Abstract
Homolog pairing is indispensable for the proper segregation of chromosomes in meiosis but the mechanism by which homologs uniquely pair with each other is poorly understood. In Drosophila, somatic chromosomes also undergo full homolog pairing by an unknown mechanism. It has been recently demonstrated that both insulator function and somatic long-distance interactions between Polycomb response elements (PREs) are stabilized by the RNAi machinery in Drosophila. This suggests the possibility that long-distance pairing interactions between homologs, either during meiosis or in the soma, may be stabilized by a similar mechanism. To test this hypothesis, we have characterized meiotic and early somatic chromosome pairing of homologous chromosomes in flies that are mutant for various components of the RNAi machinery. Despite the identification of a novel role for the piRNA machinery in meiotic progression and synaptonemal complex (SC) assembly, we have found that the components of the RNAi machinery that mediate long-distance chromosomal interactions are dispensable for homologous chromosome pairing. Thus, there appears to be at least two mechanisms that bring homologous sequences together within the nucleus: those that act between dispersed homologous sequences and those that act to align and pair homologous chromosomes.
HOMOLOGOUS chromosome pairing—defined here as the close physical association of homologous loci—mediates a host of important biological phenomena. Perhaps most importantly, the proper segregation of chromosomes during the reductional division of meiosis depends on chromosomes becoming uniquely paired with their homolog. In several species, meiotic pairing has also been recruited as a scanning and defense mechanism; in Neurospora, meiotic chromosome pairing limits the proliferation of transposons through a mechanism known as meiotic silencing of unpaired DNA (MSUD) (Shiu et al. 2001; Shiu and Metzenberg 2002) and in mice and worms unpaired sex chromosomes are silenced (Bean et al. 2004; Turner et al. 2005).
Outside of the germline, somatic pairing between homologs also plays an important role in the control of gene expression. For example, transient associations between the X chromosomes during early female mammalian development aid the establishment of the inactive X chromosome (Bacher et al. 2006; Xu et al. 2006). Somatic homolog pairing has also been implicated in the action of enhancers in trans—a phenomenon known as transvection (Lewis 1954; Wu and Morris 1999; Duncan 2002)—and pairing-sensitive silencing (Kassis 1994; Americo et al. 2002). Moreover, somatic homolog pairing can facilitate repair of DNA damage. In G1, prior to S phase when a sister chromatid is unavailable as a template, DNA damage can effectively be repaired off the homolog (Esposito 1968; Fabre 1978; Kadyk and Hartwell 1992; Rong and Golic 2003).
Despite the important nature of these interactions between homologs, the mechanism by which pairing is achieved is incompletely understood. During meiosis in yeast (Bhuiyan and Schmekel 2004; Henderson and Keeney 2004), synapsis—the formation of the proteinaceous synaptonemal complex (SC) between the homologs—depends on double-stranded breaks (DSBs) that are repaired off the homolog. This also appears to be the case in mice (Mahadevaiah et al. 2001) and Arabidopsis (Grelon et al. 2001). However, DSB-independent mechanisms are important in the establishment of pairing prior to synapsis. Cha et al. (2000), Nabeshima et al. (2001), and multiple studies have demonstrated pairing in the absence of recombination (Weiner and Kleckner 1994; Nag et al. 1995). Moreover, in some species, both meiotic pairing and synapsis are entirely independent of DSB formation. For example, Drosophila melanogaster (McKim et al. 1998) and Caenorhabditis elegans (Dernburg et al. 1998) undergo perfectly good synapsis in the absence of programmed DSBs. In fact, in the somatic tissues of broader Dipterans, homologous chromosomes are paired (Metz 1916; Hiraoka et al. 1993; Fung et al. 1998). Thus, it has been proposed that somatic and meiotic pairing in Drosophila are achieved by the same mechanism and that meiotic pairing is simply an extension of pairing established in the mitotically dividing germline (Stevens 1908; Brown and Stack 1968; Vazquez et al. 2002; Gong et al. 2005; Sherizen et al. 2005). The mechanism underlying the pairing interaction is thought to be mediated either directly by nucleic acid homology [through either direct DNA–DNA interactions (i.e., Wilson 1979, McGavin 1989) or interactions that may include transcribed RNA] or indirectly through protein–protein interactions (Comings and Riggs 1971; Gemkow et al. 1998; Phillips et al. 2005; Fritsch et al. 2006; Phillips and Dernburg 2006).
Aside from full homolog pairing, another sort of chromosomal interaction has relevance to the pairing problem. Dispersed sequences residing at heterologous sites also demonstrate long-range physical interactions within the nucleus and these interactions play an important role in gene regulation. In mice, the imprinted Igf2 locus is regulated through its physical interaction with the H19 locus (Murrell et al. 2004) and the Wsb1/Nf1 locus is also regulated by its physical interaction with Igf2 (Ling et al. 2006). In flies, regulatory elements know as Polycomb response elements (PREs) mediate the physical clustering of dispersed sequences within the nucleus and this physical clustering governs the expression of neighboring genes (Bantignies et al. 2003). A similar clustering can be seen between gypsy insulators and this clustering is proposed to mediate insulator function through the formation of looped chromosome domains (Gerasimova et al. 2000; Byrd and Corces 2003).
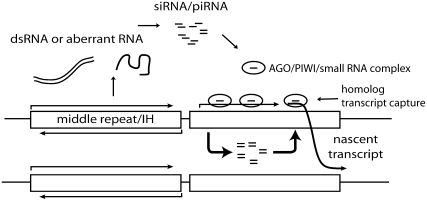
Interestingly, both PRE clustering in wing imaginal discs and insulator function have also been shown to be dependent on the RNAi machinery (Grimaud et al. 2006; Lei and Corces 2006b). The involvement of the RNAi machinery in the promotion of long-distance chromosome interactions is particularly intriguing as it suggests a model for the association of dispersed homologous sequences within the nucleus (Figure 1). In particular, since it is known that the RNAi machinery can load onto chromosomal sites in complexes that contain small RNAs (Noma et al. 2004; Verdel et al. 2004; Brower-Toland et al. 2007), these complexes could conceivably promote or stabilize pairing by capturing RNA transcripts in trans (Lei and Corces 2006a).
Figure 1.—
Model for RNAi-mediated homolog pairing. Dispersed sequences, perhaps middle repeat or intercalary heterochromatin that function as a source of small RNA could function as distributed attachment points that mediate or stabilize homolog pairing. Since small RNAs are known to assemble on chromatin, small RNA/protein complexes could potentially mediate pairing by capturing nascent transcripts in trans. In this cartoon, a nascent transcript on one homolog is captured by small RNA/protein complexes assembled on the other homolog.
