Abstract
The regulation of chemoreceptor (CR) gene expression by environmental signals and internal cues may contribute to the modulation of multiple physiological processes and behavior in Caenorhabditis elegans. We previously showed that KIN-29, a homolog of salt-inducible kinase, acts in sensory neurons to regulate the expression of a subset of CR genes, as well as sensory behaviors. Here we show that the cGMP-dependent protein kinase EGL-4 acts partly in parallel with KIN-29 to regulate CR gene expression. Sensory inputs inhibit both EGL-4 and KIN-29 functions, and KIN-29 function is inhibited in turn by cAMP-dependent protein kinase (PKA) activation. EGL-4 and KIN-29 regulate CR gene expression by antagonizing the gene repression functions of the class II HDAC HDA-4 and the MEF-2 transcription factor, and KIN-29, EGL-4, and PKA target distinct residues in HDA-4 to regulate its function and subcellular localization. While KIN-29 acts primarily via MEF-2/HDA-4 to regulate additional sensory signal-regulated physiological processes and behaviors, EGL-4 acts via both MEF-2-dependent and -independent pathways. Our results suggest that integration of complex sensory inputs via multiple signaling pathways allows animals to precisely regulate sensory gene expression, thereby appropriately modulating physiology and behavior.
CHEMOSENSORY signals regulate multiple aspects of animal development, physiology, and behavior. For example, chemical cues permit successful foraging in social insects, regulate sexual maturation in rodents, and provide information about reproductive status to potential mates (Dulac and Torello 2003; Jackson and Ratnieks 2006). Food-derived chemosensory cues signal palatability, are critical for the regulation of appetitive and feeding behaviors, and have been shown to regulate life span in both Drosophila and Caenorhabditis elegans (Apfeld and Kenyon 1999; Schwartz et al. 2000; Alcedo and Kenyon 2004; Rolls 2005; Libert et al. 2007). It is therefore essential that animals precisely regulate their ability to sense and respond to environmental chemicals.
C. elegans provides an excellent system in which to explore the neuronal and molecular mechanisms by which animals sense and respond to chemical cues. Sensory signals from food regulate nearly all aspects of C. elegans physiology, behavior, and development, including the regulation of entry/exit from the alternate dauer developmental stage, male mating, body size, locomotion, egg laying, longevity, and fat storage (Golden and Riddle 1984a,b; Apfeld and Kenyon 1999; Sawin et al. 2000; Waggoner et al. 2000; Fujiwara et al. 2002; Lanjuin and Sengupta 2002; Mak et al. 2006; Gruninger et al. 2008). As in other animals, chemosensory behaviors are plastic and are regulated by prior experience and external and internal cues (Sawin et al. 2000; Gray et al. 2005; Zhang et al. 2005; Shtonda and Avery 2006). Chemicals are sensed by a small number of chemosensory neurons, each of which expresses multiple candidate seven transmembrane domain chemoreceptors (CRs) and other signaling proteins (Bargmann 2006). The expression of each CR gene is regulated in a complex manner by multiple developmental and environmental cues, suggesting that regulated expression of CR gene subsets may partly underlie the modulation of chemosensory behaviors in C. elegans (Peckol et al. 2001; Lanjuin and Sengupta 2002; Nolan et al. 2002; van der Linden et al. 2007).
We previously showed that the KIN-29 Ser/Thr kinase acts in C. elegans sensory neurons to regulate chemosensory signal-dependent development and behavior (Lanjuin and Sengupta 2002). kin-29 mutants are small and exhibit deregulated entry into the dauer developmental stage, as well as altered food-regulated foraging behaviors (Lanjuin and Sengupta 2002; Maduzia et al. 2005). All mutant phenotypes can be rescued by expression of kin-29 only in sensory neurons (Lanjuin and Sengupta 2002). We found that KIN-29 regulates the expression of a subset of CR genes in multiple chemosensory neuron types, suggesting that altered expression of CR genes may lead to the inability of kin-29 mutants to correctly sense and transduce food signals (Lanjuin and Sengupta 2002). KIN-29 directly phosphorylates and antagonizes the HDA-4 class II histone deacetylase (HDAC), which acts via the MEF-2 transcription factor to repress gene expression (van der Linden et al. 2007). Intriguingly, KIN-29 is a member of the salt-inducible kinase (SIK) family (Wang et al. 1999), members of which have been implicated in the regulation of metabolism and gluconeogenic gene expression in response to fasting and feeding (Koo et al. 2005; Dentin et al. 2007; Wang et al. 2008). The related AMP-activated kinases (AMPKs) have also been implicated in similar food-regulated pathways, and AMPK activity in the hypothalamus has been suggested to regulate food intake and body weight (Minokoshi et al. 2004; Kahn et al. 2005; Kim and Lee 2005). Thus, the overall functions of these kinases in the regulation of food-dependent behaviors may be conserved across phyla.
