Abstract
Heterochromatin resides near yeast telomeres and at the cryptic mating-type loci, HML and HMR, where it silences transcription of the α- and a-mating-type genes, respectively. Ku is a conserved DNA end-binding protein that binds telomeres and regulates silencing in yeast. The role of Ku in silencing is thought to be limited to telomeric silencing. Here, we tested whether Ku contributes to silencing at HML or HMR. Mutant analysis revealed that yKu70 and Sir1 act collectively to silence the mating-type genes at HML and HMR. In addition, loss of yKu70 function leads to expression of different reporter genes inserted at HMR. Quantitative chromatin-immunoprecipitation experiments revealed that yKu70 binds to HML and HMR and that binding of Ku to these internal loci is dependent on Sir4. The interaction between yKu70 and Sir4 was characterized further and found to be dependent on Sir2 but not on Sir1, Sir3, or yKu80. These observations reveal that, in addition to its ability to bind telomeric DNA ends and aid in the silencing of genes at telomeres, Ku binds to internal silent loci via protein–protein interactions and contributes to the efficient silencing of these loci.
DISTINCT regions of eukaryotic genomes are packaged into different types of chromatin, which are broadly categorized as euchromatin and heterochromatin. The type of chromatin in any particular region controls the functional potential of that region. In general, euchromatic regions of the genome are replicated early in S-phase and the genes in those regions are maintained in a state that is permissive for transcription, whereas heterochromatic regions of the genome are replicated late in S-phase and transcription of the genes in heterochromatic regions is repressed (Gilbert 2002; Schwaiger and Schèubeler 2006).
In the budding yeast Saccharomyces cerevisiae, heterochromatin resides in the regions adjacent to each of the telomeres and at the silent mating-type loci HML and HMR (Rusche et al. 2003; Millar and Grunstein 2006; Shahbazian and Grunstein 2007). Heterochromatin represses transcription of the endogenous genes in these regions and can repress transcription of genes experimentally inserted into these regions (Rusche et al. 2003; Pirrotta and Gross 2005; Millar and Grunstein 2006; Talbert and Henikoff 2006). Repression of transcription by heterochromatin in yeast is typically referred to as silencing (Herskowitz et al. 1977; Haber and George 1979; Klar and Fogel 1979; Rine et al. 1979). Silencing is dependent on Sir2, Sir3, and Sir4, which bind the nucleosomes in the silent regions. The complex of these Sir proteins with the histones is thought to comprise the structural unit of heterochromatin in yeast (Grunstein 1997; Rusche et al. 2003).
Silencing is also dependent on DNA control elements associated with each of the silent regions. Two control regions are associated with HML, the HML-E and HML-I silencers, and two with HMR, the HMR-E and HMR-I silencers (Brand et al. 1985; Mahoney and Broach 1989; Rivier et al. 1999). The telomeres, which are made up of the terminal telomeric repeats and the physical end of the chromosome, are one type of control region essential for telomeric silencing (Gottschling et al. 1990; Tham and Zakian 2002). In addition, some telomeres contain short tandem repeats (STR), which also contribute to telomeric silencing (Louis 1995; Mondoux and Zakian 2007). Four of these control regions—HML-E, HML-I, HMR-E, and the telomeres—can act autonomously to direct silencing in the absence of any other control region (Loo and Rine 1995; Rusche et al. 2003). The other two, HMR-I and the STRs, contribute to the overall efficiency of silencing at their respective loci but cannot direct silencing autonomously (Rivier et al. 1999; Mondoux and Zakian 2007).
Four DNA-binding proteins—Rap1, Orc (the origin recognition complex), Ku, and Abf1—bind to silencers, telomeres, and/or STRs and play a role in silencing. Three of these proteins—Rap1, Orc and Abf1—bind specific DNA sequences (Shore 1994; Loo and Rine 1995; Grunstein 1997; Rusche et al. 2003), whereas Ku binds specifically to DNA ends (Gravel et al. 1998; Ribes-Zamora et al. 2007). The binding sites for these proteins that contribute to silencing are contained within the control regions that direct silencing. Of these four proteins, the roles of Rap1, Orc, and Ku in silencing are most well understood. Rap1 is a general regulator of silencing that acts at all three silent loci, HML, HMR, and the telomeres. Binding sites for Rap1 are found in control regions associated with each of the three silent loci HML, HMR, and the terminal telomeric repeats (Shore and Nasmyth 1987; Buchman et al. 1988; Hofmann et al. 1989; Longtine et al. 1989; Kurtz and Shore 1991). Furthermore, mutations in RAP1 result in silencing defects at HML, HMR, and the telomeres (Kyrion et al. 1993; Moretti et al. 1994).
In contrast, Ku appears to be a locus-specific regulator of silencing that acts in some silent regions but not in others. In particular, deletion of the gene encoding either subunit of the Ku heterodimer results in complete loss of telomeric silencing (Mishra and Shore 1999; Teo and Jackson 2001) but no detectable loss in silencing at HML or HMR (Boulton and Jackson 1998; Laroche et al. 1998; Nugent et al. 1998; Martin et al. 1999; Gartenberg et al. 2004; Roy et al. 2004). Also, a Ku-binding site (a DNA end) is found at the telomere but not at the internal HML or HMR loci (Gravel et al. 1998). Thus, Ku is thought to be a locus-specific regulator of silencing that acts at the telomeres but not at HML or HMR.
The focus of this work is to further understand the role of Ku in silencing. As described above, Ku is not thought to act at HML and HMR; however, the evidence that Ku does not act at these internal loci is limited. HML and HMR are internal loci that are not associated with DNA ends and therefore do not contain a DNA-binding site for Ku; however, some DNA-binding proteins have two modes of binding to DNA: one mode resulting from the DNA-binding activity intrinsic to the protein itself and another mode resulting from protein–protein interactions (Valenzuela et al. 2008). If Ku is endowed with two modes of binding DNA, then the fact that HML and HMR lack a DNA-binding site for Ku is not sufficient evidence to conclude that Ku does not bind these loci. Furthermore, although neither subunit of Ku is required for silencing HML or HMR (Boulton and Jackson 1998; Laroche et al. 1998; Nugent et al. 1998; Martin et al. 1999; Gartenberg et al. 2004; Roy et al. 2004), as they are for telomeric silencing (Mishra and Shore 1999; Teo and Jackson 2001), silencing at HML and HMR is more efficient than silencing at the telomeres and at least some DNA elements within silencers are known to be redundant. Therefore, it is possible that the increased efficiency and redundancy of silencing at HML and HMR has masked a possible role of Ku in silencing at these loci.
The goal of this work was to test directly whether Ku plays a role in silencing at HML and HMR. We show that Ku contributes to silencing at both HML and HMR, that Ku binds both these internal loci, and that binding of Ku to HML and HMR is dependent on Sir4.
MATERIALS AND METHODS
Strain construction:
Strains used in this study are isogenic to W303-1a and are listed in Table 1. Strains were constructed by cross or PCR-mediated gene disruption (Baudin et al. 1993) and confirmed by PCR or DNA blots. The kanMX4 gene of plasmid pFA6-kanMX4 (Wach et al. 1994) and the natMX4 gene of plasmid pFA6-natMX4 (Goldstein and Mccusker 1999) were integrated into pBluescript to make pDR759 and pDR1848, respectively, and to avoid any growth defects associated with auxotrophic markers. The plasmids were PCR amplified by hybrid disruption primers containing homology to both a specific gene and pUC1 or pUC2 sequences, as previously described (Replogle et al. 1999). The gene-specific confirmation primers were used in conjunction with pUC complement sequences to confirm PCR-mediated gene disruptions. All PCR-mediated gene disruption and confirmation primers are listed in Table 2.