It has been suggested that the mechanism that leads to the clustering of dispersed sequences is mechanistically related to the mechanism of homolog pairing. In fact, the observation that loss of Su(Hw) protein leads to reduced homolog pairing in wing imaginal discs supports this (Fritsch et al. 2006). If the mechanism underlying homolog pairing and the association of dispersed sequences were in fact identical, one would expect that the same components of the RNAi machinery that affect clustering would also be expected to affect homolog pairing. For this reason, we have tested this hypothesis by characterizing meiotic and early embryonic somatic pairing in such flies. By focusing on these tissues, rather than wing imaginal discs as other studies have done, we specifically ask whether defects in the same RNAi components lead to defects in pairing establishment or meiotic pairing, which is essential for proper segregation. We show here that the RNAi defects that have been previously shown to lead to defects in PRE clustering and insulator function show no obvious defects in meiotic or early somatic homolog pairing, despite other defects in synaptonemal complex formation and meiotic progression. These results indicate that either the mechanism of homolog pairing and dispersed sequence interactions are fundamentally different or homolog pairing can also be achieved by redundant mechanisms.
MATERIALS AND METHODS
Fly stocks and culture:
All flies were maintained at 25° on standard food. Stocks with the following alleles were used: dcr-2L811fsx and dcr-2R416x (Lee et al. 2004), ago2414 (Okamura et al. 2004), ago251b (Xu et al. 2004), aubHN2 and aubQC42 (Schüpbach and Wieschaus 1991), spn-E1/E616 and spn-Ehls-Δ125(Gillespie and Berg 1995), piwi1 (Cox et al. 1998) and mnk(lok)p6 (Brodsky et al. 2004). Embryos from piwi1 homozygous clones were generated using the FLP-DFS system (Chou and Perrimon 1992, 1996). Specifically, piwi1 FRT40/P{OvoD1-18}2La P{OvoD1-18}2Lb FRT40 larvae between 0 and 5 days old were heat shocked at 38° for 1 hr and females were collected and allowed to lay embryos. A near 0% hatch rate indicated that embryos were in fact defective for embryonic function of piwi (Cox et al. 1998).
FISH and immunostaining:
Prior to dissection of ovaries, female flies were kept for 2–3 days after eclosion in yeasted vials with males. Immunostaining was performed as described elsewhere (Page and Hawley 2001) with a cocktail of the following C(3)G mouse monoclonal antibodies: 1G5-2F7, 1A8-1G2, and 5G4-1F1, each used at 1:500. Either Alexa-488 or Alexa-555 conjugated anti-mouse IgG secondary antibodies (Invitrogen–Molecular Probes) were also used at 1:500 and ring canals were visualized by staining with rhodamine-phalloidin at 1:200 during staining with the secondary antibody. FISH, followed by immunostaining, was performed on germaria with a slightly altered procedure. Germaria were fixed for 4 min in 3.7% formaldehyde in 1× fix buffer (100 mm potassium cacodylate, 100 mm sucrose, 40 mm sodium acetate, and 10 mm EGTA). After fixation, ovaries were rinsed three times in 2× SSCT and hybridization was performed as previously described (Sherizen et al. 2005). Subsequent to hybridization and after the final 2× SSCT rinse, ovaries were rinsed two times in PBST and the immunostaining protocol (above) was resumed. FISH on 2- to 4-hr-old embryos was performed without immunostaining according to a previously described protocol (Fung et al. 1998). From sets of 2- to 4-hr-old embryos, pairing was assayed in embryos that had completed cellularization and just begun gastrulation (∼3 hr old).
FISH probes: All probes were generated by conjugating ARES Alexa fluors (either 488 or 647) with amine-labeled DNA probes. For euchromatic probes from region 25, DNA was prepared and aggregated from three overlapping BACs (FlyBase names: BACR05M06, BACR13M11, and BACR 06K07). For the histone probe, a portion of the histone cluster was PCR amplified with the two following primer pairs: 5′-aacctcagcggccagatatt-3′/5′-agcgccattcatcaagaagt-3′ and 5′-tgtctttgggcattatggtg-3′/5′-aattattccgcgtcatctgc-3′. For BAC- and PCR-based probes, DNA was digested into smaller fragments with the following four-cutters prior to tailing with amino-allyl-UTP with terminal transferase (Roche): AluI, HaeIII, MseI, MspI, RsaI, and Sau3AI. For the dodeca probe, an amino-conjugated oligonucleotide corresponding to the 5′-cccgtactggtcccgtactggtcccgtactcggtcccgtactcggt-3′ dodeca satellite sequence was purchased from IDT. Amino-labeled DNA (BAC, PCR product, or oligo) was dye-conjugated according to manufacturer's directions.
Microscopy:
Images were acquired on a DeltaVision microscopy system (Applied Precision) equipped with an Olympus 1×70 inverted microscope and CoolSnap CCD camera. All germarium images were acquired at 100× with auxiliary 1.5× magnification and all embryo images were acquired at 60× with auxiliary 1.5× magnification. Z slices were captured with a 0.2-μm step size and deconvolved using SoftWorx v2.5 software. All images are shown as either single z-slices or maximum intensity projections of deconvolved images, aside from ring canal images, which are single z-slices that were not deconvolved. Distances between FISH foci were measured by hand in three dimensions using the SoftWorx Explorer package.
RESULTS
Meiotic pairing is independent of RNAi components that mediate insulator function and somatic interactions between PREs:
Recent work has shown that the RNAi machinery plays an important role in mediating heterologous interchromosomal interactions that are important for proper gene regulation. Specifically, Aubergine (Aub) and PIWI, two components of the piRNA/rasiRNA pathway, Dicer-2 (Dcr-2), a component of the siRNA pathway, and Argonaute1 (AGO1), a component of the miRNA pathway, all contribute to long-distance chromosomal interactions between Polycomb response elements (PREs) (Grimaud et al. 2006). Furthermore, Aub and PIWI also play a role in the function of insulators, which are proposed to mediate intrachromosomal associations that lead to chromosome looping and the formation of unique chromosome regulatory domains (Lei and Corces 2006b).