In addition to KIN-29, the EGL-4 cGMP-dependent protein kinase (PKG) also regulates chemosensory signal-dependent behaviors in C. elegans. egl-4 mutants exhibit defects in food-regulated motor behaviors, body size, fat storage, chemosensation, and regulation of dauer entry, and these phenotypes can be largely rescued by EGL-4 activity in sensory neurons (Trent et al. 1983; Daniels et al. 2000; Fujiwara et al. 2002; L'Etoile et al. 2002; Hirose et al. 2003; Nakano et al. 2004; Raizen et al. 2006). Recently, EGL-4 has also been shown to act in sensory neurons to regulate quiescence associated with developmental stage transitions (Raizen et al. 2008), as well as food-induced quiescence behavior proposed to result from satiety (You et al. 2008). Interestingly, PKG function is also required for food-regulated behaviors in Drosophila and honeybees, suggesting that, similar to SIKs, a role for PKG in the sensation and transduction of food signals may be conserved (Osborne et al. 1997; Ben-Shahar et al. 2002; Kaun et al. 2007). Targets of EGL-4 required for the regulation of these behaviors are largely unknown.
Here we show that cGMP signaling acts via EGL-4 to regulate CR gene expression in C. elegans. We find that cAMP signaling inhibits KIN-29 activity and that the cAMP- and cGMP-signaling pathways act partly in parallel to regulate CR gene expression. Both KIN-29 and EGL-4 functions are downregulated by sensory inputs. Loss-of-function (lf) mutations in HDA-4 and MEF-2 fully suppress the CR gene expression phenotypes of both kin-29 and egl-4 mutants, and we identify potential protein kinase (PKA) and PKG target sites in HDA-4 required for its subcellular localization and function. We show that while KIN-29 acts primarily via MEF-2 and HDA-4 in the regulation of multiple physiological processes, EGL-4 acts via both MEF-2-dependent and -independent pathways. Taken together, our results suggest that regulation of distinct, but partly overlapping sets of CR and other sensory genes by KIN-29 and EGL-4 may contribute to the correct modulation of behavior and development by chemosensory cues.
MATERIALS AND METHODS
Strains:
Animals were grown as previously described (Brenner 1974). The wild-type strain used was C. elegans variety Bristol, strain N2. The following mutant strains were obtained from the Caenorhabditis Genetics Center unless indicated otherwise: MT1074 egl-4(n479) IV, DA521 egl-4(ad405sd) IV, JT195 daf-11(sa195) V (from Jim Thomas), PY1476 kin-29(oy38) X (lab collection), PY1988 kin-29 (oy39) X (lab collection), KG532 kin-2(ce179) X, DR47 daf-11(m47) V, CX2065 odr-1(n1936), PR811 osm-6(p811) V, CB1124 che-3(e1124) I, CB1265 unc-104(e1256) II, CB169 unc-31(e169) IV, KG524 gsa-1(ce94gf) I, KG522 acy-1(md1756gf) III, RB853 fkh-2(ok683) X, KM134 mef-2(gv1) I, and PY4111 hda-4(oy57) X (lab collection). Integrated strains used were CX3553 lin-15(n765ts); kyIs104[str-1p∷gfp+lin-15(+)] X (Troemel et al. 1997), PY1058 lin-15(n765ts); oyIs14[sra-6p∷gfp+lin-15(+)] V (Sarafi-Reinach et al. 2001), CX3596 lin-15(n765ts); kyIs128[str-3p∷gfp+lin-15(+)] X (Peckol et al. 2001), PY3018 oyIs41(kin-29p∷kin-29∷GFP) IV (Lanjuin and Sengupta 2002), and PY6220 oyIs68(hda-4p∷hda-4∷GFP+unc-122∷dsRed) (van der Linden et al. 2007). Double-mutant strains were constructed using standard methods, and the presence of each mutation was confirmed by amplification and/or sequencing.
