Abstract
Ongoing genomics projects of tomato (Solanum lycopersicum) and potato (S. tuberosum) are providing unique tools for comparative mapping studies in Solanaceae. At the chromosomal level, bacterial artificial chromosomes (BACs) can be positioned on pachytene complements by fluorescence in situ hybridization (FISH) on homeologous chromosomes of related species. Here we present results of such a cross-species multicolor cytogenetic mapping of tomato BACs on potato chromosomes 6 and vice versa. The experiments were performed under low hybridization stringency, while blocking with Cot-100 was essential in suppressing excessive hybridization of repeat signals in both within-species FISH and cross-species FISH of tomato BACs. In the short arm we detected a large paracentric inversion that covers the whole euchromatin part with breakpoints close to the telomeric heterochromatin and at the border of the short arm pericentromere. The long arm BACs revealed no deviation in the colinearity between tomato and potato. Further comparison between tomato cultivars Cherry VFNT and Heinz 1706 revealed colinearity of the tested tomato BACs, whereas one of the six potato clones (RH98-856-18) showed minor putative rearrangements within the inversion. Our results present cross-species multicolor BAC–FISH as a unique tool for comparative genetic studies across Solanum species.
THE first cornerstone of the International Solanaceae Genome Project (SOL) launched in November 2003 is the sequencing of the euchromatin part of the tomato (Solanum lycopersicum) genome by an international consortium of 10 countries (http://www.sgn.cornell.edu/). In The Netherlands, the Centre for BioSystems Genomics (CBSG) is in charge of the sequencing of tomato chromosome 6. This chromosome contains various genes for economically important traits, including resistance genes for Oidium neolycopersici (Ol-4 and Ol-6), Cladosporium fulvum (Cf-2 and Cf-5), root-knot nematode, aphids and whitefly (Mi-1 and Mi-9), and the tomato yellow leaf curl virus (Ty-1, Ty-3, and Ty-4) (van Daelen et al. 1993; Weide et al. 1993; van Wordragen et al. 1994, 1996; Milligan et al. 1998; Ammiraju et al. 2003; Bai et al. 2004; Seah et al. 2004; Ji et al. 2007).
The Dutch tomato sequencing project of chromosome 6 follows the bacterial artificial chromosome (BAC) walking procedure or BAC-by-BAC approach, which involves the anchoring of a limited number of BAC clones (seed BACs) to the genome and further BAC contig building via BAC extension (Peters et al. 2006). For this approach, genetic mapping data, fluorescence in situ hybridization (FISH), and BAC sequencing analysis provide a framework, in which 84 anchored seed and extension BACs were positioned, covering a total of 2.4 Mb for the short arm and 10.2 Mb for the long arm (our unpublished data). Similarly, a BAC–FISH map has been published for tomato chromosome 1 showing the relation of the linkage map to pachytene chromosome structure (Chang et al. 2007). Moreover, a BAC–FISH map is being completed at http://www.sgn.cornell.edu/cview/ to illustrate the orders and locations of tomato BACs on pachytene chromosomes. Such a full set of BACs forms a chromosomal scaffold along the chromosome and can be used to compare chromosomal colinearity between related species and to unravel chromosomal rearrangements by cross-species BAC–FISH painting. Large-scale genomic changes involving chromosomal inversions and/or interchanges can be important for species isolation and might also contribute to the phenotypic differences between species through possible effects on gene structure or expression (Tanksley et al. 1992; Livingstone et al. 1999; Donganlar et al. 2002).
FISH allows the simultaneous localization of different target sequences on chromosomes, depending on the number of fluorochromes with different excitation and emission wavelengths and the use of combined binary ratio (COBRA) as well as related labeling technologies (Raap and Tanke 2006). For basic FISH only red and green fluorochromes for probe detection are used, together with 4′,6-diamidino-2-phenylindole (DAPI) for counterstaining of chromosomal DNA. Advanced multicolor FISH can involve up to 12 different fluorescent dyes together with DAPI as a counterstain in a single experiment (Muller et al. 2002). In tomato, five-color high-resolution FISH mapping has been successfully applied to process a large number of BACs on chromosome 6 (Szinay et al. 2008).
Cross-species FISH painting was first applied to mammalian chromosomes and human chromosome probes have now been hybridized to metaphases of >100 species (Rens et al. 2006b). In plants, it has been accomplished in Arabidopsis thaliana and related species of the Brassicaceae family (Lysak et al. 2003, 2005, 2006). In addition, the small genome of Sorghum has been used as a basis for integrating genetic and physical maps across grass genera with larger genomes (Draye et al. 2001; Koumbaris and Bass 2003).
Although the Solanaceae represents one of the best-studied and attractive plant systems for comparative genetics (Bonierbale et al. 1988; Tanksley et al. 1992; Grube et al. 2000; Donganlar et al. 2002; Fulton et al. 2002), applications of BAC–FISH for studying chromosomal evolutionary processes and chromosomal rearrangements have not been undertaken except for the very recent study by Iovene et al. (2008, accompanying article, this issue), due to the lack of defined BAC libraries. So far, genomewide colinearity within Solanaceae has been studied only with genetic maps. For example, comparative maps (Tanksley et al. 1992; Donganlar et al. 2002; Fulton et al. 2002) have revealed that tomato (S. lycopersicum) and potato (S. tuberosum) are differentiated by a series of paracentric inversions (inversions that do not involve the centromere) of chromosomes 5, 9, 10, 11, and 12 (Bonierbale et al. 1988; Grube et al. 2000). The inversion on chromosome 10 demonstrated that S. lycopersicoides and S. sitiens are colinear with S. tuberosum (Pertuze et al. 2002), suggesting that this inversion was fixed in the common ancestor of the tomato lineage. The limitations in such comparative genetic linkage mapping studies are that (1) mapping populations are needed, (2) deviations can occur between genetic and physical chromosome maps, and (3) the large pericentromere regions contain markers with low genetic resolution in mapping because of the absence of crossovers. The latter two limitations were encountered in the tomato genome sequencing project, in which BACs were selected on the basis of genetic markers and the physical positions of these BACs were validated by FISH prior to sequencing. Discrepancies have been observed between the actual chromosomal positions of some of these BACs and the positions of their corresponding markers on the genetic map (Chang et al. 2007 and our unpublished data). This was most notable in the repeat-rich domains in highly condensed pericentromere heterochromatin where crossovers were almost absent (Sherman and Stack 1995).