These results suggest that DSB independent pairing between homologs during meiosis in Drosophila could be mediated by a related mechanism. To determine whether this is the case, we assayed pairing in meiotic nuclei of the germarium in female flies that were mutant for two components of the siRNA machinery, dcr-2 and ago-2, as well as aubergine. In addition, we also examined pairing in flies defective for spindle-E (spn-E), also known to be important for piRNA/rasiRNA function. Meiotic pairing in females mutant for piwi was not characterized due to gross defects in the germarium that made identification of meiotic nuclei difficult.
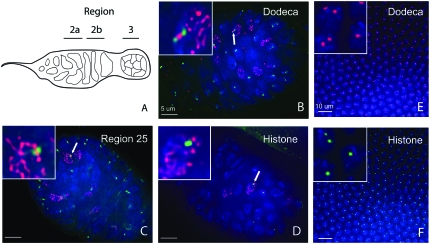
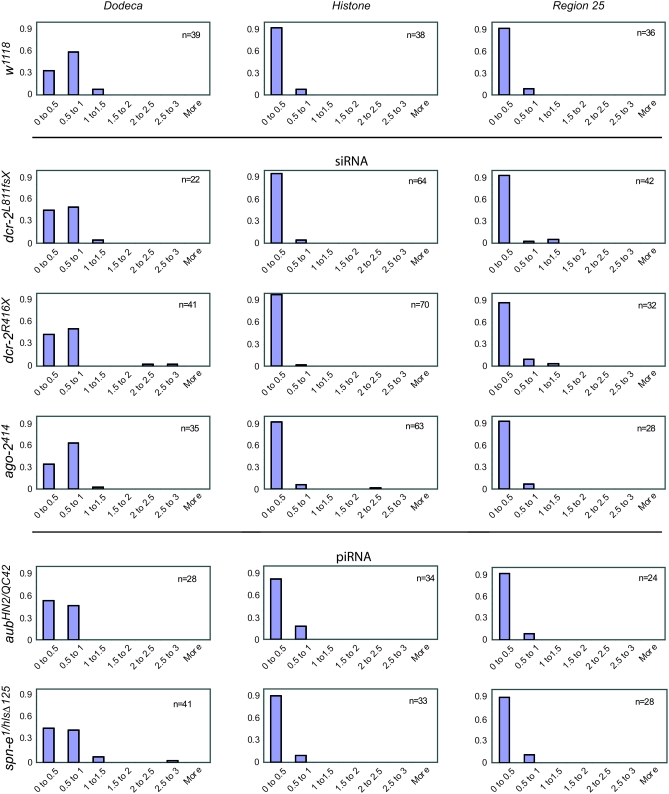
Pairing was assayed for three unique loci. For a heterochromatic sequence, we used the dodeca satellite, residing in the pericentric heterochromatin of chromosome 3. For euchromatic regions, we used the histone locus, which demonstrates an especially strong affinity for its homolog (Hiraoka et al. 1993) and resides near the base of 2L in region 39, and a normal euchromatic locus on the arm of 2L in region 25. This set of loci was chosen to exhibit a range of characteristics with regard to chromatin state and nature of repetitiveness. Meiotic nuclei within the germarium were identified by performing immunofluorescence with antibodies to C(3)G—a protein that forms the transverse filaments within the synaptonemal complex (Page and Hawley 2001) (Anderson et al. 2005). Synapsis is known to maintain pairing in the meiotic nuclei of Drosophila. Thus, our meiotic pairing assay specifically determined whether the pairing that is maintained by synapsis is disrupted. Figure 2 shows results of pairing experiments in the germarium of control w1118 flies with C(3)G staining indicating meiotic nuclei. Figure 3 shows the distribution of distance between foci in meiotic nuclei, as defined by the presence of synaptonemal complex visualized by immunostaining of C(3)G, in regions 2a and 2b. In control w1118 flies, the vast majority of histone foci and euchromatic foci are within 0.5 μm of each other. Interestingly, dodeca foci are typically farther apart—a large fraction of foci are between 0.5 and 1 μm apart. This is consistent with the fact that the synaptonemal complex does not appear as robust in the centromeric heterochromatin (Carpenter 1979; Mehrotra and McKim 2006). Thus, while heterochromatic regions are paired in meiosis, this loose SC structure may not facilitate as tight an association of homologous sequences in the centromere.
Figure 2.—
FISH pairing assay in germaria and embryos of control w1118 flies. (A) Cartoon diagram of the germarium. Pairing was assayed collectively in C(3)G staining nuclei of regions 2a and 2b. (B) Paired dodeca foci (green), partially separated between a strand of SC, indicated by immunofluorescence (red) to C(3)G in region 2a. (C) A single focus (green) using a region 25 BAC probe indicating pairing in a meiotic nucleus of 2a. (D) A single focus (green) using a histone cluster probe indicating pairing in a meiotic nucleus of 2a. (E and F) FISH in early embryos. FISH experiments were performed on 2- to 4-hr-old embryos. Pairing was assayed in embryos that had completed cellularization and just begun gastrulation (∼3 hr). (E) Dodeca probe. (F) Histone cluster probe. In all panels blue indicates DAPI. Each panel is a single z-section.
Figure 3.—
Distribution of distances between foci (in μm) of C(3)G staining nuclei of regions 2a and 2b of the germarium in w1118, siRNA defective and piRNA/rasiRNA defective females. Rows 2, 3, and 4 indicate females that were homozygous for defects in the siRNA machinery. Rows 5 and 6 indicate females that were trans-heterozygous for defects in the piRNA/rasiRNA machinery.
Overall, we found a striking consistency in the distribution of pairing distances for all loci across mutants for different RNAi mutants. This was the case for flies that were defective for components of the siRNA machinery as well as the piRNA/rasiRNA machinery. If anything, dodeca foci are closer together in flies that are RNAi defective, especially in the case of the piRNA/rasiRNA mutants. This is consistent with the observation that defects in the RNAi machinery can reduce chromatin compaction of heterochromatic sequences (Peng and Karpen 2007)—reduction in chromatin compaction of heterochromatic sequences may increase the degree to which homologous sequences tightly pair with one another.
Embryonic establishment of somatic pairing is also independent of RNAi components that mediate insulator function and somatic interactions between PREs:
In addition to examining pairing in meiotic tissues, we also examined pairing in embryonic nuclei just as pairing is established. Somatic pairing is established in early embryogenesis on a locus-by-locus basis soon after the early rapid mitotic divisions begin to slow—around cycle 13 (Fung et al. 1998). This occurs at about the same time as robust zygotic transcription begins, suggesting that pairing may be mediated by residual maternally contributed factors. Thus, it was necessary to examine pairing in embryos whose mothers were also homozygous for the given mutation. Components of the siRNA machinery are dispensable for both fertility and viability but defects in the piRNA/rasiRNA pathway lead to sterility. Thus, pairing in embryos of dcr-2 or AGO2 mothers was directly assayed but pairing in piRNA/rasiRNA defective embryos was assayed using additional measures (see below).