Real-time qRT–PCR:
Total RNA was isolated from a growth-synchronized population of adult animals and reverse transcribed using oligo(dT) primers. Real-time quantitative reverse transcription–PCR (qRT–PCR) was performed with a Corbett Research Rotor-Gene 3000 real-time cycler, Platinum Taq polymerase (Invitrogen), and primers specific for str-1 and odr-10 (Sengupta et al. 1996) coding sequences. Primer sequences for str-1 are 5′-TCAATAACGCCTTCTTCAGG-3′ and 5′-GTGGAATGCTTGGATGTTCA-3′.Primer sequences for odr-10 are 5′-GAGAATTGTGGATTACCCTAG-3′ and 5′-CTCAATATGCATTATAGGTCGTAATATG-3′.
Expression constructs and generation of transgenic animals:
Site-directed mutagenesis (Statagene QuikChange site-directed mutagenesis kit) was used to generate the constructs pSL204 odr-4p∷kin-29(S517A)∷gfp and pSL287 hda-4p∷hda-4(S363 462A)∷gfp. Mutations were confirmed by sequencing. Expression vectors of Kv1.2, egl-4, and kin-2 were generated by fusing 3.6 and 2.8 kb of upstream regulatory sequences of lim-4 or srd-23, respectively, to a Kv1.2 cDNA (gift from C. Bargmann) (Peckol et al. 1999), egl-4 cDNA plus 0.6 kb of 3′-untranslated region, or a kin-2 cDNA (gift from Kenneth Miller). This resulted in the generation of the constructs pSL137 lim-4p∷Kv1.2, pSL294 lim-4p∷egl-4 and pSL293 srd-23p∷kin-2. In all cases, amplified products were sequenced to confirm the absence of errors generated via the amplification procedure.
For each promoter–gfp fusion, expression from the same extrachromosomal array was examined in wild-type, kin-29, or egl-4 mutant animals. Transgenic animals were generated using either the unc-122p∷dsRed (50–100 ng/μl) or the pRF4 rol-6(su1006) co-injection markers injected at 100 ng/μl. All other plasmids were injected at 20 or 50 ng/μl.
Growth in 8-bromo-cGMP:
NGM agar plates were made containing 8-bromo-cGMP (8-Br-cGMP) (Sigma) added to a final concentration of 5 mm from a freshly made 250 mm stock. On the following day, plates were seeded with 30 μl of a HB101 bacterial suspension and allowed to dry. Approximately 10 animals were picked to each plate and allowed to lay eggs for 2–3 hr. Parents were then removed and eggs were allowed to develop into adults at 20°. str-1p∷gfp expression levels in wild-type and daf-11 animals were quantified at ×50 magnification. As shown previously, addition of 8-bromo-cGMP resulted in smaller body size and rescued the dauer formation defects of daf-11 mutants (Birnby et al. 2000; Nakano et al. 2004).
Feeding/quiescence behavior:
Quiescence without fasting was measured as described (You et al. 2008). In brief, L4 larvae were placed on a plate seeded with Escherichia coli HB101 for 12 hr before the assay. After 12 hr, each young adult was individually isolated into HB101-seeded plates and fed for 6 hr without disturbance. Six hours later worms were observed individually through a dissecting microscope at ×50 magnification. Quiescence duration was measured from the time when the worms were found quiescent until either continuous feeding or continuous locomotion occurred.
Measurement of body size:
Body-length measurements were carried out as described (Raizen et al. 2006). In short, digital images of adult worms 24 hr after the L4 larval molt were acquired at ×100 magnification. The length of the worm was traced using four short line segments using OpenLab software (Improvision), and the sum of the line lengths was calculated. The tail was not included in the measurements (Raizen et al. 2006).