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| DRY705 | MATα his3-11 leu2-3,112 lys2Δ∷hisG trp1-1 ura3-1 |
| DRY5430 | MATaADE2 lys2Δ∷hisG |
| DRY5431 | MATayku70Δ∷natMX4 ADE2 lys2Δ∷hisG |
| DRY5432 | MATasir1Δ∷kanMX4 ADE2 lys2Δ∷hisG |
| DRY5433 | MATayku70Δ∷natMX4 sir1Δ∷kanMX4 ADE2 lys2Δ∷hisG |
| DRY5438 | MATα yku70Δ∷natMX4 ADE2 lys2Δ∷hisG |
| DRY5439 | MATα sir1Δ∷kanMX4 ADE2 lys2Δ∷hisG |
| DRY5440 | MATα yku70Δ∷natMX4 sir1Δ∷kanMX4 ADE2 lys2Δ∷hisG |
| DRY829 | MATα HMR∷ADE2 ade2∷HIS3 his3-11 leu2-3,112 trp1-1, ura3-1 |
| DRY5406 | sir1Δ∷kanMX4 in DRY829 |
| DRY5408 | yku70Δ∷natMX4 in DRY829 |
| DRY826 | MATα HMR∷ADE2 ΔI ade2∷HIS3 his3-11 leu2-3,112 trp1-1, ura3-1 |
| DRY5414 | sir1Δ∷kanMX4 in DRY826 |
| DRY5416 | yku70Δ∷natMX4 in DRY826 |
| DRY815 | MATα HMR-SS∷ADE2 ade2∷HIS3 his3-11 leu2-3,112 trp1-1, ura3-1 |
| DRY5418 | sir1Δ∷kanMX4 in DRY815 |
| DRY5420 | yku70Δ∷natMX4 in DRY815 |
| DRY640 (JRY2334)a | MATα ade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can1-100 |
| DRY707 | MATaADE2 his3-11 leu2-3,112 lys2∷hisG trp1-1 ura3-1 |
| DRY1667 | MATα HMR∷URA3pr-ADE2 ade2∷HIS3 |
| DRY1665 | MATα HMR∷URA3pr-ADE2 ΔI ade2∷HIS3 |
| DRY5404 | MATα HMR∷URA3pr-ADE2 yku70Δ∷natMX4 ade2∷HIS3 |
| DRY5402 | MATα HMR∷URA3pr-ADE2 sir1Δ∷kanMX4 ade2∷HIS3 |
| DRY3348 | MATaTELVIIL∷URA3 ADE2 LYS2 ura3Δ0∷kanMX4 |
| DRY3762 | MATaYKU70-MYC9-TRP1 in DRY3348 |
| DRY5539 | MATα sir4Δ∷natMX4 YKU70-MYC9-TRP1 ADE |
| DRY2805 (PJ69-4A)b | MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7-lacZ |
| DRY5119 | sir1Δ∷kanMX4 in DRY2805 |
| DRY5122 | sir3Δ∷kanMX4 in DRY2805 |
| DRY5125 | sir4Δ∷kanMX4 in DRY2805 |
| DRY5129 | yku70Δ∷kanMX4 in DRY2805 |
| DRY5136 | sir2Δ∷kanMX4 in DRY2805 |
| DRY5137 | yku80Δ∷kanMX4 in DRY2805 |
| DRY5447 | pDR1344 (pGBD-YKU70) and pDR1473 (pGAD-Sir4, aa 1205–1348) in DRY2805 |
| DRY5448 | pDR1473 (pGAD-Sir4, aa 1205–1348) and pDR1406 (pGBD-TRP1-empty) in DRY2805 |
| DRY5449 | pDR1344 (pGBD-YKU70) and pDR1405 (pGAD-LEU2-empty) in DRY2805 |
| DRY5456 | pDR1344 (pGBD-YKU70) and pDR1473 (pGAD-Sir4, aa 1205–1348) in DRY5129 |
| DRY5465 | pDR1344 (pGBD-YKU70) and pDR1473 (pGAD-Sir4, aa 1205–1348) in DRY5137 |
| DRY5474 | pDR1344 (pGBD-YKU70) and pDR1473 (pGAD-Sir4, aa 1205–1348) in DRY5119 |
| DRY5483 | pDR1344 (pGBD-YKU70) and pDR1473 (pGAD-Sir4, aa 1205–1348) in DRY5136 |
| DRY5492 | pDR1344 (pGBD-YKU70) and pDR1473 (pGAD-Sir4, aa 1205–1348) in DRY5122 |
| DRY5501 | pDR1344 (pGBD-YKU70) and pDR1473 (pGAD-Sir4, aa 1205–1348) in DRY5125 |
All strains are isogenic to W303-1a.
From J. Rine.
From E. Craig.
TABLE 2.
Primers used in this study
| Region | Primer |
|---|---|
| Chromatin immunoprecipitation | |
| HMR-E | 1. gttttaacattacgtatcttgtaccc |
| 2. tgcaaatgtggaggaaaagaaatgcg | |
| HMR-I | 1. gaagagacttatgatcaacataattttgc |
| 2. catatacgaaaatgttggtgacatgtaatc | |
| HML-E | 1. taaagttttcggcacggacttatttgg |
| 2. atgtgcgctagatataaaaatcttattgtg | |
| HML-I | 1. cgaaatttctctaatgccagctgagt |
| 2. tgaaaataatcgggtgaaaaagagga | |
| GIT1 | 1. ctaggttagctatggtaacgag |
| 2. gggaatggaaatatatggtatagcg | |
| 0.5 TEL | 1. gacaaataaaaattcagctttttcaag |
| 2. gttcgaatccttaagtaaaacacattc | |
| 7.5 TEL | 1. gtggaaagtatcgagttatgtgtacct |
| 2. gtcattcaaatacagtgggaagtctac | |
| PCR-mediated gene disruption | |
| KU70 disruption | 1. tcaacagtaaagctatgatttgttaagtgactctaagcctgattttaaaacgggaatatt- |
| 2. ataccctaccctaccaaatattgtatgtaacgttatagatatgaaggatttcaatcgtct- | |
| KU70 confirmation | 1. ccgactgaatgcggccgcctcaatttcatgtattaggg |
| 2. ccgactgaatgcggccgcaggttttttgagaatgccgc | |
| KU80 disruption | 1. aaaacctaattaacgagagtgcaggacatatgcacaaataatatatctcacaccataata- |
| 2. tttttttttctctttaactgtggtgacgaaaacataactcaaaggatgttagacctttt- | |
| KU80 confirmation | 1. ccgactgaatgcggccgcgctgcatacataattctc |
| 2. ccgactgaatgcggccgcgcagtcatccagattctg | |
| SIR1 disruption | 1. aaagtttgtcgcgagaatttgggcacatgtgacccggaatgtatattgagtaatataaga- |
| 2. tgaaatgagacatcacccgcttatatgttggtatccataactgataatcttaccaactat- | |
| SIR1 confirmation | 1. ccgactgaatgcggccgcactaagaagccggacctagg |
| 2. ccgactgaatgcggccgcccacccacgcattattgtcgg | |
| SIR2 disruption | 1. aggcatcgcttcggtagacacattcaaaccatttttccctcatcggcacattaaagctgg- |
| 2. tgccatactatgtaaattgatattaatttggcacttttaaattattaaattgccttctac- | |
| SIR2 confirmation | 1. ccgactgaatgcggccgcggtccaggacagccaggacc |
| 2. ccgactgaatgcggccgcgctgttccacctgcccttc | |
| SIR3 disruption | 1. atcaccttccttacaggggtttaagaaagttgttttgttctaacaattggattagctaaa- |
| 2. gaagagactgcatgtgtacataggcatatctatggcggaagtgaaaatgaatgttggtgg- | |
| SIR3 confirmation | 1. ccgactgaatgcggccgccaggggaacaaagtattcggg |
| 2. ccgactgaatgcggccgcgagtcctggaatttccagcgg | |
| SIR4 disruption | 1. gggataaaaaaaaaaaggaagcttcaacccacaataccaaaaaagcgaagaaaacagcca- |
| 2. aaaacgacaaagaaaaacagggtacacttcgttactggtcttttgtagaatgataaaaag- | |
| SIR4 confirmation | 1. ccgactgaatgcggccgccgtccttaaacatgtgcac |
| 2. ccgactgaatgcggccgcggcaaggtcggtttggatgac | |
| pUC1a | -ccggctcgtatgttgtgtgg (attach to 3′-end disruption primer 1) |
| pUC2 a | -cgacgttgtaaaacgacggcc (attach to 3′-end disruption primer 2) |
| Complement to pUC1 | ccacacaacatacgagccgg |
| Complement to pUC2 | ggccgtcgttttacaacgtcg |
| Yeast two-hybrid construction | |
| DHR195 | gcccggaattcatgcgctcagtcactaatgc |
| DHR196 | ctcgcggatccttatatattgaatttcggc |
| DHR239 | gcccggatccgatcgtcgagtgaaacaactc |
| DHR240 | gcccgctgcagtcaatacggttttatctcc |
| Myc epitope tag for YKU70 | |
| DHR265 | gataacatttcgataaaagaagaaaagaagccctttgataaaaagccgaaattcaatatacgtacgctgcaggtcgac |
| DHR266 | ataccctaccctaccaaatattgtatgtaacgttatagatatgaaggatttcaatcgtctatcgatgaattcgagctcg |
All primers are described in the 5′ to 3′ direction.