Pairing state is substantially more transparent and binary in the nuclei of the embryo than in meiotic nuclei. For example, paired histone loci are virtually always seen as a single dot, whereas unpaired foci are seen as two (Figure 2). As others have shown (Hiraoka et al. 1993; Fung et al. 1998; Gemkow et al. 1998), this allows for a binary classification of whether loci were paired or unpaired within each nucleus (though it should be stated that for the dodeca satellite, this was achieved with some additional difficulty due to the fact that sister chromatids often appeared separated, giving the occasional appearance of three or four foci). The distribution of distances between foci is included as supplemental material and shows similar results (supplemental Figure S2).
To determine whether there were any pairing defects in flies whose mothers were defective for components of the siRNA pathway, FISH was performed in 2- to 4-hr-old embryos. To ensure consistency of staging for these embryos whose age ranged over a total of 2 hr, pairing was assayed in a subset of these embryos—specifically, those that were cellularized and just prior to or at the beginning of gastrulation. In each of the multiple embryos, at least 50 nuclei were counted. Pairing of canonical euchromatic regions is very low at this developmental stage (Fung et al. 1998, data not shown). Since the power to detect reduced pairing levels is weak when wild-type pairing levels are already low, we only analyzed pairing state for the histone locus and dodeca satellite (Figure 2). Furthermore, while pairing for canonical euchromatic regions is higher at larval stages, embryos that are defective for components of the piRNA/rasiRNA machinery do not progress to this stage. Thus, this study of pairing establishment in embryos is limited to loci that are not representative of canonical euchromatic regions. Interestingly, as suggested by others (Hiraoka et al. 1993), we found a nonrandom clustering of nuclei that shared the same pairing state (data not shown), suggesting that either local domains within the embryo can effect pairing state (perhaps based on relative timing of nuclear division) or neighboring nuclei are more likely to share pairing state since they are more likely derived from the same mother nucleus (indicating that pairing status can be transmitted through nuclear division).
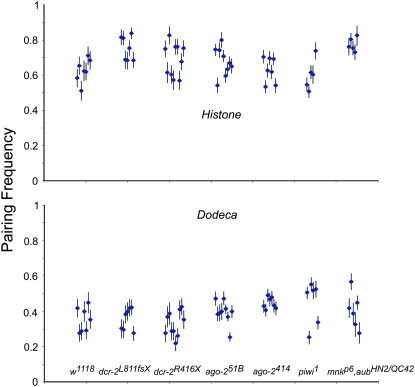
As in the case of meiotic pairing, we found no obvious defects in pairing in embryos whose mothers were defective in components of the siRNA machinery (Figure 4). If anything, pairing frequencies for the dodeca satellite are higher for ago-2414 and, for the histone locus, higher for dcr-2L811fsx.
Figure 4.—
Pairing frequencies for the histone cluster and dodeca satellite in ∼3-hr-old siRNA and piRNA/rasiRNA defective embryos. siRNA defective embryos were the progeny of homozygous mothers and fathers. Piwi1 embryos were generated with the FLP-DFS technique (see text). Piwi1 and mnk p6,aubHN2/mnk p6,aubQC42 embryos were the product of mutant females mated with w1118 males. Each point indicates the pairing frequency of a single embryo in which the pairing state in at least 50 nuclei was determined. Error bars are standard errors for estimates of the pairing frequency.
While mothers that are defective for components of the piRNA/rasiRNA machinery are sterile, associated defects can be partially rescued by a mutation in mnk, the fly homolog of the DNA damage signaling Chk2 (Klattenhoff et al. 2007). In particular mnk p6,aubHN2/mnk p6,aubQC42 mothers lay embryos that partially develop, enough to potentially assay pairing state. Although these embryos are defective, with grossly misshapen nuclei (Figure 5), pairing can be assayed in embryos that partially escape this phenotype. Upon first observation, it appeared that pairing frequencies were substantially greater for the dodeca satellite in such embryos. However, after quantifying the fluorescence intensity of the dodeca probe in embryos that appeared to have high levels of pairing, it was clear that the apparent high numbers of nuclei with a single focus was associated with a substantial reduction in fluorescence intensity (supplemental Figure S1). It is known that defects in the RNAi machinery can lead to decondensation of heterochromatin and this decondensation can lead to ectopic exchange and loss of genetic material (Peng and Karpen 2007). Thus, it appears that for embryos that show a large number of nuclei with single dodeca foci, this can be attributed to either loss of a single third chromosome or loss of a substantial portion of the dodeca satellite, perhaps during an early embryonic nuclear division. In embryos that do not show this excess of single dodeca foci (cutoff >60% nuclei showing a single dot), pairing rates are not reduced (Figure 4). Together, these data indicate that even maternally supplied Aubergine protein is not necessary for the establishment of chromosome pairing in embryos. To assay pairing in piwi1/piwi1 embryos we generated germline clones using the FLP-DFS method (Chou and Perrimon 1992, 1996). As in mnk,aub, embryos were highly defective with many misshapen nuclei. Nonetheless, in embryos for which this phenotype did not prohibit analysis, pairing of the histone and dodeca sequences was not reduced (Figure 4).