Chemosensory behaviors and dauer formation assays:
Chemosensory behavioral assays were carried out essentially as previously described (Bargmann et al. 1993). For dauer formation assays, five well-fed and growth-synchronized adult animals were allowed to lay eggs at 25° for 4–5 hr on a 3.5-cm assay plate. Adult animals were then removed, and the plates containing ∼100 eggs each were placed at 25° for 48 hr. Dauer and nondauer animals were identified by visual inspection and confirmed by selecting for dauer animals that survived 1% SDS treatment for 45 min (Riddle 1988). Assay plates were made with Noble agar (BD Biosciences) lacking peptone and seeded with OP50 bacteria. Five microliters of crude dauer pheromone extract (Vowels and Thomas 1994) was added to each plate prior to adding agar. For each experiment, all strains were assayed in parallel in at least four independent experiments. The conditions used here for the dauer assay differ from those used previously (Daniels et al. 2000) and may account for the observed differences in the overall number of dauers formed.
Microscopy:
Levels of chemoreceptor gene expression were quantified in adult animals 24 hr after the L4 stage and grown at 20° under well-fed conditions. Levels of gfp expression were scored using a dissection microscope equipped with epifluorescence. For quantitation, gfp expression was scored as “strong” if levels of gfp fluorescence allowed visualization of the neuronal cell bodies and processes, as “reduced” if gfp expression allowed visualization of the cell bodies but not processes, and as “weak/none” if gfp expression could not be detected or could be detected weakly only under higher magnifications.
Statistical analyses:
Statistical analyses presented in Tables 1–5 were performed using the χ2 test of independence to test for statistically significant differences between proportions in the categories “strong,” “reduced,” and “weak/none” for different genotypes (d.f. = 2). A proportion of 0% was set to a default of 1. Significances presented in Figures 1–4 were determined using one-way ANOVA. When appropriate, significances were adjusted for multiple comparisons using Tukey's HSD post-hoc test.
TABLE 1.
cGMP signaling and EGL-4 regulate str-1p∷gfp gene expression
| % expressing str-1p∷gfp in at least one AWB neuron at the indicated levelb
|
||||
|---|---|---|---|---|
| Straina | Strong | Reduced | Weak/none | P-valuesc |
| Wild type | 100 | 0 | 0 | |
| kin-29(oy38) | 0 | 0 | 100 | <0.001d |
| PKG mutants | ||||
| egl-4(n479) | 0 | 92 | 8 | <0.001d |
| egl-4(ad450sd) | 100 | 0 | 0 | d |
| egl-4(n479); Ex[lim-4p∷egl-4] | 94 | 5 | 1 | <0.001e |
| Guanylyl cyclase mutants | ||||
| daf-11(m47) | 0 | 0 | 100 | <0.001d |
| daf-11(sa195) | 0 | 0 | 100 | <0.001d |
| 5 mm 8-Br-cGMP | 100 | 0 | 0 | d |
| daf-11(sa195) + 5 mm 8-Br-cGMP | 39 | 38 | 23 | <0.001f |
| odr-1(n1936) | 0 | 53 | 47 | <0.001d |
| Double mutants | ||||
| egl-4(n479); odr-1(n1936) | 0 | 90 | 10 | <0.001g |
| egl-4(ad450sd); daf-11(sa195) | 99 | 1 | 0 | <0.001f |
| egl-4(ad450sd); odr-1(n1936) | 100 | 0 | 0 | <0.001g |
n = 100–250.
Adult animals grown at 20° were examined in all cases, except for daf-11 mutant animals, which were examined at 15° to permit non-dauer development. All strains contain stably integrated copies of str-1p∷gfp fusion genes.
Expression of str-1p∷gfp was examined at ×50 magnification. “Strong” expression was defined as expression levels similar to those of wild-type animals allowing visualization of the cell bodies and processes; “reduced” expression was defined as expression levels that allowed visualization of the cell bodies but not neuronal processes; “weak/none” was defined as expression that was not visible at ×50 magnification but could be observed weakly at higher magnifications.
P-values were determined using a χ2 test of independence (see materials and methods). Only differences at P < 0.001 are indicated.
Compared to wild type.
Compared to egl-4(n479).
Compared to daf-11(sa195).
Compared to odr-1(n1936).