From D. Rivier.
Strains DRY5430-5433 and DRY5438-5440, used in quantitative mating analysis, were derived from a cross of DRY5397 and DRY5401 (W303-1a; hdf1Δ∷natMX4 sir1Δ∷kanMX4 lys2Δ∷hisG). Strains DRY5406 and DRY5408, DRY5414 and DRY5416, and DRY5418 and DRY5420, used in HMR∷ADE2 color assays, were generated by PCR-mediated gene disruption in strains DRY829, DRY826, and DRY815, respectively. Strains DRY5402 and DRY5404, used in the HMR∷URA3pr-ADE2 growth assays, were generated by PCR-mediated gene disruption in DRY1667. Strains DRY5119, DRY5136, DRY5122, DRY5125, DRY5129, and DRY5137, used in two-hybrid analysis, were generated by PCR-mediated gene disruption of PJ69-4A (DRY2805) (James et al. 1996). These strains were transformed with the two-hybrid plasmids pDR1344 (pGBD-KU70) and pDR1473 (pGAD-SIR4, aa1205–1348) to produce strains DRY5447, DRY5456, DRY5474, DRY5483, DRY5492, and DRY5501. Strain DRY3762 (KU70-MYC9-TRP1), used in chromatin immunoprecipitation (ChIP), was generated by the addition of a sequence that encoded by nine copies of the Myc epitope to the C-terminal end of the native YKU70 gene. In particular, a segment of pYM6 (Knop et al. 1999) was PCR amplified using primers DHR265 and DHR266 and the resting product was transformed into strain DRY3348. PCR confirmation for correct integration was as previously described (Knop et al. 1999). Strain DRY5539 was generated by disrupting SIR4 in DRY5532, a derivative of DRY3762.
Media and genetic manipulations:
Rich medium (YPD) and minimal medium (YM) were as described (Sherman 1991). Medium for red/white colony assays was as described (Gottschling et al. 1990), except no l-aspartic acid was added. The kanMX4 genes were selected for on YPD containing 200 mg/liter G418 and natMX4 genes were selected for on YPD containing 100 mg/liter nourseothricin. Transformation was by a modified lithium–acetate method (Gietz and Schiestl 1991).
Quantitative and patch mating analysis:
Quantitative matings were performed as described previously (Xu et al. 1999). Strains JRY2726 and JRY2728 were used as tester strains. Values reported are the average of a minimum of three independent trials.
Two-hybrid analysis:
The two-hybrid analysis was as described previously (James et al. 1996). An EcoRI/BamHI HDF1 fragment was amplified by PCR using primers DHR195 and DHR196 and cloned into plasmid pDR1406 [pGBD (c1)] to generate plasmid pDR1344. A BamHI/PstI Sir4 (aa 1205–1348) fragment was amplified by PCR using primers DHR239 and DHR240 and cloned into plasmid pDR1405 [pGAD (c1)] to generate plasmid pDR1473. The pGBD and pGAD plasmids were sequentially transformed into the prepared two-hybrid deletion strains.
Chromatin immunoprecipitation:
Chromatin immunoprecipitations of strains DRY3762 and DRY5539 were performed as previously described (Meluh and Broach 1999). Crosslinking was performed at a final concentration of 1% formaldehyde for 15 min. Genomic DNA was sonicated to fragments in a 200- to 400-bp range using both a Diagenode Bioruptor (20; 30-sec pulses with 1-min intervals on ice) and a Branson Sonifier 450 (sonicate 5 min, 20 sec on high, stopping to change the ice bath every 80 sec). A monoclonal mouse 9E11 Myc antibody from Abcam (ab56) was used to precipitate the DNA. Bound DNA was eluted from a Calibiochem Protein G Plus/Protein A Agarose suspension using a 10% BioRad Chelex-100 molecular grade resin solution.
Real-time PCR and data analysis:
Quantitative ChIP analysis was performed on a Corbett Research Rotor-Gene RG3000A instrument using Invitrogen Platinum SYBR Green qPCR SuperMix-UDG as the detection dye. The primers used in the real-time PCR are listed in Table 2 and were screened prior to use for similar melting temperatures and amplification efficiencies. Real-time PCR was carried out as follows: step 1—50° for 2 min; step 2—95° for 2 min; step 3—95° for 15 sec; step 4—53° for 15 sec; step 5—68° for 20 sec (40 cycles: steps 3–5); step 6—melt curve from 68° to 95° and then hold at 4°. The annealing temperature in step 4 was increased to 58° for the HML primers in comparison to the HMR primers to decrease the number of primer dimers. The same amount of DNA, 2.5 μl of each sample and 2.5 μl of 1/1000 dilution of input, was analyzed in duplicate for each primer set to determine a relative increase in DNA from the immunoprecipitation (IP) compared to the input DNA. The CT values were between 20 and 30 cycles, which indicated that the values were within the linear range. The fold increase was then normalized to the 7.5 TEL primer set. The standard error for a minimum of three crosslinks and five IPs was calculated for each strain and primer set.
RESULTS
Ku contributes to silencing at the internal HML and HMR loci:
As an initial test of whether Ku contributes to silencing at HML or HMR, we performed quantitative mating-type assays. Wild-type MATα cells can mate with MATa cells; however, disruption of silencing in MATα cells results in the nonmating phenotype due to the simultaneous expression of the MATα genes and the HMRa genes. Similarly, disruption of silencing in MATa cells results in transcription of the MATa genes and the HMLα genes, also resulting in the nonmating phenotype. To test whether Ku contributes to silencing at HMR, we constructed a MATα strain in which the entire coding region of YKU70 was deleted (yku70Δ) and performed quantitative mating analysis (Table 3). This yku70Δ strain had the same mating efficiency as an isogenic wild-type strain, confirming previous observations that Ku is not required for silencing at HMR. In contrast, an isogenic sir1Δ strain had a slightly reduced mating efficiency of 0.72 relative to the wild-type strain, consistent with previous results (Pillus and Rine 1989). To test whether Ku contributes to the overall efficiency of silencing at HMR, we compared the mating efficiency of a strain lacking both YKU70 and SIR1 (yku70Δ sir1Δ) to the isogenic wild-type strain and to the two isogenic yku70Δ and sir1Δ strains. The mating efficiency of the yku70Δ sir1Δ strain relative to the wild-type strain was 0.036, a 28-fold reduction in silencing relative to the wild-type strain and a 20-fold reduction relative to the sir1Δ strain. These results suggest that yKu70 normally contributes to silencing at the wild-type HMRa locus and, furthermore, that Sir1 and Ku are collectively required for efficient silencing at HMR.