With the aims to study chromosomal colinearity between tomato and potato, we developed a cross-species multicolor BAC–FISH technique for the Solanum species (Szinay et al. 2008). By applying this technique, we painted tomato BACs of chromosome 6 on potato chromosomes and vice versa and discovered a new paracentric inversion in the short arm euchromatin. Our results show that the cross-species multicolor FISH strategy provides a powerful tool with the potential application for comparative genetics in the genus Solanum.
MATERIALS AND METHODS
Plant materials and BAC clones:
For preparing cell spread preparations we used anthers of the tomato (S. lycopersicum) cultivars Cherry VFNT (LA1221) and Heinz 1706, the potato (S. tuberosum) diploid genotype G254, and five diploid potato clones, RH88-025-50, RH98-856-18, RH90-038-21, RH97-654-15, and CD1015. Detailed genetic background of these plant materials is presented in Table 1. For tomato, 25 tomato BACs were included in this study (Table 2). At the time of this study only 6 potato BACs were available for the short arm (Table 2).
TABLE 1.
Plant materials used in this study
| Genotype | Genetic background | |
|---|---|---|
| Tomato | Cherry VFNT (LA1221) | Solanum lycopersicum with introgressed S. peruvianum |
| Heinz 1706 | S. lycopersicum | |
| Potato | G254 | Diploid Gineke |
| RH88-025-50 | F1S. tuberosum × S. phureja | |
| RH98-856-18 | F1S. sparsipilum × S. tuberosum | |
| RH90-038-21 | BC1 (S. tuberosum × S. microdontum) × S. tuberosum | |
| RH97-654-15 | F1S. tuberosum × S. spegazzinii | |
| CD1015 | (S. phureja × S. tuberosum) × (S. tuberosum × (S. phureja × S. tuberosum)) |
TABLE 2.
Overview of the tomato and potato chromosome 6 BACs used in this study
| BACa | Genetic map positionb (cM) | Molecular markers | BAC size (kb) | Chromosome position by FISHd | |
|---|---|---|---|---|---|
| Tomato | H107A05 | 3 | T1188 | 166 | 6S/EU |
| H054K13 | 3 | T1182 | 160 | 6S/EU | |
| H153O03 | 5 | T1198 | NAc | 6S/EU | |
| H251G05 | 5 | T1198 | 98 | 6S/EU | |
| H112G05 | 5.5 | Mi | 91 | 6S/EU | |
| H073H07 | 5 | Mi | 82 | 6S/EU | |
| H250I21 | 5.5 | Mi | 148 | 6S/EU | |
| H024L21 | 6.5 | TG436/SSR47 | 75 | 6S/EU | |
| H288L16 | 10 | cLET-2-H1 | 112 | 6S/EU-PC | |
| H304P16 | 10 | cLET-2-H1 | 124 | 6S/EU-PC | |
| H309K01 | 10 | cLET-5-A4 | 102 | 6L/PC | |
| H295L11 | 10 | T0244 | 110 | 6L/PC | |
| H003K02 | 10 | TG178 | 110 | 6L/PC | |
| H242H19 | 12 | T1063 | 98.2 | 6S/PC | |
| H023B17 | 25 | FER | 111.9 | 6L/PC | |
| H261A18 | 28 | cLET-4-G2 | 106 | 6L/EU | |
| H059K09 | 41.3 | NA | NA | 6S/PC | |
| H106K23 | 44 | C2_At4g10030 | 89 | 6L/EU | |
| H194N16 | 45 | cLET-5-C8 | 93 | 6L/EU | |
| H176D13 | 45.6 | NA | NA | 6S/PC | |
| H026E06 | 47 | P27 | 130.3 | 6L/EU | |
| H097D13 | 47.7 | NA | NA | 6S/PC | |
| H012O10 | 48 | C2_At1g73885 | 80 | 6L/EU | |
| H309D09 | 50 | TG365 | 142 | 6L/EU | |
| H060A01 | 101 | Ct_At1g20050 | 168 | 6L/EU | |
| Potato | 112M11 | NAc | NA | NA | 6S/EU |
| RH026H24 | 1.6 | EACAMAGG_94 | NA | 6S/EU | |
| 67P23 | NA | CT119 | NA | 6S/EU | |
| RH034P18 | 7 | EACCMACT_286 | NA | 6S/EU | |
| RH069B12 | 10.7 | EAACMCCT_377 | NA | 6S/PC | |
| RH084A13 | 12.2 | EAGAMAGG_152 | NA | 6S/PC | |
| EACGMCTA_215 |
All tomato BACs are from the Heinz 1706 HindIII library; the four RH potato BACs are from the RHPOTKEY BAC library; the other two potato BACs were kindly donated by Edwin A. G. van der Vossen (van der Vossen et al. 2005).
The tomato map position was adopted from the tomato-EXPEN 1992 map (Tanksley et al. 1992); the potato map position was adopted from the ultradense RH genetic map (van Os et al. 2006).
Not available.
FISH, fluorescence in situ hybridization; S, short arm; L, long arm; PC, pericentromere heterochromatin; EU, euchromatin; Cent, centromere; EU-PC, border between euchromatin and pericentromere heterochromatin.
Cot-100 DNA:
Cot-100 fractions of tomato genomic DNA were prepared according to Zwick et al. (1997) with some modifications. Total genomic DNA was isolated and sonicated to a fragment size of ∼1 kb. The fragmented DNA (0.5 μg/μl) was denatured in 0.3 m NaCl at 95° for 10 min and then allowed to reanneal at 65° for 37 hr 40 min. The remaining single-strand DNA (ssDNA) was digested with S1 endonuclease (Fermentas, final concentration 1 unit/μg) for 90 min at 37°. The reaction was stopped and extracted by adding 300 μl chloroform:isoamylalcohol (24:1). Then the DNA solution layer was transferred to a new tube, 2.5 vol of ice-cold absolute alcohol was added to precipitate DNA, and the dry pellet was resuspended in 20 μl HB50 (pH 8.0).
FISH:
Pachytene chromosome preparations were made as described by Zhong et al. (1996a) with few minor modifications. BAC DNA was isolated using a standard alkaline extraction and labeled by standard digoxigenin or biotin nick translation mix according to the instructions of the manufacturer (Roche Diagnostics, Indianapolis). Two-color FISH of BAC clones to pachytene chromosomes was performed according to the FISH protocols (Zhong et al. 1996b). Probes labeled with digoxigenin-dUTP, which were detected by digoxigenin-FITC, gave the green color, biotin-dUTP-labeled probes, detected by Avidin-Tex-Red showed the red color, and streptavidin-Cy5 showed the purple color. For direct labeling in multicolor FISH, five fluorescent nucleotides were used. They are fluorescein-12-dUTP (FITC), Cy3-dUTP, Cy3.5-dCTP, Cy5-dUTP, and diethylaminocoumarin-5-dUTP (DEAC). Cy5 was also used in an indirect labeling with biotin-dUTP-streptavidin-Cy5 detection (see below). The labeling methods followed the protocols of Amersham Bioscience (GE Healthcare, Sweden).