Figure 5.—
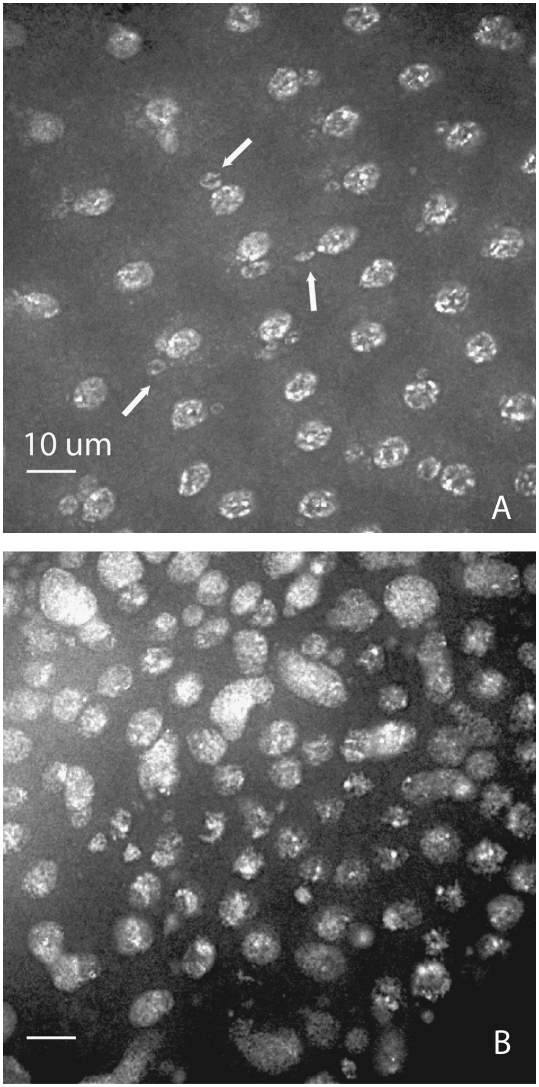
Embryos of mnk p6,aubHN2/mnk p6,aubQC42 mothers are highly disorganized: DAPI staining of 2- to 4-hr-old embryos. The exact developmental point in early embryonic development is difficult to determine due to gross defects. (A) DAPI nuclei show looped-out circles of DNA. (B) A highly disorganized field of nuclei.
Mutations in aubergine lead to defects in SC assembly and oocyte determination:
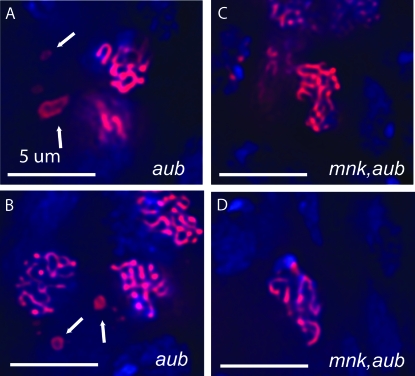
Although we found no obvious defects in either somatic or meiotic full homolog pairing in flies defective for components of the RNAi machinery, we did observe two obvious meiotic defects. For example, in meiotic nuclei of aubHN2/aubQC42 females, C(3)G displays the normal threadlike morphology, suggesting that gross SC structure is normal. However, large donut-shaped aggregations of C(3)G protein were often observed flanking the meiotic nuclei (Figure 6). While these donut structures are reminiscent of ring canals, they do not colocalize with them (supplemental Figure S3). In wild-type flies, the proportion of cysts in region 3 of the germarium with aggregates was 10% (n = 21) and in stage 2, they were observed 14% (n = 21) of the time. In aub flies, the proportion of region 3 and stage 2 cysts with aggregates was 80% (n = 15) and 56% (n = 18), respectively (P < 0.01 and P < 0.01, Fisher's exact test comparing respective regions). These aggregates of C(3)G resemble the polycomplexes observed when the C(3)G protein lacks the C terminus that mediates attachment to the lateral elements of the SC (Jeffress et al. 2007). However, the complexes seen here differ from those by Jeffress et al. (2007) in that they were seen outside of the DAPI staining nuclei. The fact that normal SC structure is observed alongside these complexes suggests that a mutation in aubergine leads to an accumulation of free C(3)G protein in the cytoplasm rather than simply prohibiting incorporation into the SC. In turn, this accumulation of cytoplasmic protein forms aggregates. Interestingly, the presence of these complexes depended on activation of the Chk2 DNA damage response. In mnk p6,aubHN2/mnk p6,aubQC42 flies, the occurrence of these aggregates was equal to that in wild-type flies (region 3: 10%, n = 21; stage 2: 0%, n = 15). Thus, in mnk flies where the DNA damage response fails to proceed, C(3)G protein no longer accumulates and forms aggregates outside the nucleus. This suggests that activation of the DNA damage checkpoint governs either the production of C(3)G protein or access of C(3)G protein to the meiotic nucleus. Furthermore, since a large excess of DSBs are observed to form in aub flies (Klattenhoff et al. 2007), DSB excess may also play a role in regulating C(3)G function or expression.
Figure 6.—
Aggregates of C(3)G (red) that resemble polycomplexes form flanking the meiotic nuclei within the germarium of aubHN2/aubQC42 flies. (A and B) Donut-shaped aggregates of C(3)G in aubHN2/aubQC42 flies. (C and D) Aggregates fail to form in mnk p6,aubHN2/mnk p6,aubQC42 flies.
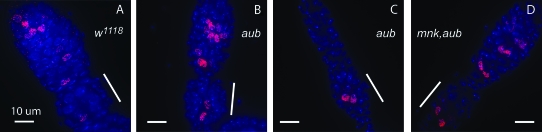
Mutations in aubergine also lead to a delay in final oocyte determination (Figure 7). In wild-type flies, region 3 cysts of the germarium typically display a single oocyte nucleus, as defined by threadlike C(3)G staining. Occasionally two nuclei show threadlike C(3)G staining, but in one nucleus the threadlike structure is midway toward disassembly. We established an “oocyte index” by counting within cysts the number of nuclei that show maintenance of threadlike C(3)G staining and designating those nuclei that have partially dissembled SC as 0.5 nuclei. In wild-type flies, this gave an oocyte index of 1.40 (n = 21) in region 3 and 1.05 (n = 21) in stage 2. However, aub flies frequently display two to four oocytes in region 3 of the germaria (oocyte index: 2.40, n = 15) and frequently two oocytes in stage 2 egg chambers (oocyte index: 1.5, n = 18). Since aub germaria are much reduced in size, it is conceivable that region 3 cysts in aub flies may be actually younger and less developmentally mature than region 3 cysts from wild-type flies. Thus, a direct comparison between oocyte indices of region 3 cysts may not be reasonable. Nonetheless, stage 2 egg chambers in aub flies show a higher oocyte index than region 3 wild-type cysts, indicating that this delay in oocyte determination is in fact real. And while a mutation in mnk rescues the formation of the C(3)G aggregates, a similar delay in final oocyte determination is also observed in mnk p6,aubHN2/mnk p6,aubQC42 females (region 3 oocyte index: 2.0, n = 21; stage 2 oocyte index: 1.41, n = 16). This indicates that while activation of the DNA damage checkpoint promotes the formation of C(3)G aggregates, it does not lead to the delay in oocyte determination in aubergine flies.
Figure 7.—
A defect in aub leads to a delay in meiotic progression. White bars indicate region 3 of the germarium. (A) w1118 control flies show one C(3)G staining nucleus in region 3. (B and C) aubHN2/aubQC42 flies frequently show two or more C(3)G staining nuclei in region 3, indicating a delay in the decision to form a single oocyte. (D) This defect is not alleviated in mnk p6,aubHN2/mnk p6,aubQC42 flies.