TABLE 3.
Mating efficiencies of strains with different alleles of SIR1 and YKU70
| Mating efficiencies
|
||
|---|---|---|
| Relevant genotype | HMR | HML |
| Wild type | 1.0 | 1.0 |
| yku70Δ∷natMX4 | 1.05 ± 0.11 | 0.81 ± 0.14 |
| sir1Δ∷kanMX4 | 0.72 ± 0.13 | 0.65 ± 0.10 |
| yku70Δ∷natMX4 sir1Δ∷kanMX4 | 3.6 × 10−2 ± 4.0 × 10−3 | 0.26 ± 0.14 |
All strains were isogenic to W303. At least three independent mating assays were performed on all strains and the averages and standard error are reported. MATα strains were used to determine mating efficiencies for HMR, and MATa strains were used to determine mating efficiencies for HML. Strains tested were DRY705 (wild type), DRY5438 (yku70Δ), DRY5439 (sir1Δ), and DRY5440 (yku70Δ sir1Δ) at HMR and DRY5430 (wild type), DRY5431 (yku70Δ), DRY5432 (sir1Δ), and DRY5433 (yku70Δ sir1Δ) at HML.
To test for a role of Ku in silencing HML, quantitative mating analysis was performed on an isogenic set of MATa strains: wild-type, yku70Δ, sir1Δ, and yku70Δ sir1Δ (Table 3). The yku70Δ strain mated with an efficiency of 0.81 relative to the wild-type strain, suggesting that loss of yKu70 function alone is sufficient to result in a slight silencing defect at HML. The mating efficiency of the yku70Δ sir1Δ strain was 0.26 relative to the wild-type strain and was less than either the yku70Δ strain or the sir1Δ strain, further suggesting that yKu70 contributes to silencing at the wild-type HMLα locus.
Silencing is a general mechanism of repression that can inhibit transcription directed by a variety of different promoters. In principle, the results presented above could be due to a role for Ku in specifically acting on the mating-type gene promoters, rather than due to a role for Ku in silencing per se at HML and HMR. As a second test for a role of Ku in silencing at an internal locus, we determined whether loss of yKu70 function resulted in an increase in expression of a reporter gene. The yeast ADE2 gene can serve as a reporter gene that sensitively detects a reduction in the efficiency of silencing when it is inserted into HMR (HMR∷ADE2) (Gottschling et al. 1990; Sussel et al. 1993; Maillet et al. 2001). Yeast cells that do not transcribe ADE2 form red colonies on media containing adenine in contrast to wild-type cells that form white colonies. HMR∷ADE2 cells form red colonies on media containing adenine, whereas HMR∷ADE2 strains in which silencing is disrupted form white colonies, or pink colonies if silencing is partially disrupted (Sussel et al. 1993; Rivier et al. 1999). In addition, we previously determined that this reporter assay is more sensitive to silencing defects than quantitative mating assays (Rivier et al. 1999). As observed previously, an HMR∷ADE2 strain displays the red color phenotype, whereas an isogenic strain in which the SIR1 coding region was deleted (sir1Δ HMR∷ADE2) displays a white phenotype (Figure 1). In contrast, an isogenic strain lacking the entire YKU70 coding region displayed a pink color phenotype, indicating that Ku contributes to silencing at HMR and that Ku's contribution to the overall efficiency of silencing at HMR is less than that of Sir1.
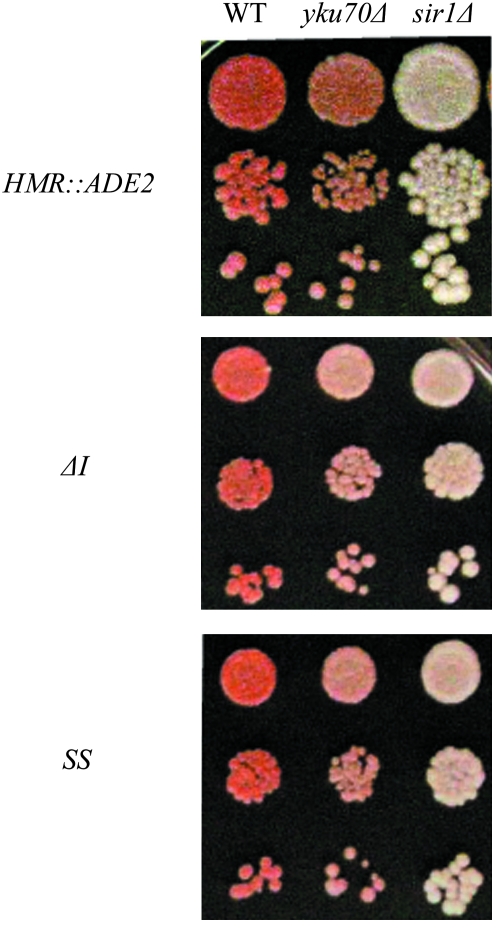
Figure 1.—
yKu70 contributes to silencing of the HMR∷ADE2 reporter gene in three genetic contexts: in the presence of the wild-type (WT) HMR silencers (top), in the absence of the HMR-I silencer (middle), and when HMR is replaced by the synthetic silencer (bottom). (Top) A 10-fold dilution series of strains DRY829 (HMR∷ADE2), DRY5408 (yku70Δ HMR∷ADE2), and DRY5406 (sir1Δ HMR∷ADE2). (Middle) A 10-fold dilution series of strains DRY826 (HMR∷ADE2 ΔI), DRY5416 (yku70Δ HMR∷ADE2 ΔI), and DRY5414 (sir1Δ HMR∷ADE2 ΔI). (Bottom) A 10-fold dilution series of strains DRY815 (HMR-SS∷ADE2), DRY5420 (yku70Δ HMR-SS∷ADE2), and DRY5418 (sir1Δ HMR-SS∷ADE2).
HMR∷ADE2 strains that also have a mutant version of the HMR silencers also provide a sensitive background for monitoring silencing. One such strain lacks the HMR-I silencer (HMR∷ADE2 ΔI) and another contains a reduced function allele of the HMR-E, known as the synthetic silencer (HMR-SS∷ADE2), which has been studied extensively (McNally and Rine 1991). Both the HMR∷ADE2 ΔI strain and the HMR-SS∷ADE2 strain display a lighter color phenotype than wild type, indicating that silencing is partially disrupted in each case. However, both strains can display a red color phenotype if given sufficient time (Figure 1). Deletion of YKU70 from the HMR∷ADE2 ΔI strain resulted in a lighter color phenotype than the isogenic wild-type strain but not as light a color phenotype as an isogenic strain in which SIR1 was deleted (Figure 1). Deletion of YKU70 from the HMR-SS∷ADE2 strain resulted in a lighter color phenotype than the isogenic wild-type strain but not as light a color phenotype as an isogenic strain in which SIR1 was deleted (Figure 1). Collectively, these three related assays indicate that Ku contributes to silencing at HMR, that Ku is not required for silencing at HMR, and that the contribution of Ku to the overall efficiency of silencing at HMR is less than the contribution of Sir1.