Cross-species was adapted with some minor modifications following the published protocol for cross-species chromosome painting (Rens et al. 2006a). For those BACs inside heterochromatin, 2 μg (100× probe concentration) of Cot-100 DNA are sufficient for blocking if 20 ng of a BAC probe are used per slide. Slides were examined under a Zeiss Axioplan 2 imaging photomicroscope equipped with epifluorescence illumination and filter sets for DAPI, FITC, Cy3, Cy5, DEAC, and Cy3.5 fluorescence. Selected images were captured by a Photometrics Sensys 1305 × 1024-pixel CCD camera. Image processing and thresholding was performed with the Genus Image Analysis Workstation software (Applied Imaging). DAPI images were separately sharpened with a 7 × 7 Hi-Gauss high-pass spatial filter to accentuate minor details and heterochromatin differentiation of the chromosomes. The different FISH signals were captured consecutively by double or multiple exposures and combined in a multichannel mode. Fluorescence images were displayed in dark gray for DAPI and pseudocolored for the other colors. Further brightness and contrast improvement were done on the whole image in Adobe Photoshop. We used ImageJ (http://rsb.info.nih.gov/ij) for measurements and for straightening of the chromosomes (plug in of Kocsis et al. 1991).
RESULTS
Chromosome 6 of tomato and potato at pachytene stage:
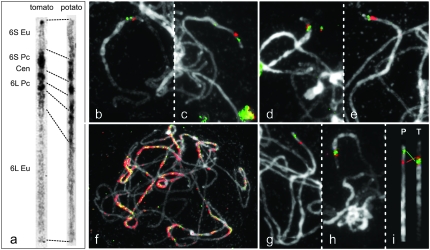
We first compared the morphology of the DAPI-stained pachytene chromosomes 6 of tomato and potato. Figure 1a displays converted black-and-white images of these chromosomes, which were straightened and stretched to equal length and slightly sharpened for better heterochromatin differentiation. The tomato chromosome 6 has an asymmetric centromere position and characteristic heterochromatin blocks in the long and short arms (Figure 1a). In addition, the short arm has the shortest euchromatin region in the complement, constituting ∼4.1 Mb of euchromatin (Chang et al. 2008). Potato chromosome 6 has a submedian centromere and its diagnostic heterochromatin blocks are less condensed than those of tomato (Figure 1a). The borders between euchromatin and heterochromatin are also gradual in the short and long arms of the potato chromosome. Besides, many tiny chromomeres in the euchromatin were observed. The short arm has a small distal knob that was seen in most chromosomes (Figure 1, c and d), but sometimes was absent in the straightened chromosome of potato (Figure 1a). A second small knob just below the distal knob in the short arm euchromatin of potato, which was described as a diagnostic heterochromatic knob for chromosome 6 by Ramanna and Wagenvoort (1976), was not visible here. Conceivably, the knob is polymorphic and not visible in the potato clones that we used for our work, or it could not be detected in our DAPI-stained preparations. Similarly, we did not find a knob on the long arm of potato chromosome 6 as reported by Iovene et al. (2008).
Figure 1.—
(a) Chromosomes 6 of tomato and potato at the pachytene stage. S, short arm; L, long arm; Eu, euchromatin; Pc, pericentromere heterochromatin; Cen, centromere. (b) FISH of the tomato BACs H153O03 (red) and H073H07 (green) on tomato pachytene chromosome 6. (c) Cross-species FISH of the same BACs on potato chromosome 6. (d) FISH of H112G05 (red) and H24L21 (green) on tomato chromosome 6. (e) Cross-species FISH of the same BACs on potato chromosome 6. (f) Cross-species FISH of the tomato BACs H003K02 (green) and H309K01 (red) on potato chromosomes without Cot-100 blocking. (g and h) FISH of the potato BACs 67P23 (red) and 112M11 (green) on potato RH98-856-18 chromosome 6 (g) and tomato chromosome 6 (h). (i) The straightened part of chromosome 6 of potato (P) and tomato (T), showing the orientation and relative distance of the two potato BACs 67P23 (red) and 112M11 (green).
Selection of BACs and cytogenetic maps for tomato and potato BACs on chromosome 6:
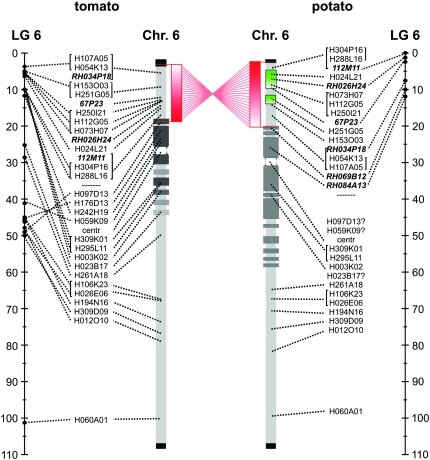
We focused on the regions of tomato chromosome 6 that are rich for resistance genes, as preliminary comparative mapping studies within the Solanaceae genera showed that resistance genes occurred at syntenic positions in cross-generic gene clusters more frequently than expected by chance (Grube et al. 2000). Moreover, we selected potato BACs that are in the chromosomal region for which previously published data on genetic colinearity were controversial or doubtful, for example, the chromosomal region where the Mi-1 gene is located (Tanksley et al. 1992; van Wordragen et al. 1994; van der Vossen et al. 2005). For tomato we selected 14 BACs for the short arm (6S) and 11 for the long arm (6L) (Table 2 and Figure 3), on the basis of known positions on the chromosome maps (our unpublished data). The physical positions of the BACs in the 6S euchromatin were in agreement with their relative orders on the genetic map, except for H153O03 that was genetically mapped at 5 cM but was located closer to the distal telomere knob of tomato 6S than other BACs that had the same genetic positions of 5 cM (Figure 1b). BACs H250I21, H112G05, and H073H07, which had been mapped genetically around 5 cM and assembled in the BAC contig containing the Mi-1 gene, were partly overlapped in the euchromatin region of pachytene chromosome 6 [Figures 2b (T) and 3]. Although the signal of H073H07 showed a single focus in most of the pachytene complements, we also observed cases of clear double signals (Figure 1b). BACs H107A05 and H054K13 overlapped by FISH mapping and were identified as the most distal BACs in the euchromatin region so far [Figures 2, a and c (T), and 3 (tomato)]. BACs H288L16 and H304P16 colocalized at the border of 6S euchromatin and heterochromatin, which were shown to be the most proximal BACs on 6S euchromatin [Figures 2, a and c (T), and 3 (tomato)]. In the heterochromatin region of tomato 6S, four BACs were selected. Three of them (H097D13, H176D13, and H059K09) have been genetically mapped on the long arm and the fourth one (H242H19) mapped around the centromere (Table 2 and Figure 3). On the tomato 6L, we positioned four BACs in the pericentromere heterochromatin and seven in the euchromatin. The pericentromere BACs, except H023B17, harbor markers that are genetically located at the same locus as the 6S BACs H288L16 and H304P16. Clearly, discrepancies between genetic positions and relative physical positions exist, especially when the candidate BACs come from chromosomal regions near or in heterochromatin (Figure 3). For potato, only six BACs were available for the short arm (Table 2). BAC 112M11 was located in the very distal and RH034P18 was in the proximal euchromatic region of the short arm of potato [Figures 1g and 2d (P)]. The cytogenetic and genetic map orders of all the potato BACs were in agreement (Figure 3).