DISCUSSION
In sexual species, proper segregation of chromosomes during meiosis requires homologs to become paired. Ironically, sexual reproduction itself fosters the conditions for proliferation of selfish genetic elements, such as transposons (Hickey 1982) that might pose a challenge for the pairing process due to their dispersed repetitive nature. The RNAi machinery is thought to have evolved to limit the spread of such selfish repeats and viruses (Matzke et al. 2000; Plasterk 2002; Zamore 2002; Cerutti and Casas-Mollano 2006; Ding and Voinnet 2007) and subsequently, the RNAi machinery has also evolved a wide range of secondary functions. In Drosophila, the RNAi machinery has been shown to have several different roles in producing the higher order structure of chromosomes in the nucleus (Pal-Bhadra et al. 2004; Matzke and Birchler 2005; Kavi et al. 2006; Savitsky et al. 2006; Peng and Karpen 2007). In particular, the RNAi machinery mediates both insulator function and the long-distance clustering of Polycomb response elements (Grimaud et al. 2006; Lei and Corces 2006b). These observations have suggested a mechanism for how long-range chromosomal associations are mediated by the RNAi machinery. An important feature of this model is that insulator looping and PRE clustering depends on both the RNAi machinery and proteins that are specific to particular loci. In the case of insulator looping, a complex of CP190/Mod(mdg4)2.2/Su(HW) proteins is proposed to bind to insulator sequences and loops may be stabilized by the small RNAs that are recruited to the complex. In the case of Polycomb response elements it is proposed that bound Polycomb proteins, together with small RNAs, stabilize long-range interactions.
Nonetheless, we have shown that defects in the RNAi machinery that mediate these functions have little effect on pairing establishment both in the embryo and in the germline. This is consistent with work that has shown no reduction in crossing over frequency in flies defective in piRNA/rasiRNA components (Cross and Simmons 2008), contrary to what would be expected if there were a pairing defect. In light of Su(Hw)'s effect on homolog pairing in the soma (Fritsch et al. 2006), the association of dispersed sequences may in fact facilitate whole chromosome pairing, but this function appears to be independent of the RNAi machinery. Altogether, this suggests a hierarchical model for the association of homologous sequences—between homologs and at heterologous locations—within the nucleus. First, particular chromatin-bound proteins dispersed along the chromosomes may facilitate chromosome pairing on a locus-by-locus basis. For example, the histone cluster, which shows great affinity between the homologs, may pair in a very efficient manner due to the fact that it is uniquely contained within the histone locus body (Liu et al. 2006; Liu and Gall 2007). Full-length homolog pairing may thus be mediated by a large assemblage of various chromatin-bound proteins, including insulator and Polycomb proteins, distributed along the length of the chromosome arms. Overlaid on this system, the RNAi machinery may contribute, in a cooperative fashion, to long-distance interactions by binding distinct classes of chromatin-bound proteins into larger domains. Consistent with this hierarchical model, Grimaud et al. (2006) found that the localization of Polycomb proteins to endogenous PREs did not depend on components of the RNAi machinery and that the RNAi machinery more likely plays a role in stabilizing long-distance interactions rather than initiating them. A hierarchical model is also suggested by the observation that the RNAi machinery, in conjunction with heterochromatic proteins, has an inhibitory effect on ectopic exchange between dispersed heterochromatic repeat sequences (Peng and Karpen 2007). In a combinatorial fashion, the RNAi machinery, interacting with one class of chromatin-bound proteins, may thus mediate dispersed interactions, but in the context of another set of chromatin-bound proteins, lead to the inhibition of dispersed interactions, presumably by leading to condensation of heterochromatin and exclusion from the homology search initiated after DNA damage.
One caveat to this interpretation is that functional redundancy between different components of the RNAi machinery may limit our conclusion that the RNAi machinery does not play a role on meiotic or somatic homolog pairing. Our results show that the same components that mediate long-range interactions are not individually necessary for homolog pairing. However, full proof of a complete lack of involvement of the RNAi machinery in pairing requires an analysis of pairing in a genetic background that is simultaneously defective for primary components of each of the three branches of the RNAi system—the miRNA, siRNA, and piRNA/rasiRNA pathways. Unfortunately, such analysis is difficult since these components are essential for viability and fertility.
In spite of these results, we have uncovered two new functions for the RNAi machinery in oocyte determination and regulation of synaptonemal complex formation during Drosophila female meiosis. Components of the piRNA/rasiRNA machinery—aubergine, piwi, spindle-E, and armitage—have previously been shown to be essential for silencing transposons in the germline. In the face of transposon mobilization, DNA damage will lead to activation of the ATR/Chk2 DNA damage checkpoint, leading to the classic spindle defects that include fused dorsal appendages and abnormal karysome formation (Chen et al. 2007; Klattenhoff et al. 2007). We have now found that defects in the piRNA/rasiRNA machinery also lead to defects in the assembly of the synaptonemal complex as measured by C(3)G immunostaining. In particular, a mutation in the piRNA/rasiRNA component aubergine leads to the formation of extranuclear C(3)G aggregates as well as a delay in resolving the identity of the single oocyte among the 16 mitotic nuclei. The C(3)G aggregates resemble those polycomplexes observed in flies whose sole sources of C(3)G protein is truncated at the C-terminal end that interacts with the lateral elements of the synaptonemal complex (Jeffress et al. 2007). However, the fact that the aubergine C(3)G aggregates reside outside the nucleus and occur alongside SC with normal C(3)G morphology indicates that they likely arise due to a failure for all C(3)G protein to enter the nucleus rather than any intrinsic failure to form SC. Interestingly, the formation of the extranuclear C(3)G aggregates is rescued by a mutation in mnk/Chk2. This suggests a potential feedback between activation of the DNA damage checkpoint and regulation of C(3)G.
Unlike C(3)G aggregate formation, the delay in oocyte determination in aubergine flies is not dependent on activation of the Chk2 DNA damage cascade—similar numbers of two-oocyte region 3 germarium were found in aub and aub,mnk flies. This two-oocyte phenotype has also been observed in flies defective for the rasiRNA component spn-E, as well as mus301/spn-C (Huynh and St Johnston 2000; McCaffrey et al. 2006). Mus301 has been shown to play a role in DSB repair, but the associated two-oocyte phenotype in mus301 flies was also observed in flies that lack meiotic double-stranded breaks programmed by mei-W68 (McCaffrey et al. 2006). This result suggested that mus301 had a role in oocyte specification independent of programmed DSBs. It remains to be determined whether the mechanism underlying the two-oocyte phenotype is the same in the case of DNA repair defective flies that lack programmed DSBs and piRNA/rasiRNA defective flies with abundant DSBs that arise in the face of transposon mobilization and are thus W68 independent.