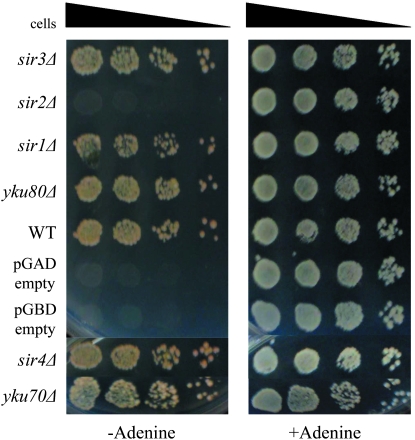
The URA3pr-ADE2 chimeric gene provides an assay for silencing that complements those described above (Rivier et al. 1999). In the case of this chimeric gene, transcription of the ADE2 coding region is driven by the promoter region of the URA3 gene; hence this gene provides the opportunity to study silencing of another type of promoter that is distinct from the promoters of the a-genes, the α-genes, and the ADE2 gene described above. Previously, we determined that the URA3pr-ADE2 reporter gene is silenced when inserted into HMR and that it provides a more sensitive metric of silencing than quantitative mating-type assays (Rivier et al. 1999). Wild-type yeast cells can grow on minimal media that lacks adenine whereas cells that do not express ADE2 cannot. HMR∷URA3pr-ADE2 cells cannot grow on media lacking adenine because the HMR∷URA3pr-ADE2 gene is silenced. Disruption of silencing results in transcription of HMR∷URA3pr-ADE2 and the ability to grow on media lacking adenine. Thus, HMR∷URA3pr-ADE2 provides a gain-of-function phenotype for disruption of silencing. As a third test of whether Ku contributes to silencing at HMR, the entire YKU70 coding region was deleted from an HMR∷URA3pr-ADE2 strain (yku70Δ HMR∷URA3pr-ADE2). As can be seen in Figure 2, the yku70Δ HMR∷URA3pr-ADE2 strain is capable of growth on media lacking adenine, whereas the wild-type HMR∷URA3pr-ADE2 strain is not, providing a third line of evidence that Ku contributes to silencing at HMR.
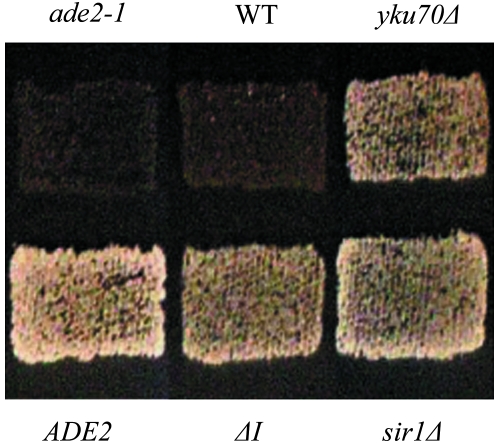
Figure 2.—
Loss of yKu70 function results in expression of the HMR∷URA3pr-ADE2 reporter gene as revealed by growth on media lacking adenine. Strains shown are DRY640 (HMRa ade2-1), DRY707 (HMRa ADE2), DRY1667 (HMR∷URA3pr-ADE2), DRY1665 (HMR∷URA3pr-ADE2 ΔI), DRY5404 (HMR∷URA3pr-ADE2 yku70Δ), and DRY5402 (HMR∷URA3pr-ADE2 sir1Δ).
Collectively, our results indicate that Ku contributes to silencing at HML and HMR. In conjunction with previous observations that Ku is required for telomeric silencing, these results also reveal that Ku makes a contribution to silencing at HML and HMR different from the one it does at the telomeres. In particular, Ku is required for telomeric silencing, whereas Ku contributes to the overall efficiency of silencing at HML and HMR but is not absolutely required for silencing of these internal loci.
Ku binds HML and HMR:
In principle, Ku could contribute to silencing at HML and HMR by acting directly as a component of the silencing machinery. Alternatively, since Ku regulates multiple chromosomal processes, it is possible that loss of Ku function disrupts the overall physiology of the cell in such a way that silencing is compromised indirectly (Bertuch and Lundblad 2003; Daley et al. 2005; Fisher and Zakian 2005). These two possibilities make distinct predictions. If Ku acts directly in silencing at HML and HMR, it is expected that Ku would bind these loci. In contrast, if loss of Ku function disrupts silencing at HML and HMR indirectly, it is predicted that Ku would exert its effect on silencing by acting at some other chromosomal locus or independently of DNA. In this case, Ku is not expected to bind to HML or HMR.
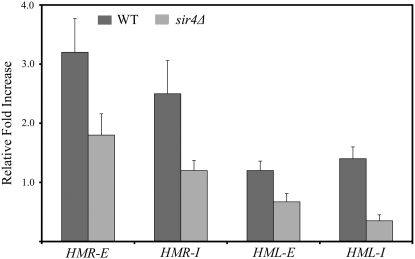
To determine whether Ku binds to HML or HMR, we performed quantitative ChIP experiments. The endogenous copy of the YKU70 gene was modified so that the yKu70 protein produced would contain nine copies of the myc epitope at the C terminus (Knop et al. 1999). The resulting strain (YKU70-MYC9) displayed no growth defects and a telomeric reporter gene was silenced to the same extent in the YKU70-MYC9 strain as in an isogenic wild-type strain, indicating that the epitope tag did not compromise Ku function (data not shown). To determine whether Ku bound to HMR, we performed quantitative ChIP analysis of binding of yKu70-9myc to HMR relative to GIT1, a gene that is adjacent to HMR but is outside the silenced region and is not bound by the silencing machinery. Each value reported was the average of a minimum of three independent crosslinking reactions and five immunoprecipitations. Using a set of primers specific for HMR-E, we found that the HMR-E DNA was enriched 3.2-fold (±0.56) in the immunoprecipitated DNA relative to GIT1 (Figure 3). That HMR-E DNA is enriched to a statistically significant extent relative to GIT1 reveals that yKu70 binds specifically to HMR in the vicinity of the HMR-E silencer. However, the absolute magnitude of the enrichment, 3.2-fold in this case, which is modest relative to quantitative enrichment values for some DNA-binding proteins, has no unique interpretation. For example, modest enrichment values could result from transient association of a protein with a specific region or could be the result of the protein being tethered to a region via protein–protein interactions rather than by directly contacting the DNA. Alternatively, the epitope could be partially masked when the protein is bound to DNA, resulting in inefficient immunoprecipition. Therefore, while the data presented here reveal that Ku binds HMR in the vicinity of HMR-E, they do not provide a detailed insight into the nature or dynamics of binding. Using the set of primers specific for HMR-I, we found that HMR-I DNA was enriched 2.5-fold (±0.56) relative to GIT1 (Figure 3). This observation indicates that the enrichment of HMR DNA observed in the immunoprecipitated DNA is independent of the set of primers used and provides further evidence that Ku binds to HMR. As described above, we selected GIT1 as a control site because GIT1 is not silenced. However, since Ku has been implicated in chromosomal processes other than silencing, it is not formally known that Ku does not associate with GIT1. We therefore repeated each of the quantitative ChIP experiments described above using primers to a second control site that is 7.5 kb from the chromosome 6R telomere, which does not bind Ku or other silencing proteins (Martin et al. 1999). In the case of both HMR-E and HMR-I, we detected a modest but statistically significant enrichment of HMR DNA in the immunoprecipitate relative to the chromosome 6R control site (Figure 4). The observation that enrichment HMR DNA in the immunoprecipitate relative to both a GIT1 and the chromosome 6R site indicates that our results are independent of the control site used and provide another line of evidence that Ku binds to HMR.
Figure 3.—
yKu70 is recruited to HMR and HMR silencer regions in a Sir4-dependent manner as revealed by chromatin immunoprecipitation and real-time quantitative PCR. Fold enrichment values are normalized to the 3′ GIT1 gene adjacent to HMR for the wild-type strain DRY3762 (darkly shaded bar) and the sir4Δ strain DRY5539, (lightly shaded bar).