Figure 3.—
A comparison of genetic and physical maps for tomato BACs and cross-species FISH of tomato and potato BACs on pachytene chromosomes. The schematic drawings of the chromosomes are based on pachytene morphology. Black and dark gray blocks are heterochromatin regions; the dark blocks represent the dense brightly fluorescing heterochromatin regions, whereas the lighter regions are lighter and more variable; the white blocks are the centromeres. The BACs are positioned in sequence of FISH position. Brackets on the left of the BACs have the same genetic map positions; brackets on the right have overlapping FISH signals on the chromosome. The dotted lines show the position of their markers on the genetic map. BACs in bold italics are the potato BACs. The red bar between the tomato and potato chromosomes represents the short arm paracentric inversion; the green block indicates the positions of minor chromosome rearrangements between potato clone RH98-856-18 and the other five remaining potato lines. The question marks point at weak, variable, or no signals of the tomato BACs on potato.
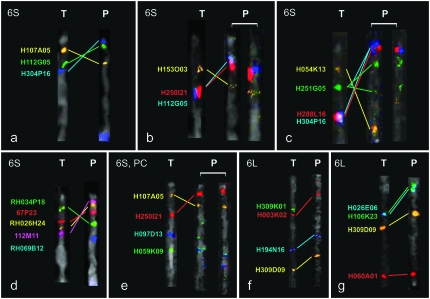
Figure 2.—
Examples of FISH and cross-species FISH of tomato BACs on pachytene chromosome 6 of tomato (T) and potato (P). The chromosome regions of interest were straightened and oriented with the signals close to the short arm telomere upward. (a) FISH of H107A05 (orange), H112G05 (green), and H304P16 (blue) on the short arms of tomato and potato showed a clear inverted arrangement of the BAC signals. (b) FISH of H153O03 (orange), H250I21 (red), and H112G05 (blue) showed an inverted order between the homeologs. (c) FISH of H054K13 (orange), H251G05 (green), H288L16 (red), and H304P16 (blue) showed an inverted order between the homeologs. Notably, here we used potato clone RH98-856-18, and the H251G05 (green) BAC produced a large and a small focus on the potato chromosome, suggesting a breakpoint in this BAC for a putative chromosomal rearrangement. (d) FISH of RH034P18 (green), 67P23 (red), RH026H24 (orange), 112M11 (pink), and RH69B12 (blue) showed an inverted order between the homeologs; RH069B12 did not give a signal on tomato. (e) FISH of H107A05 (orange), H250I21 (red), H097D13 (blue), and H059K09 (green) on the short arm. The two pericentromere heterochromatin BACs H097D13 (blue) and H059K09 (green) showed weak and variable foci on the potato short arm. (f) FISH of H309K01 (green), H003K02 (red), H194N16 (blue), and H309D09 (orange) on the long arm showed the same order of the BACs except H309K01 (green) hybridization that gave no signal in potato. (g) FISH of H026E06 (blue), H106K23 (green), H309D09 (orange), and H060A01 (red) on the long arm showed the same order on the tomato and potato chromosomes.
Within- and cross-species BAC–FISH in tomato and potato:
A crucial factor for cross-species FISH is the posthybridization controlling stringency. When the same washing stringency (50% formamide, 2× SSC at 42° for 15 min) as within-species FISH was used, the signals were found to be not highly specific or reproducible (data not shown). To enhance hybridization efficiency, posthybridization washes under conditions of low stringency were carried out for 3 × 5 min in 20% formamide, 2× SSC at 42°. With this stringency, nearly every tested BAC could be painted across species.
FISH signal intensity depends on the chromosomal target size and repeat content. The tomato BAC clones in this study have genomic inserts of 75–168 kb (Table 1), which are large enough to produce bright fluorescent foci on the pachytene chromosomes. However, BAC clones harboring high amounts of tandem and dispersed sequences produce abundant fluorescence signals over all chromosomes, mostly in the pericentromeric regions. We therefore used Cot-100 to suppress hybridization by the highly and middle repetitive DNA sequences of the BAC probes when the BACs were hybridized to chromosome targets. Examples of such tomato BACs FISH with Cot-100 are shown in Figures 1, b and d, and 2, a–c (T) and e–g (T). However, the potato BACs in this study produced specific foci even in the absence of Cot-DNA, suggesting a very low repeat content in these BACs [Figures 1g and 2d (P)].
Initial cross-species FISH with tomato BACs on potato chromosomes at a low hybridization stringency and without Cot-100 blocking demonstrated excessive cross-hybridization signals (Figure 1f). Then repeat signals could effectively be suppressed in the presence of tomato Cot-100 to paint tomato BACs on potato chromosomes [Figures 1, c and e, and 2, a–c (P) and e–g (P)]. To hybridize the potato BACs on tomato chromosomes, potato Cot-100 was still not needed to produce clear signals [Figures 1h and 2d (T)].
Colinearity and rearrangements of BACs between tomato and potato chromosomes 6:
Upon mapping of several 6S euchromatin tomato BACs to potato, we found that the orders of any two BACs were inverted between tomato and potato (Figure 1, b–c and d–e). The most distal tomato 6S BAC H107A05 (or H054K13) flipped to the proximal euchromatin of potato 6S, whereas H304P16 (or H288L16), a tomato 6S euchromatin/heterochromatin boundary BAC, mapped at the most distal region on potato 6S nearly covering the telomere knob [Figure 2, a (P) and c (P)]. Similarly, painting the four potato 6S euchromatin BACs on tomato (Figure 2d) also showed an inverted order of these BACs between tomato and potato. The most distal potato BAC 112M11 was clearly positioned at the border of euchromatin and heterochromatin on tomato 6S [Figures 1h and 2d (T)]. Further FISH mapping of 112M11 together with H304P16 (or H288L16) demonstrated that they colocalized. In addition, H107A05 (or H054K13) showed an overlap with RH034P18 (Figure 3). These results suggest that an inversion involving the whole 6S euchromatin and possibly even involving the telomere exists between tomato and potato.