Acknowledgments
Many thanks to Jack Bateman for discussions, suggestions, and insight and Danny Stark for assistance in microscopy. Flies were kindly provided by the Carthew, Gao, Schedl, Elgin, and Berg laboratories. This work was funded by an American Cancer Society Postdoctoral Fellowship to J.P.B. and an American Cancer Society Professor Award to R.S.H. Additional funding was provided by the Stowers Institute for Medical Research.
References
- Americo, J., M. Whiteley, J. Lesley Brown, M. Fujioka, J. B. Jaynes et al., 2002. A complex array of DNA-binding proteins required for pairing-sensitive silencing by a Polycomb group response element from the Drosophila engrailed gene. Genetics 160 1561–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L. K., S. M. Royer, S. L. Page, K. S. McKim, A. Lai et al., 2005. Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. USA 102 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher, C. P., M. Guggiari, B. Brors, S. Augui, P. Clerc et al., 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8 293–299. [DOI] [PubMed] [Google Scholar]
- Bantignies, F., C. Grimaud, S. Lavrov, M. Gabut and G. Cavalli, 2003. Inheritance of polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 17 2406–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean, C. J., C. E. Schaner and W. G. Kelly, 2004. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet. 36 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan, H., and K. Schmekel, 2004. Meiotic chromosome synapsis in yeast can occur without Spo11-induced DNA double-strand breaks. Genetics 168 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, M. H., B. T. Weinert, G. Tsang, Y. S. Rong, N. M. McGinnis et al., 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 24 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland, B., S. D. Findley, L. Jiang, L. Liu, H. Yin et al., 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 21 2300–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W. V., and S. M. Stack, 1968. Somatic pairing as a regular preliminary to meiosis. Bull. Torrey Bot. Club 95 369–378. [Google Scholar]
- Byrd, K., and V. G. Corces, 2003. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A. T., 1979. Synaptonemal complex and recombination nodules in wild-type Drosophila melanogaster females. Genetics 92 511–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti, H., and J. A. Casas-Mollano, 2006. On the origin and functions of RNA-mediated silencing: from protists to man. Curr. Genet. 50 81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha, R. S., B. M. Weiner, S. Keeney, J. Dekker and N. Kleckner, 2000. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 14 493–503. [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., A. Pane and T. Schüpbach, 2007. cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr. Biol. 17 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, T. B., and N. Perrimon, 1992. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics 131 643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, T. B., and N. Perrimon, 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comings, D. E., and A. D. Riggs, 1971. Molecular mechanisms of chromosome pairing, folding and function. Nature 233 48–50. [DOI] [PubMed] [Google Scholar]
- Cox, D. N., A. Chao, J. Baker, L. Chang, D. Qiao et al., 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, E. W., and M. J. Simmons, 2008. Does RNA interference influence meiotic crossing over in Drosophila melanogaster? Genet. Res. 90 253–258. [DOI] [PubMed] [Google Scholar]
- Dernburg, A. F., K. McDonald, G. Moulder, R. Barstead, M. Dresser et al., 1998. Meiotic recombination in C-elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94 387–398. [DOI] [PubMed] [Google Scholar]
- Ding, S. W., and O. Voinnet, 2007. Antiviral immunity directed by small RNAs. Cell 130 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, I. W., 2002. Transvection effects in Drosophila. Annu. Rev. Genet. 36 521–556. [DOI] [PubMed] [Google Scholar]
- Esposito, R. E., 1968. Genetic recombination in synchronized cultures of Saccharomyces cerevisiae. Genetics 59 191–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre, F., 1978. Induced intragenic recombination in yeast can occur during the G1 mitotic phase. Nature 272 795–798. [DOI] [PubMed] [Google Scholar]
- Fritsch, C., G. Ploeger and D. J. Arndt-Jovin, 2006. Drosophila under the lens: imaging from chromosomes to whole embryos. Chromosome Res. 14 451–464. [DOI] [PubMed] [Google Scholar]
- Fung, J. C., W. F. Marshall, A. Dernburg, D. A. Agard and J. W. Sedat, 1998. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J. Cell Biol. 141 5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemkow, M. J., P. J. Verveer and D. J. Arndt-Jovin, 1998. Homologous association of the bithorax-complex during embryogenesis: consequences for transvection in Drosophila melanogaster. Development 125 4541–4552. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T. I., K. Byrd and V. G. Corces, 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6 1025–1035. [DOI] [PubMed] [Google Scholar]
- Gillespie, D. E., and C. A. Berg, 1995. homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 9 2495–2508. [DOI] [PubMed] [Google Scholar]
- Gong, W. J., K. S. McKim and R. S. Hawley, 2005. All paired up and no place to go: pairing, synapsis and DSB formation in a balancer heterozygote. PLoS Genet. 1 e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon, M., D. Vezon, G. Gendrot and G. Pelletier, 2001. AtSPO11–1 is necessary for efficient meiotic recombination in plants. EMBO J. 20 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud, C., F. Bantignies, M. Pal-Bhadra, P. Ghana, U. Bhadra et al., 2006. RNAi components are required for nuclear clustering of polycomb group response elements. Cell 124 957–971. [DOI] [PubMed] [Google Scholar]
- Henderson, K. A., and S. Keeney, 2004. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 101 4519–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey, D. A., 1982. Selfish DNA: a sexually transmitted nuclear parasite. Genetics 101 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka, Y., A. F. Dernburg, S. J. Parmelee, M. C. Rykowski, D. A. Agard et al., 1993. The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J. Cell Biol. 120 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, J. R., and D. St Johnston, 2000. The role of BicD, Egl, Orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development 127 2785–2794. [DOI] [PubMed] [Google Scholar]
- Jeffress, J. K., S. L. Page, S. M. Royer, E. D. Belden, J. P. Blumenstiel et al., 2007. The formation of the central element of the synaptonemal complex may occur by multiple mechanisms: the roles of the N- and C-terminal domains of the Drosophila C(3)G protein in mediating synapsis and recombination. Genetics 177 2445–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk, L. C., and L. H. Hartwell, 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis, J. A., 1994. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6-kb region. Genetics 136 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi, H. H., H. R. Fernandez, W. Xie and J. A. Birchler, 2006. Polycomb, pairing and PIWI: RNA silencing and nuclear interactions. Trends Biochem. Sci. 31 485–487. [DOI] [PubMed] [Google Scholar]