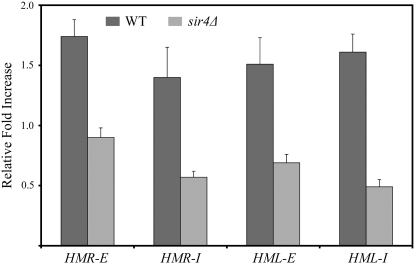
Figure 4.—
yKu70 is recruited to HMR and HML silencer regions in a Sir4-dependent manner as revealed by chromatin immunoprecipitation and real-time quantitative PCR. Fold enrichment values are normalized to the 7.5 TEL on chromosome 6R for the wild-type strain DRY3762 (darkly shaded bar) and the sir4Δ strain DRY5539 (lightly shaded bar).
To determine whether Ku binds HML in vivo, we also used two sets of primers, one specific for HML-E and the other specific for HML-I. These two sets of primers were individually used in quantitative PCR on the same DNA samples used for HMR above and were run in parallel to the reactions described above. For both HML-E and HML-I, the ratio of the immunoprecipitated DNA relative to the input DNA was calculated and normalized independently to the two control sites described above. We found that HML-E DNA was enriched in the immunoprecipitated DNA to a statistically significant extent relative to both control sites, revealing that yKu70 binds specifically to HML in the vicinity of the HML-E silencer (Figure 3; Figure 4). Similarly, HML-I DNA was enriched in the immunoprecipitated DNA to a statistically significant extent relative to both control sites, providing another line of evidence that yKu70 binds specifically to HML (Figures 3 and 4). Collectively, these results reveal that yKu70, and presumably the Ku heterodimer, bind specifically to the two internal silent loci HML and HMR.
Binding of yKu70 to HML and HMR is dependent on Sir4:
In principle, there are two mechanisms by which Ku could bind HML and HMR. Ku could bind DNA directly or, alternatively, Ku could bind HML and HMR as a result of binding to a protein or protein complex that directly binds DNA at HML and HMR. If Ku binds the internal silent loci via protein–protein interactions, it is likely that Sir4 plays a central role in that process since Ku interacts with Sir4 in two-hybrid assays. To test this possibility, we performed quantitative ChIP experiments on the YKU70-MYC9 strain described above and on an isogenic strain in which the entire SIR4 coding region was deleted (YKU70-MYC9 sir4Δ). In analyzing HMR, we found that HMR-E DNA was enriched 3.2-fold (±0.56) in the immunoprecipitate from the wild-type YKU70-MYC9 strain relative to GIT1 but had an ∼2-fold reduced enrichment in the YKU70-MYC9 sir4Δ strain (1.85-fold ± 0.36) relative to GIT1, revealing that the binding of Ku to HMR is dependent to a large extent on Sir4 (Figure 3). Furthermore, HMR-I DNA was enriched 2.5-fold (±0.56) in the immunoprecipitate from the wild-type YKU70-MYC9 strain but was not enriched in the YKU70-MYC9 sir4Δ strain (1.16-fold ± 0.17) relative to GIT1, providing further evidence that binding of Ku to HMR is dependent on Sir4 (Figure 3). Similarly, HMR-E and HMR-I DNA was enriched in the immunoprecipitate from the wild-type YKU70-MYC9 strain but was not enriched in the YKU70-MYC9 sir4Δ strain when the chromosome 6R site was used as the negative control (Figure 4).
To determine whether binding of Ku to HML is dependent on Sir4 as it is at HMR, we performed quantitative PCR using HML primers on this same set of immunoprecipitated DNA samples. Again, we compared the fold enrichment of each of the HML silencers relative to both the GIT1 and the telomere 6R control sites in the wild-type YKU70-MYC9 strain and the YKU70-MYC9 sir4Δ strain. As can be seen in Figures 3 and 4, the enrichment of both HML-E and HML-I DNA in the immunoprecipitate from the sir4Δ strain is reduced to a statistically significant extent relative to the wild-type strain when either GIT1 (Figure 3) or the chromosome 6R site (Figure 4) is used as the control. Collectively, analysis of Ku binding to each of the four silencers relative to two control sites in the wild-type and sir4Δ strains provide eight lines of evidence that Ku binds to HML and HMR and that binding of Ku to these loci is dependent on Sir4. In these experiments a value of 1.0 for the ratio of enrichment in the wild-type strain relative to the sir4Δ strain is expected if Sir4 is required for Ku binding to HML or HMR, whereas a ratio that is statistically >1.0 but less than the enrichment in the wild-type strain would indicate that Sir4 contributes to the overall efficiency of Ku binding to HML or HMR but is not absolutely required for Ku binding. All eight experimental observations support the conclusion that binding of Ku to HML and HMR is more efficient in the presence of Sir4 than in its absence, and seven of the eight observations support the model that Ku does not bind HML or HMR in the absence of Sir4. Thus, the preponderance of evidence presented here suggests that Sir4 is required for binding of Ku to HML and HMR; however, we cannot rule out the possibility that there is some residual binding of Ku to HML and HMR that is independent of Sir4.
The two-hybrid interaction between yKu70 and Sir4 is dependent on Sir2 but not on Sir1, Sir3, or yKu80:
Our discovery that Ku plays a role in silencing of HML and HMR, that Ku binds HML and HMR, and that binding of Ku to these internal loci is dependent on Sir4 provides additional evidence that the two-hybrid interaction between Sir4 and Ku described previously (Tsukamoto et al. 1997; Roy et al. 2004) reflects bona fide protein–protein interactions that occur in vivo. To gain further insight into the nature of the Sir4–yKu70 interaction, we tested whether any of the other Sir proteins influence the interaction between yKu70 and Sir4. We constructed one plasmid that contained YKU70 fused to the coding region of the Gal4 DNA-binding domain (pGBD-YKU70) and a second plasmid in which SIR4 was fused to the GAL4 activation domain (pGAD-SIR4). Introduction of both pGBD-YKU70 and pGAD-SIR4 into a yeast two-hybrid reporter strain resulted in expression of an ADE2 two-hybrid reporter gene as indicated by adenine prototrophy (Figure 5). The ADE2 reporter gene was not expressed if the pGBD-YKU70 plasmid and the pGAD plasmid lacking SIR4 was introduced into the reporter strain, nor was the reporter gene expressed if the pGBD plasmid lacking the YKU70 fusion was introduced into the reporter strain along with the pGAD-SIR4 plasmid, indicating that expression of the reporter gene is dependent on a specific interaction between yKu70 and Sir4. These results confirm the previously observed two-hybrid interaction between yKu70 and Sir4, and since the plasmids and strains used here differ from those used previously, it indicates that the yKu70–Sir4 two-hybrid interaction is not dependent on any particular strain, any particular set of plasmids, or any particular reporter gene.
Figure 5.—
The two-hybrid interaction of yKu70 and Sir4 is dependent on Sir2, but not on Sir1, Sir3, yKu80, or the endogenous copies of Sir4 or yKu70. Strains shown are DRY5492 (sir3Δ), DRY5483 (sir2Δ), DRY5474 (sir1Δ), DRY5465 (yku80Δ), DRY5447 (wild-type pGBD-HDF1 and pGAD-Sir4, aa 1205–1348), DRY5449 (pGAD-empty), DRY5448 (pGBD-empty), DRY5492 (sir4Δ), and DRY5456 (yku70Δ).
To determine whether any of the Sir proteins were required for the interaction of yKu70 with Sir4, we created a series of isogenic two-hybrid strains, each lacking one of the SIR genes (sirΔ strains), and determined whether the pGBD-YKU70 and pGAD-SIR4 plasmids were capable of driving expression of the reporter gene in those strains. As shown in Figure 5, the reporter gene was expressed in sir1Δ and sir3Δ strains at the same level as in the wild-type strain, indicating that neither Sir1 nor Sir3 contributed significantly to the two-hybrid interaction between yKu70 and Sir4. In contrast, expression of the two-hybrid reporter gene was abolished in the sir2Δ strain. This observation revealed that Sir2 is required for the in vivo two-hybrid interaction between yKu70 and Sir4.