To further investigate whether the 6S inversion involved also the pericentromere heterochromatin, we analyzed 4 tomato and 2 potato BACs from the pericentromere region. BACs H097D13 and H059K09, which produced single foci on the tomato chromosome 6, appeared in two or more copies on the potato homeolog with weak and not reproducible signals in separate experiments (Figure 2e). Moreover, 2 more tomato and 2 potato pericentromere BACs gave no signals on their potato and tomato homeologs, respectively (Figure 2d). As for the long arm, we observed that the 11 used tomato BACs mapped to tomato and potato at comparable positions, with the exception of H023B17, a pericentromere heterochromatin BAC of 6L, which gave no signal in potato. This reflects that the long arm of chromosome 6 is well conserved between these two species (Figures 2, f and g, and 3).
Sequence feature of BACs in the heterochromatin region:
Of the seven BACs that could not be efficiently painted cross-species, three have been sequenced. These are the tomato BACs H023B17 (6L) and H242H19 (6S) and potato BAC RH069B12 (6S). For the long arm, sequence analysis of H023B17 revealed a lower gene content compared to BACs (H309K01, H295L11, and H003K02) that did paint the heterochromatin of both tomato and potato 6L. H023B17 contains only one putative gene, covering 1.8% of the BAC sequence. In contrast, there are four putative genes (16.3%) in H309K01, three (11.4%) in H295L11, and two (6.8%) in H003K02. Since Cot-100 was used to block the repeat sequences in painting the tomato BACs on potato, a high repeat content in H023B17 is likely the cause of the loss of its signal in potato. Alternatively, this might be caused by the lack of homolog sequences from this BAC in the corresponding potato region.
In the 6S heterochromatin, potato BAC RH069B12 and tomato BAC H242H19 were found to be highly repetitive. The majority of repeats in these two BACs were similar to the Gypsy-type GYPSODE1_I retrotransposon. The putative gene content of BACs RH069B12 and H242H19 was low (3.1 and 3.2% of the BAC sequence length, respectively). Since Cot-100 was not applied to paint potato BACs on tomato, it seems that gene content and homology as well as repeat sequences played a role in the failure of cross-species FISH of BACs in the 6S heterochromatin region. Alternatively, this region could be involved in the 6S inversion that has led to chromosomal rearrangements and/or loss of chromosome fragments.
Colinearity of chromosome 6 BACs within tomato and potato:
Most of our tomato FISH work was done with the Cherry VFNT (LA1221) cultivar containing an introgression of S. peruvianum in 6S; while the BACs were from the Heinz 1706 cultivar that does not have this introgression. To verify the colinearity of the chromosome 6 BACs between VFNT and Heinz 1706, we used 26 tomato BACs covering the whole linkage group from 0 to 101 cM (supplemental table). The results showed that the order of the BACs on chromosome 6 of Cherry tomato is the same as that of the Heinz 1706 (data not shown), demonstrating that the S. peruvianum introgression does not contain large-scale chromosomal rearrangements.
As to the question of colinearity of the selected BACs within potato, we compared the potato (S. tuberosum) diploid genotype G254 with the diploid clones RH88-025-50, RH98-856-18, RH90-038-21, RH97-654-15, and CD1015 (Table 1). In general, all BACs that we used for the comparison displayed comparable positions on the short arms of chromosome 6. However, clone RH98-856-18 containing an introgression of S. sparsipilum showed a striking difference in the relative distance between the potato BACs 67P23 and 112M11 covering the Mi region in the inversion between tomato and potato (Figure 1, g–i). Measurements of the BAC distances in five pachytene complements demonstrated that the distance between the two potato BACs is about one-third shorter in tomato than in potato clone RH98-856-18, while such differences do not exist with the other potato clones. We also observed that tomato BAC H251G05 produces two signals in this potato clone (Figure 2c) in contrast to one signal in the other clones. Both observations suggest the existence of a second nested inversion or other minor rearrangement in the middle of the short arm of RH98-856-18 with probably one breakpoint in the chromosomal target area of BAC H251G05.
DISCUSSION
Cross-species multicolor BAC–FISH—a powerful tool for comparative genomics across Solanum:
Cross-species BAC–FISH was previously applied to Arabidopsis and related Brassicaceae species for demonstrating chromosomal evolutionary processes and rearrangements (Fransz et al. 2000; Jackson et al. 2000; Lysak et al. 2005, 2006, 2007). In Solanum species chromosomal rearrangements have not been cytologically studied so far. Within the scope of the ongoing tomato and potato sequencing projects, chromosome-specific BACs have been obtained by genetic and physical mapping and by contig construction. In this study, we present a multicolor cross-species BAC–FISH painting for directly displaying synteny between related Solanum species on the chromosomal level. It facilitates the simultaneous detection of more than two BACs in one pachytene preparation and enables colinearity studies on BACs while avoiding laborious reprobing of FISH experiments. We applied this technology successfully to paint a set of tomato and potato BACs onto chromosome 6 of potato and tomato. These experiments revealed both agreements and discrepancies between genetic and physical locations of tomato BACs, the paracentric inversion on 6S, and the colinearity of BACs on 6L between tomato and potato as well as some minor rearrangements in one of the potato lines. Our results show that cross-species multicolor BAC–FISH is a powerful tool to connect genetic and physical maps. This tool can be used for comparative genetic and evolution studies to reveal genome colinearity between tomato and potato and most likely also among different genomes across the Solanum species. Without using mapping populations, high-density BAC maps can be readily obtained for many species and accessions within one species.
Structural chromosome rearrangements may exist among different species (interspecific) as well as within accessions of the same species (intraspecific). Examples are the 7S paracentric inversion between a distant wild relative of tomato S. pennellii and S. esculentum/S. pimpinellifolium (van der Knaap et al. 2004) and an inversion of the two clusters of Mi-1 homologs between S. esculentum and S. peruvianum (Seah et al. 2004). The possible minor chromosomal rearrangements in one of the potato lines used in this study and the absence in our potato lines of an interstitial heterochromatin knob of 6L as described in Iovene et al. (2008) indicate the existence of intraspecific variation. The extent of such rearrangements may parallel morphological diversity or taxonomical groupings and would contribute to our understanding of the importance of this type of mutation in the evolution of fertility barriers in the whole Solanaceae family (Perez et al. 1999). Currently, we have ongoing projects to construct such a BAC-synteny map for potato and tomato (including several related wild species). This BAC-synteny map will enable us to identify chromosomal linearity/rearrangement(s) between tomato and potato as well as among different wild species. The latter will help to identify chromosomal regions showing deviation and to provide a basis for breeding strategies to introgress genes from wild Solanum species into cultivated crops. Furthermore, this BAC map can be used to study chromosomal evolutionary processes within Solanum at a variety of taxonomic levels and to understand biodiversity with genomics data.
