. Plant J. (in press). [DOI] [PubMed]
- Tang, X., D. Szinay, C. Lang, M. S. Ramanna, E. A. G. van der Vossen et al., 2008. Cross-species bacterial artificial chromosome–fluorescence in situ hybridization painting of the tomato and potato chromosome 6 reveals undescribed chromosomal rearrangements. Genetics 180 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S. D., M. W. Ganal, J. P. Prince, M. C. de Vicente, M. W. Bonierbale et al., 1992. High-density molecular linkage maps of the tomato and potato genomes. Genetics 132 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Os, H., S. Andrzejewski, E. Bakker, I. Barrena, G. J. Bryan et al., 2006. Construction of a 10,000-marker ultradense genetic recombination map of potato: providing a framework for accelerated gene isolation and a genomewide physical map. Genetics 173 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wordragen, M. F., R. Weide, T. Liharska, A. Vandersteen, M. Koornneef et al., 1994. Genetic and molecular organization of the short arm and pericentromeric region of tomato chromosome 6. Euphytica 79 169–174. [Google Scholar]
- Walling, J. G., R. C. Shoemaker, N. D. Young, J. Mudge and S. A. Jackson, 2006. Chromosome level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics 172 1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.-J. R., L. Harper and Z. W. Cande, 2006. High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 18 529–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. H., W. W. Jin, K. Nagaki, S. Tian, S. Ouyang et al., 2005. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell 17 3227–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H. H., H. Ito, K. Nobuta, S. Ouyang, W. W. Jin et al., 2006. Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell 18 2123–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, B. P., and S. J. Peloquin, 1965. Pachytene chromosomes of the potato (Solanum tuberosum, group andigena). Am. J. Bot. 52 1014–1020. [Google Scholar]
- Zhang, H. N., J. P. T. Valkonen and K. N. Watanabe, 2003. A bacterial artificial chromosome (BAC) library for potato and identification of clones related to the potato Y potyvirus resistance gene Ryadg. Breed. Sci. 53 155–161. [Google Scholar]
- Zhu, W., S. Ouyang, M. Iovene, K. O'Brien, H. Vuong et al., 2008. Analysis of 90 Mb of the potato genome reveals conservation of gene structures and order with tomato but divergence in repetitive sequence composition. BMC Genomics 9 286. [DOI] [PMC free article] [PubMed] [Google Scholar]