To determine whether Ku80 was required for the interaction of Ku70 with Sir4, we created an isogenic two-hybrid reporter strain that lacked the entire coding region of YKU80 (yku80Δ). Introduction of the pGBD-YKU70 and pGAD-SIR4 plasmids into the yku80Δ strain resulted in expression of the two-hybrid reporter gene at a level similar to that of the wild-type strain; hence, yKu70 can interact with Sir4 independently of yKu80 (Figure 5). Since yKu80 is required for telomeric silencing and can contribute to nucleation when tethered to a defective silencer (Martin et al. 1999; Mishra and Shore 1999), the simplest interpretation of these observations is that yKu70 and yKu80 form the classical Ku heterodimer, which plays a role in silencing in wild-type cells and that each of the yKu70 and yKu80 subunits of the dimer provide sufficient protein–protein contacts to interact with Sir4 in the two-hybrid assay in the absence of the other subunit.
We also reasoned that if the interaction between yKu70 and Sir4 involved multimers of either yKu70 or Sir4, it might be possible that the endogenous yKu70 or Sir4 proteins influence the interaction of GBD-yKu70 with GAD-Sir4. To test this possibility, we created an isogenic set of two-hybrid strains that lacked either YKU70 (yku70Δ) or SIR4 (sir4Δ). As shown in Figure 5, introduction of the pGBD-YKU70 and pGAD-SIR4 into either the yku70Δ strain or the sir4Δ strain resulted in the same level of expression of the reporter gene as in wild-type cells, suggesting that either yKu70 and Sir4 function as monomers or the GBD–yKu70 and GAD–Sir4 fusion proteins are capable of efficient multimerization.
DISCUSSION
The key discoveries presented here are that the DNA end-binding protein Ku contributes to silencing at HML and HMR, that Ku binds these internal loci, and that binding of Ku to HML and HMR is dependent on Sir4. The observation that Ku binds HML and HMR suggests that Ku plays a direct role in silencing at both of these loci as it does at the telomeres. While these conclusions were drawn from our analysis of yKu70, recent evidence indicates that yKu80 also contributes to silencing of HML and HMR and physically associates with both of these loci (Patterson and Fox 2008). We therefore propose that the Ku heterodimer is a general regulator of silencing that acts directly at each of the known silent loci in yeast rather than as a locus-specific regulator that acts only at the telomeres. It remains to be determined exactly what role Ku plays in silencing at HML and HMR. Perhaps the simplest model is that Ku is a subunit of the protein complexes that bind the silencers and plays a role in the nucleation of silencing at HML and HMR as it does at the telomeres; however, we cannot rule out the possibility that Ku is a structural component of silent chromatin at HML and HMR.
Our observation that binding of Ku to HML and HMR is dependent on Sir4 suggests that Ku is tethered to these loci via protein–protein interactions rather than by contacting the DNA directly as a result of any sequence-specific or structure-specific binding property intrinsic to Ku itself. Taken together with previous observations, our results suggest that Ku is endowed with two modes of binding to silent regions of DNA: it can bind telomeric regions directly via its DNA end-binding activity and it can bind HML and HMR as a result of protein–protein interactions. Previous observations have also implicated Ku in binding to internal chromosomal loci and suggest that Ku may play a role in the activation of transcription and possibly in initiation of replication (Barnes and Rio 1997; Ruiz et al. 1999; Novac et al. 2001; Walker et al. 2001; Schild-Poulter et al. 2003; Sibani et al. 2005; Grote et al. 2006; Shi et al. 2007; Rampakakis et al. 2008). The data presented here support and extend the data indicating that Ku binds to internal chromosomal loci and broaden our knowledge of the number of processes in which Ku plays a role at internal loci to include silencing.
Our observation that Ku binds HML and HMR also provides a plausible resolution to a paradox associated with the internal silent loci. Each of the three silent regions in yeast—HML, HMR, and the telomeres—localize to the nuclear periphery, as do regions of heterochromatin in other eukaryotes. Ku plays a role in the nuclear localization of HML, HMR, and the telomeres (Gartenberg et al. 2004; Taddei and Gasser 2004; Taddei et al. 2004). Since Ku binds telomeres, it is thought that Ku plays a direct and central role in nuclear localization of the telomeres. However, given the previous idea that Ku does not bind HML or HMR, it was not clear how Ku could mediate nuclear localization of these loci. Our discovery that Ku binds HML and HMR suggests that Ku directly participates in localization of these loci to the nuclear periphery.
Acknowledgments
We thank K. Replogle for help with initial stages of this work and L. Valenzuela and N. Dhillon for technique instruction, helpful comments, and suggestions. This work was supported in part by a National Institutes of Health (NIH) grant (GM078068 to R.T.K.). The initial stages of this work were also supported by an NIH grant (GM52103 to D.H.R.).
References
- Barnes, G., and D. Rio, 1997. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94 867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute and C. Cullin, 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch, A. A., and V. Lundblad, 2003. Which end: dissecting Ku's function at telomeres and double-strand breaks. Genes Dev. 17 2347–2350. [DOI] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., L. Breeden, J. Abraham, R. Sternglanz and K. Nasmyth, 1985. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 41 41–48. [DOI] [PubMed] [Google Scholar]
- Buchman, A. R., W. J. Kimmerly, J. Rine and R. D. Kornberg, 1988. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol. 8 210–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley, J. M., P. L. Palmbos, D. Wu and T. E. Wilson, 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39 431–451. [DOI] [PubMed] [Google Scholar]
- Fisher, T. S., and V. A. Zakian, 2005. Ku: a multifunctional protein involved in telomere maintenance. DNA Rep. 4 1215–1226. [DOI] [PubMed] [Google Scholar]
- Gartenberg, M. R., F. R. Neumann, T. Laroche, M. Blaszczyk and S. M. Gasser, 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119 955–967. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., and R. H. Schiestl, 1991. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast 7 253–263. [DOI] [PubMed] [Google Scholar]
- Gilbert, D. M., 2002. Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell Biol. 14 377–383. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian, 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63 751–762. [DOI] [PubMed] [Google Scholar]
- Gravel, S., M. Larrivâee, P. Labrecque and R. J. Wellinger, 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280 741–744. [DOI] [PubMed] [Google Scholar]
- Grote, J., S. Kèonig, D. Ackermann, C. Sopalla, M. Benedyk et al., 2006. Identification of poly(ADP-ribose)polymerase-1 and Ku70/Ku80 as transcriptional regulators of S100A9 gene expression. BMC Mol. Biol. 7 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein, M., 1997. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 9 383–387. [DOI] [PubMed] [Google Scholar]
- Haber, J. E., and J. P. George, 1979. A mutation that permits the expression of normally silent copies of mating-type information in Saccharomyces cerevisiae. Genetics 93 13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz, I., J. N. Strathern, J. B. Hicks and J. Rine, 1977. Mating type interconversion in yeast and its relationship to development in higher eucaryotes, pp. 193–202 in ICN-UCLA Symposia on Molecular and Cellular Biology: Molecular Approaches to Eucaryotic Genetic Systems, edited by G. Wilcox, J. Abelson and C. F. Fox. Academic Press, New York.