The 6S inversion encompasses a hotspot of resistance genes:
Initial macrosynteny studies of genetic linkage maps between tomato and potato did until now not clearly show an inversion in the 6S chromosome arm, although the marker order between GP164 and GP79 was reported to be inverted (Tanksley et al. 1992; van Wordragen et al. 1994). Recently, the Rpi-blb2 gene conferring late blight resistance in potato was mapped as a Mi-1 gene homolog on 6S of potato (van der Vossen et al. 2005). Both Mi-1 and Rpi-blb2 are tightly linked to a common RFLP marker CT119, with Mi-1 proximal to CT119 in tomato and Rpi-blb2 distal to CT119 in potato (van der Vossen et al. 2005), indicating that a hidden inversion may exist between tomato and potato. In this study, by using cross-species multicolor BAC–FISH analysis, we provide firm evidence for a conspicuous paracentric inversion between tomato and potato covering the entire short arm of chromosome 6.
In general, inversions do not change the phenotype of the individual unless a breakpoint of the inversion lies within the regulatory or structural region of a gene. However, with the reshuffling of gene order within chromosome arms, the changed context may also affect the functions of genes involved in the inversion to some extent (Hoffmann et al. 2004). The paracentric inversion of 6S that differentiates tomato and potato probably has moved genetic loci from regions of low recombination (e.g., centromeres) to regions of higher recombination (and vice versa) and therefore has changed the evolutionary perspective for those loci. Interestingly, in tomato, 6S is a chromosomal region where many resistance (R) genes reside. This R gene hotspot contains the Cf-2/Cf-5, Ol-4/Ol-6, Mi-1/Mi-9, and Ty-1 genes, which confer resistance to several unrelated pathogens. R genes in plants are most frequently members of multigene families and locate in tandem arrays, like the Mi or Cf genes. Inter- and intragenic recombination at R gene loci has been described extensively and is thought to be a major mechanism for generating novel resistance specificities (reviewed in Hulbert et al. 2001). Previously, the physical position of the Mi-1 gene and its six homologs was mapped also using FISH at the border of euchromatin and heterochromatin regions of tomato chromosome 6S (Zhong et al. 1999). Suppression of recombination frequency is apparent in the Mi-1 gene region in tomato. However, the Rpi-blb2 gene in potato is mapped on the euchromatin of potato 6S and resides in a gene cluster that is twice as big as the Mi-1 gene cluster in tomato (van der Vossen et al. 2005). We speculate that the 6S inversion and observed expansion of the locus reflect the opposite evolutionary potentials of the interacting pathogens. Root-knot nematodes have little potential for gene flow and thus exert little evolutionary pressure on the host to generate new specificities. Phytophthora infestans on the other hand is a high-risk pathogen with both sexual and asexual reproduction systems, resulting in a dynamic spread of genetic variation (McDonald and Linde 2002). Availability of more genome sequences of both tomato and potato in the near future will allow us to study the sequence composition near the breakpoints, possibly by virtue of cross-species microarray painting (Ferguson-Smith et al. 2005). This will shed light on the molecular mechanisms underlying chromosome rearrangements in plant genomes and will thus form a basis for investigating mechanisms of gene regulation, evolution, signaling, disease resistance, and defense, the phenotypic diversity and comparative biology in the Solanaceae.
The molecular nature of the chromosomal inversion:
Studies of chromosomal inversions were pioneered in Drosophila >60 years ago (Sturtevant 1919; Dobzhansky 1970). Of the two types of inversions, paracentric and pericentric inversion, it has been assumed that the former affects fitness less and thus may be the most likely form of chromosomal rearrangements to survive through evolutionary time. Although the number of BACs was insufficient to establish the precise breakpoints of the inversion, the 6S inversion discovered in this study is most likely a paracentric inversion covering the entire euchromatin region and possibly involves the telomere. This can be deduced from the physical position of potato BAC 112M11 and tomato BAC H304P16 (or H288L16). These three BACs hybridized to positions close to the border of euchromatin and heterochromatin of tomato 6S, while on potato 6S, they were found at the most distal region of the short arm euchromatin and nearly covering the telomere (Figure 2, a, c, and d). The putative proximal breakpoint of this inversion likely occurs in the short arm pericentromeric heterochromatin as tomato/potato BACs in these regions produced weak and variable signals or no signals at all in the cross-species FISH. Evidence on chromosome breakpoints in these regions came from Khush and Rick (1963, 1968) and Liharska et al. (1997) on radiation-induced deletion mapping studies.
Whether the inversion involves the most distal short arm euchromatin and heterochromatin block is still to be confirmed as BACs or sequence data between the most distal BACs (potato BAC 112M11 or tomato BAC H304P16/H288L16) and the subtelomere repeats (Figure 2, a, c, and d) are lacking. Although telomere sequences have been shown in (peri)centromere regions of tomato and potato (Ganal et al. 1991; Presting et al. 1996; Tek and Jiang 2004; our unpublished data), it is not clear as to whether these telomere sequences resulted from inversion events. So, the 6S inversion between tomato and potato may be explained in two ways, either as the result of a single break in the pericentromere where the broken end formed a new telomere de novo (Blackburn 1991; Werner et al. 1992) or through the simultaneous incidence of breaks in the distal part of the short arm euchromatin and in the pericentromere itself. Further, it has to be proved in material showing the subdistal heterochromatic knob as mentioned by Ramanna and Wagenvoort (1976) whether this knob represents one of the breakpoints of the inversion, as is the case with the short arm heterochromatic knob in Arabidopsis representing a paracentric inversion between the accessions Col and WS and the knobless accessions Ler, C24, Zh, and NoO (Fransz et al. 2000; Lysak et al. 2002).
Acknowledgments
This research was done in the framework of the European Union-Solanaceae Project PL 016214 (financed by the European Commission), of the Centre for BioSystems Genomics that is part of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research, and of the Dutch Potato Genome Sequencing Consortium projects (funded by Netherlands Genomics Initiative and Ministry of LnV via the Fonds Economische Structuurverstersterking).