- Klattenhoff, C., D. P. Bratu, N. McGinnis-Schultz, B. S. Koppetsch, H. A. Cook et al., 2007. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12 45–55. [DOI] [PubMed] [Google Scholar]
- Lee, Y. S., K. Nakahara, J. W. Pham, K. Kim, Z. He et al., 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117 69–81. [DOI] [PubMed] [Google Scholar]
- Lei, E. P., and V. G. Corces, 2006. a A long-distance relationship between RNAi and polycomb. Cell 124 886–888. [DOI] [PubMed] [Google Scholar]
- Lei, E. P., and V. G. Corces, 2006. b RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat. Genet. 38 936–941. [DOI] [PubMed] [Google Scholar]
- Lewis, E. B., 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88 225–239. [Google Scholar]
- Ling, J. Q., T. Li, J. F. Hu, T. H. Vu, H. L. Chen et al., 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312 269–272. [DOI] [PubMed] [Google Scholar]
- Liu, J. L., M. Buszczak and J. G. Gall, 2006. Nuclear bodies in the Drosophila germinal vesicle. Chromosome Res. 14 465–475. [DOI] [PubMed] [Google Scholar]
- Liu, J. L., and J. G. Gall, 2007. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc. Natl. Acad. Sci. USA 104 11655–11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah, S. K., J. M. A. Turner, F. Baudat, E. P. Rogakou, P. de Boer et al., 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27 271–276. [DOI] [PubMed] [Google Scholar]
- Matzke, M. A., and J. A. Birchler, 2005. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6 24–35. [DOI] [PubMed] [Google Scholar]
- Matzke, M. A., M. F. Mette and A. J. M. Matzke, 2000. Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol. Biol. 43 401–415. [DOI] [PubMed] [Google Scholar]
- McCaffrey, R., D. St Johnston and A. Gonzalez-Reyes, 2006. Drosophila mus301/spindle-C encodes a helicase with an essential role in double-strand DNA break repair and meiotic progression. Genetics 174 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavin, S., 1989. Four strand recombination models. J. Theor. Biol. 136 135–150. [DOI] [PubMed] [Google Scholar]
- McKim, K. S., B. L. Green-Marroquin, J. J. Sekelsky, G. Chin, C. Steinberg et al., 1998. Meiotic synapsis in the absence of recombination. Science 279 876–878. [DOI] [PubMed] [Google Scholar]
- Mehrotra, S., and K. S. McKim, 2006. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2 1883–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz, C. W., 1916. Chromosome studies on the Diptera. II. The paired association of chromosomes in the Diptera, and its significance. J. Exp. Zool. 21 213–279. [Google Scholar]
- Murrell, A., S. Heeson and W. Reik, 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 36 889–893. [DOI] [PubMed] [Google Scholar]
- Nabeshima, K., Y. Kakihara, Y. Hiraoka and H. Nojima, 2001. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 20 3871–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, D. K., H. Scherthan, B. Rockmill, J. Bhargava and G. S. Roeder, 1995. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics 141 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma, K., T. Sugiyama, H. Cam, A. Verdel, M. Zofall et al., 2004. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 36 1174–1180. [DOI] [PubMed] [Google Scholar]
- Okamura, K., A. Ishizuka, H. Siomi and M. C. Siomi, 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2001. (3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra et al., 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303 669–672. [DOI] [PubMed] [Google Scholar]
- Peng, J. C., and G. H. Karpen, 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 9 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, C. M., and A. F. Dernburg, 2006. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell 11 817–829. [DOI] [PubMed] [Google Scholar]
- Phillips, C. M., C. Wong, N. Bhalla, P. M. Carlton, P. Weiser et al., 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk, R. H. A., 2002. RNA silencing: the genome's immune system. Science 296 1263–1265. [DOI] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky, M., D. Kwon, P. Georgiev, A. Kalmykova and V. Gvozdev, 2006. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 20 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach, T., and E. Wieschaus, 1991. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129 1119–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherizen, D., J. K. Jang, R. Bhagat, N. Kato and K. S. McKim, 2005. Meiotic recombination in Drosophila females depends on chromosome continuity between genetically defined boundaries. Genetics 169 767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, P. K. T., and R. L. Metzenberg, 2002. Meiotic silencing by unpaired DNA: properties, regulation, and suppression. Genetics 161 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, P. K. T., N. B. Raju, D. Zickler and R. L. Metzenberg, 2001. Meiotic silencing by unpaired DNA. Cell 107 905–916. [DOI] [PubMed] [Google Scholar]
- Stevens, N. M., 1908. A study of the germ cells of certain Diptera, with reference to the heterochromosomes and the phenomena of synapsis. J. Exp. Zool. 5 359–374. [Google Scholar]
- Turner, J. M. A., S. K. Mahadevaiah, O. Fernandez-Capetillo, A. Nussenzweig, X. Xu et al., 2005. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 37 41–47. [DOI] [PubMed] [Google Scholar]
- Vazquez, J., A. S. Belmont and J. W. Sedat, 2002. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12 1473–1483. [DOI] [PubMed] [Google Scholar]
- Verdel, A., S. T. Jia, S. Gerber, T. Sugiyama, S. Gygi et al., 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner, B. M., and N. Kleckner, 1994. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell 77 977–991. [DOI] [PubMed] [Google Scholar]
- Wilson, J. H., 1979. Nick-free formation of reciprocal heteroduplexes: a simple solution to the topological problem. Proc. Natl. Acad. Sci. USA 76 3641–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. T., and J. R. Morris, 1999. Transvection and other homology effects. Curr. Opin. Genet. Dev. 9 237–246. [DOI] [PubMed] [Google Scholar]
- Xu, K., B. A. Bogert, W. Li, K. Su, A. Lee et al., 2004. The fragile X-related gene affects the crawling behavior of Drosophila larvae by regulating the mRNA level of the DEG/ENaC protein Pickpocket1. Curr. Biol. 14 1025–1034. [DOI] [PubMed] [Google Scholar]
- Xu, N., C. L. Tsai and J. T. Lee, 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science 311 1149–1152. [DOI] [PubMed] [Google Scholar]
- Zamore, P. D., 2002. Ancient pathways programmed by small RNAs. Science 296 1265–1269. [DOI] [PubMed] [Google Scholar]