- Hofmann, J. F., T. Laroche, A. H. Brand and S. M. Gasser, 1989. RAP-1 factor is necessary for DNA loop formation in vitro at the silent mating type locus HML. Cell 57 725–737. [DOI] [PubMed] [Google Scholar]
- James, P., J. Halladay and E. A. Craig, 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar, A. J., and S. Fogel, 1979. Activation of mating type genes by transposition in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 76 4539–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor et al., 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15 963–972. [DOI] [PubMed] [Google Scholar]
- Kurtz, S., and D. Shore, 1991. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 5 616–628. [DOI] [PubMed] [Google Scholar]
- Kyrion, G., K. Liu, C. Liu and A. J. Lustig, 1993. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 7 1146–1159. [DOI] [PubMed] [Google Scholar]
- Laroche, T., S. G. Martin, M. Gotta, H. C. Gorham, F. E. Pryde et al., 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8 653–656. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., N. M. Wilson, M. E. Petracek and J. Berman, 1989. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr. Genet. 16 225–239. [DOI] [PubMed] [Google Scholar]
- Loo, S., and J. Rine, 1995. Silencing and heritable domains of gene expression. Annu. Rev. Cell Dev. Biol. 11 519–548. [DOI] [PubMed] [Google Scholar]
- Louis, E. J., 1995. The chromosome ends of Saccharomyces cerevisiae. Yeast 11 1553–1573. [DOI] [PubMed] [Google Scholar]
- Mahoney, D. J., and J. R. Broach, 1989. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol. Cell. Biol. 9 4621–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet, L., F. Gaden, V. Brevet, G. Fourel, S. G. Martin et al., 2001. Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep. 2 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. G., T. Laroche, N. Suka, M. Grunstein and S. M. Gasser, 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97 621–633. [DOI] [PubMed] [Google Scholar]
- McNally, F. J., and J. Rine, 1991. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 11 5648–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P. B., and J. R. Broach, 1999. Immunological analysis of yeast chromatin. Methods Enzymol. 304 414–430. [DOI] [PubMed] [Google Scholar]
- Millar, C. B., and M. Grunstein, 2006. Genome-wide patterns of histone modifications in yeast. Nature Rev. Mol. Cell Biol. 7 657–666. [DOI] [PubMed] [Google Scholar]
- Mishra, K., and D. Shore, 1999. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr. Biol. 9 1123–1126. [DOI] [PubMed] [Google Scholar]
- Mondoux, M. A., and V. A. Zakian, 2007. Subtelomeric elements influence but do not determine silencing levels at Saccharomyces cerevisiae telomeres. Genetics 177 2541–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti, P., K. Freeman, L. Coodly and D. Shore, 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8 2257–2269. [DOI] [PubMed] [Google Scholar]
- Novac, O., D. Matheos, F. D. Araujo, G. B. Price and M. Zannis-Hadjopoulos, 2001. In vivo association of Ku with mammalian origins of DNA replication. Mol. Biol. Cell 12 3386–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger et al., 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8 657–660. [DOI] [PubMed] [Google Scholar]
- Patterson, E. E., and C. A. Fox, 2008. The Ku complex in silencing the cryptic mating-type loci of Saccharomyces cerevisiae. Genetics 180 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus, L., and J. Rine, 1989. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell 59 637–647. [DOI] [PubMed] [Google Scholar]
- Pirrotta, V., and D. S. Gross, 2005. Epigenetic silencing mechanisms in budding yeast and fruit fly: different paths, same destinations. Mol. Cell 18 395–398. [DOI] [PubMed] [Google Scholar]
- Rampakakis, E., D. Di Paola and M. Zannis-Hadjopoulos, 2008. Ku is involved in cell growth, DNA replication and G1-S transition. J. Cell Sci. 121 590–600. [DOI] [PubMed] [Google Scholar]
- Replogle, K., L. Hovland and D. H. Rivier, 1999. Designer deletion and prototrophic strains derived from Saccharomyces cerevisiae strain W303-1a. Yeast 15 1141–1149. [DOI] [PubMed] [Google Scholar]
- Ribes-Zamora, A., I. Mihalek, O. Lichtarge and A. A. Bertuch, 2007. Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat. Struct. Mol. Biol. 14 301–307. [DOI] [PubMed] [Google Scholar]
- Rine, J., J. N. Strathern, J. B. Hicks and I. Herskowitz, 1979. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics 93 877–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier, D. H., J. L. Ekena and J. Rine, 1999. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics 151 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, R., B. Meier, A. D. McAinsh, H. M. Feldmann and S. P. Jackson, 2004. Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J. Biol. Chem. 279 86–94. [DOI] [PubMed] [Google Scholar]
- Ruiz, M. T., D. Matheos, G. B. Price and M. Zannis-Hadjopoulos, 1999. OBA/Ku86: DNA binding specificity and involvement in mammalian DNA replication. Mol. Biol. Cell 10 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72 481–516. [DOI] [PubMed] [Google Scholar]
- Schild-Poulter, C., D. Matheos, O. Novac, B. Cui, W. Giffin et al., 2003. Differential DNA binding of Ku antigen determines its involvement in DNA replication. DNA Cell Biol. 22 65–78. [DOI] [PubMed] [Google Scholar]
- Schwaiger, M., and D. Schèubeler, 2006. A question of timing: emerging links between transcription and replication. Curr. Opin. Genet. Dev. 16 177–183. [DOI] [PubMed] [Google Scholar]
- Shahbazian, M. D., and M. Grunstein, 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76 75–100. [DOI] [PubMed] [Google Scholar]
- Sherman, F., 1991. Getting started with yeast. Methods Enzymol. 194 3–21. [DOI] [PubMed] [Google Scholar]
- Shi, L., D. Qiu, G. Zhao, B. Corthesy, S. Lees-Miller et al., 2007. Dynamic binding of Ku80, Ku70 and NF90 to the IL-2 promoter in vivo in activated T-cells. Nucleic Acids Res. 35 2302–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore, D., 1994. RAP1: a protean regulator in yeast. Trends Genet. 10 408–412. [DOI] [PubMed] [Google Scholar]
- Shore, D., and K. Nasmyth, 1987. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51 721–732. [DOI] [PubMed] [Google Scholar]
- Sibani, S., G. B. Price and M. Zannis-Hadjopoulos, 2005. Decreased origin usage and initiation of DNA replication in haploinsufficient HCT116 Ku80+/− cells. J. Cell Sci. 118 3247–3261. [DOI] [PubMed] [Google Scholar]
- Sussel, L., D. Vannier and D. Shore, 1993. Epigenetic switching of transcriptional states: cis- and trans-acting factors affecting establishment of silencing at the HMR locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 13 3919–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei, A., and S. M. Gasser, 2004. Multiple pathways for telomere tethering: functional implications of subnuclear position for heterochromatin formation. Biochim. Biophys. Acta 1677 120–128. [DOI] [PubMed] [Google Scholar]
- Taddei, A., F. Hediger, F. R. Neumann, C. Bauer and S. M. Gasser, 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P. B., and S. Henikoff, 2006. Spreading of silent chromatin: inaction at a distance. Nat. Rev. Genet. 7 793–803. [DOI] [PubMed] [Google Scholar]
- Teo, S. H., and S. P. Jackson, 2001. Telomerase subunit overexpression suppresses telomere-specific checkpoint activation in the yeast yku80 mutant. EMBO Rep. 2 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham, W. H., and V. A. Zakian, 2002. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene 21 512–521. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, Y., J. Kato and H. Ikeda, 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388 900–903. [DOI] [PubMed] [Google Scholar]
- Valenzuela, L., N. Dhillon, R. N. Dubey, M. R. Gartenberg and R. T. Kamakaka, 2008. Long-range communication between the silencers of HMR. Mol. Cell. Biol. 28 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pèohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Walker, J. R., R. A. Corpina and J. Goldberg, 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412 607–614. [DOI] [PubMed] [Google Scholar]
- Xu, E. Y., S. Kim, K. Replogle, J. Rine and D. H. Rivier, 1999. Identification of SAS4 and SAS5, two genes that regulate silencing in Saccharomyces cerevisiae. Genetics 153 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]