References
- Ammiraju, J. S. S., J. C. Veremis, X. Huang, P. A. Roberts and I. Kaloshian, 2003. The heat-stable root-nematode resistance gene Mi-9 from Lycopersicon peruvianum is localized on the short arm of chromosome 6. Theor. Appl. Genet. 106 478–484. [DOI] [PubMed] [Google Scholar]
- Bai, Y., R. van der Hulst, C. C. Huang, L. Wei, P. Stam et al., 2004. Mapping Ol-4, a gene conferring resistance to Oidium neolycopersici and originating from Lycopersicon peruvianum LA2172, requires multi-allelic, single-locus markers. Theor. Appl. Genet. 109 1215–1223. [DOI] [PubMed] [Google Scholar]
- Blackburn, E. H., 1991. Structure and function of telomeres. Nature 350 569–573. [DOI] [PubMed] [Google Scholar]
- Bonierbale, M., R. L. Plaisted and S. D. Tanksley, 1988. RFLP maps of potato and tomato based on a common set of clones reveal modes of chromosomal evolution. Genetics 120 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S. B., L. K. Anderson, J. D. Sherman, S. M. Royer and S. M. Stack, 2007. Predicting and testing physical locations of genetically mapped loci on tomato pachytene chromosome 1. Genetics 176 2131–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S. B., T. J. Yang, E. Datema, J. van Vugt, B. Vosman et al., 2008. FISH mapping and molecular organization of the major repetitive sequences of tomato. Chromosome Res. (in press). [DOI] [PubMed]
- Dobzhansky, T., 1970. Genetics of the Evolutionary Process. Columbia University Press, New York.
- Donganlar, S., A. Frary, M. C. Daunay, R. N. Lester and S. D. Tanksley, 2002. A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the Solanaceae. Genetics 161 1697–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draye, X., Y. R. Lin, X. Y. Qian, J. E. Bowers, G. B. Burow et al., 2001. Toward integration of comparative genetic, physical, diversity, and cytomolecular maps for grasses and grains, using the Sorghum genome as a foundation. Plant Physiol. 125 1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith, M. A., F. Yang, W. Rens and P. C. M. O. Brien, 2005. The impact of chromosome sorting and painting on the comparative analysis of primate genomes. Cytogenet. Genome Res. 108 112–121. [DOI] [PubMed] [Google Scholar]
- Fransz, P. F., S. Armstrong, J. H. De Jong, L. D. Parenell, C. van Drunen et al., 2000. Integrated cytogenetic map of chromosome arms 4S of A. thalinana: structural organization of heterochromatic knob and centromere region. Cell 100 367–376. [DOI] [PubMed] [Google Scholar]
- Fulton, T. M., R. van der Hoeven, N. T. Eanetta and S. D. Tanksley, 2002. Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14(7): 1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganal, M. W., N. L. V. Lapitan and S. D. Tanksley, 1991. Macrostructure of the tomato telomeres. Plant Cell 3 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grube, R. C., E. R. Radwanski and M. Jahn, 2000. Comparative genetics of disease resistance within the Solanaceae. Genetics 155 873–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A. A., C. M. Sgro and A. R. Weeks, 2004. Chromosomal inversion polymorphisms and adaptation. Trends Ecol. Evol. 19 482–488. [DOI] [PubMed] [Google Scholar]
- Hulbert, S. H., C. A. Webb, S. M. Smith and Q. Sun, 2001. Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. 39 285–312. [DOI] [PubMed] [Google Scholar]
- Iovene, M., S. M. Wielgus, P. W. Simon, C. R. Buell and J. Jiang, 2008. Chromatin structure and physical mapping of chromosome 6 of potato and comparative analyses with tomato. Genetics 180 1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S. C., Z. Cheng, M. Wang, H. M. Goodman and J. Jiang, 2000. Comparative fluorescence in situ hybridization mapping of a 431-kb Arabidopsis thaliana bacterial artificial chromosome contig reveals the role of chromosomal duplication in the expansion of the Brassica rapa genome. Genetics 156 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Y., D. J. Schuster and J. W. Scott, 2007. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol. Breed. 20 271–284. [Google Scholar]
- Khush, G. S., and C. M. Rick, 1963. Meiosis in hybrids between Lycopersicon esculentum and Solanum pennellii. Genetica 33 167–183. [Google Scholar]
- Khush, G. S., and C. M. Rick, 1968. Cytogenetic analysis of the tomato genome by means of induced deficiencies. Chromosoma 23 452–484. [Google Scholar]
- Kocsis, E., B. L. Trus, C. J. Steer, M. E. Bisher and A. C. Steven, 1991. Image averaging of flexible fibrous macromolecules: the clathrin triskelion has an elastic proximal segment. J. Struct. Biol. 107 6–14. [DOI] [PubMed] [Google Scholar]
- Koumbaris, G. L., and H. W. Bass, 2003. A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected Sorghum (S. propinquum L) BAC clones. Plant J. 35 647–659. [DOI] [PubMed] [Google Scholar]
- Liharska, T. B., J. Hontelez, A. Kammen, P. Zabel and M. Koornneef, 1997. Molecular mapping around the centromere of tomato chromosome 6 using irradiation-induced deletions. Theor. Appl. Genet. 95 969–974. [Google Scholar]
- Livingstone, K. D., V. K. Lackney, J. R. Blauth, R. van Wijk and M. K. Jahn, 1999. Genome mapping in Capsicum and the evolution of genome structure in the Solanaceae. Genetics 152 1183–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak, M. A., P. F. Fransz, H. B. M. Ali and I. Schubert, 2002. Chromosome painting in Arabidopsis thaliana. Plant J. 28 689–697. [DOI] [PubMed] [Google Scholar]
- Lysak, M. A., A. Pecinka and I. Schuert, 2003. Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Res. 11 195–204. [DOI] [PubMed] [Google Scholar]
- Lysak, M. A., M. A. Koch, A. Pecinka and I. Schuert, 2005. Chromosome triplication found across the tribe Brassiceae. Genome Res. 15 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak, M. A., A. Berr, A. Pecinka, R. Schmidt, K. Mcbreen et al., 2006. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 103(13): 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak, M. A., K. Cheung, M. Kitschke and P. Bures, 2007. Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol. 145 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, B. A., and C. Linde, 2002. The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 124(2): 163–180. [Google Scholar]
- Milligan, S. B., J. Bodeau, J. Yaghoobi, I. Kaloshian, P. Zabel et al., 1998. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, S., M. Neusser and J. Wienberg, 2002. Towards unlimited colors for fluorescence in-situ hybridization (FISH). Chromosome Res. 10 223–232. [DOI] [PubMed] [Google Scholar]
- Perez, F., A. Menendez, P. Dehal and C. F. Quiros, 1999. Genomic structural differentiation in Solanum: comparative mapping of the A- and E-genomes. Theor. Appl. Genet. 98 1183–1193. [Google Scholar]
- Pertuze, R. A., Y. Ji and R. T. Chetelat, 2002. Comparative linkage map of the Solanum lycopersicoides and S. sitiens genomes and their differentiation from tomato. Genome 45 1003–1012. [DOI] [PubMed] [Google Scholar]
- Peters, S. A., J. C. van Haarst, T. P. Jesse, D. Woltinge, K. Jansen et al., 2006. TOPAAS, a tomato and potato assembly assistance system for selection and finishing of bacterial artificial chromosomes. Plant Physiol. 140 805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presting, G. G., A. Frary, K. Pillen and S. D. Tanksley, 1996. Telomere-homologous sequences occur near the centromeres of many tomato chromosomes. Mol. Gen. Genet. 251 526–531. [DOI] [PubMed] [Google Scholar]
- Raap, A. K., and H. J. Tanke, 2006. Combined binary ratio fluorescence in situ hybridiziation (COBRA-FISH): development and applications. Cytogenet. Genome Res. 114 222–226. [DOI] [PubMed] [Google Scholar]
- Ramanna, M. S., and M. Wagenvoort, 1976. Identification of the trisomic series in diploid Solanum tuberosum L., group tuberosum. I. Chromosome identification. Euphytica 25 233–240. [Google Scholar]
- Rens, W., B. Fu, P. C. M. O. Brien and M. A. Ferguson-Smith, 2006. a Cross-species chromosome painting. Nat. Protoc. 1(2): 783–790. [DOI] [PubMed] [Google Scholar]
- Rens, W., K. Moderegger, H. Skelton, O. Clarke, V. Trifonov et al., 2006. b A procedure for image enhancement in chromosome painting. Chromosome Res. 14 497–503. [DOI] [PubMed] [Google Scholar]
- Seah, S., J. Yaghoobi, M. Rossi, C. A. Gleason and V. M. Williamson, 2004. The nematode-resistance gene, Mi-1, is associated with an inverted chromosomal segment in susceptible compared to resistant tomato. Theor. Appl. Genet. 108 1635–1642. [DOI] [PubMed] [Google Scholar]
- Sherman, J. D., and S. M. Stack, 1995. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., 1919. Contributions to the genetics of Drosophila melanogaster. III. Inherited linkage variations in the second chromosome. Carnegie Inst. Wash. Publ. 278 305–341. [Google Scholar]
- Szinay, D., S. B. Chang, L. Khrustaleva, S. Peters, E. Schijlen et al., 2008. High-resolution chromosome mapping of BACs using multi-colour FISH and pooled-BAC FISH as a backbone for sequencing tomato chromosome 6. Plant J. (in press). [DOI] [PubMed]
- Tanksley, S. D., M. W. Ganal, J. P. Prince, M. C. De Vicente, M. W. Bonierbale et al., 1992. High density molecular linkage maps of the tomato and potato genomes. Genetics 132 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tek, A. L., and J. Jiang, 2004. The centromeric regions of potato chromosomes contain megabase-sized tandem arrays of telomere-similar sequence. Chromosoma 113 77–83. [DOI] [PubMed] [Google Scholar]
- van Daelen, R. A., F. Gerbens, F. van Ruissen, J. Aarts, J. Hontelez et al., 1993. Long-range physical maps of two loci (Aps-1 and GP79) flanking the root-knot nematode resistance gene (Mi) near the centromere of tomato chromosome 6. Plant Mol. Biol. 23 185–192. [DOI] [PubMed] [Google Scholar]
- van Der Knaap, E., A. Sanyal, S. A. Jackson and S. D. Tanksley, 2004. High-resolution fine mapping and fluorescence in situ hybridization analysis of sun, a locus controlling tomato fruit shape, reveals a region of the tomato genome prone to DNA rearrangements. Genetics 168 2127–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen, E., J. Gros, A. Sikkema, M. Muskens, D. Wouter et al., 2005. The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 44 208–222. [DOI] [PubMed] [Google Scholar]
- van Os, H., S. Andrzejewski, E. Bakker, I. Barrena and G. J. Bryan, 2006. Construction of a 10,000-marker ultradense genetic recombination map of potato: providing a framework for accelerated gene isolation and a genomewide physical map. Genetics 173 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wordragen, M. F., R. Weide, T. Liharska, A. van der Steen, M. Koornneef et al., 1994. Genetic and molecular organization of the short arm and pericentromeric region of tomato chromosome 6. Euphytica 79 169–174. [Google Scholar]
- van Wordragen, M. F., R. Weide, E. Coppoolse, M. Koornneef and P. Zabel, 1996. Tomato chromosome 6: a high resolution map of the long arm and construction of a composite integrated marker-order map. Theor. Appl. Genet. 92 1065–1072. [DOI] [PubMed] [Google Scholar]
- Weide, R., M. F. van Wordragen, R. K. Lankhorst, R. Verkerk, C. Hanhart et al., 1993. Integration of the classical and molecular linkage maps of tomato chromosome 6. Genetics 135 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, J. E., R. S. Kota, B. S. Gill and T. R. Endo, 1992. Distribution of telomeric repeats and their role in the healing of broken chromosome ends in wheat. Genome 35 844–848. [Google Scholar]
- Zhong, X. B., J. H. De Jong and P. Zabel, 1996. a Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridisation (FISH). Chromosome Res. 4 24–28. [DOI] [PubMed] [Google Scholar]
- Zhong, X. B., P. F. Fransz, J. W. van Eden, P. Zabel, A. van Kammen et al., 1996. b High resolution mapping by fluorescence in situ hybridisation to pachytene chromosomes and extended DNA fibres. Plant Mol. Biol. Rep. 14 232–242. [Google Scholar]
- Zhong, X. B., J. Bodeau, P. F. Fransz, V. M. Williamson, A. van Kammen et al., 1999. FISH to meiotic pachytene chromosomes of tomato locates the root knot nematode resistance gene Mi-1 and the acid phosphatase gene Aps-1 near the junction of euchromatin and pericentromeric heterochromatin of chromosome arms 6S and 6L, respectively. Theor. Appl. Genet. 98 365–370. [Google Scholar]
- Zwick, M. S., R. E. Hanson, T. D. Mcknight, M. N. Islam-Faridi, M. N. Stelly et al., 1997. A rapid procedure for the isolation of Cot-1 DNA from plants. Genome 40 138–142. [DOI] [PubMed] [Google Scholar]