TABLE 2.
cAMP signaling via KIN-2 inhibits KIN-29
| % expressing str-1p∷gfp in at least one AWB neuron at the indicated levelb
|
||||
|---|---|---|---|---|
| Straina | Strong | Reduced | Weak/none | P-valuesc |
| Wild typed | 100 | 0 | 0 | |
| kin-29(oy38)d | 0 | 0 | 100 | <0.001f |
| gsa-1 Gαs, acy-1 AC and kin-2 PKA mutants | ||||
| gsa-1(ce94gf) | 0 | 31 | 69 | <0.001f |
| acy-1(md1756gf) | 0 | 46 | 54 | <0.001f |
| kin-2(ce179) | 0 | 3 | 97 | <0.001f |
| kin-2(ce179); Ex[srd-23p∷kin-2]e | 93 | 4 | 2 | <0.001g |
| Double mutants with kin-2 and gsa-1 | ||||
| gsa-1(ce94gf); kin-2(ce179) | 0 | 1 | 99 | g |
| gsa-1(ce94gf); kin-29(oy38) | 0 | 6 | 94h | i |
| Expression of KIN-29[S517A] | ||||
| kin-29(oy38); Ex[odr-4p∷kin-29]e | 95 | 5 | 0 | <0.001i |
| kin-29(oy38); Ex[odr-4p∷kin-29(S517A)]e | 93 | 6 | 1 | <0.001i |
n = 30–250.
Adult animals grown at 20° were examined in all cases. All strains contain stably integrated copies of str-1p∷gfp fusion genes.
Expression of str-1p∷gfp was examined at ×50 magnification. Expression levels were defined as in Table 1.
P-values were determined as in Table 1.
Data are from Table 1.
Data shown are from one to two transgenic lines for each. The srd-23 promoter drives expression in the AWB and ASK chemosensory neurons (Colosimo et al. 2004). odr-4 regulatory sequences drive expression in a subset of chemosensory neurons (Dwyer et al. 1998).
Compared to wild type.
Compared to kin-2(ce179).
Weak expression of str-1p∷gfp in kin-29(oy38) animals can be detected at ×400 magnification (Lanjuin and Sengupta 2002; van der Linden et al. 2007). This weak expression was not further decreased in gsa-1(gf); kin-29 double mutants.
Compared to kin-29(oy38).
TABLE 3.
EGL-4 may act partly in parallel to KIN-2 and KIN-29 to regulate str-1p∷gfp gene expression
| % expressing str-1p∷gfp in at least one AWB neuron at the indicated levelb
|
||||
|---|---|---|---|---|
| Straina | Strong | Reduced | Weak/none | P-valuesc |
| Wild typed | 100 | 0 | 0 | |
| kin-29(oy38)d | 0 | 0 | 100 | <0.001e |
| kin-29(oy39) | 0 | 0 | 100 | <0.001e |
| egl-4(n479)d | 0 | 92 | 8 | <0.001e |
| egl-4(ad450sd)d | 100 | 0 | 0 | e |
| daf-11(sa195)d | 0 | 0 | 100 | <0.001e |
| kin-2(ce179)d | 0 | 3 | 97 | <0.001e |
| Double mutants with kin-29 | ||||
| egl-4(n479); kin-29(oy38) | 0 | 0 | 100 | f |
| egl-4(ad450sd); kin-29(oy38) | 0 | 96 | 4 | <0.001f |
| daf-11(sa195); kin-29(oy38) | 0 | 0 | 100 | f |
| Double mutants with kin-2 | ||||
| egl-4(n479); kin-2(ce179) | 0 | 0 | 100 | g |
| egl-4(ad450sd); kin-2(ce179) | 100 | 0 | 0 | <0.001g |
n = 30–250.
Adult animals grown at 20° were examined in all cases, except for daf-11 mutants, which were grown at 15°. All strains contain stably integrated copies of str-1p∷gfp fusion genes.
Expression of str-1p∷gfp was examined at ×50 magnification. Expression levels were defined as in Table 1.
P-values were determined as in Table 1.
Compared to wild type.
Compared to kin-29(oy38).
Compared to kin-2(ce179).
TABLE 4.
Sensory inputs inhibit both KIN-29 and EGL-4
| % expressing str-1p∷gfp in at least one AWB neuron at the indicated levelb
|
||||
|---|---|---|---|---|
| Straina | Strong | Reduced | Weak/none | P-valuesc |
| Wild typed | 100 | 0 | 0 | |
| kin-29(oy38)d | 0 | 0 | 100 | <0.001e |
| kin-29(oy39)d | 0 | 0 | 100 | <0.001e |
| egl-4(n479)d | 0 | 92 | 8 | <0.001e |
| egl-4(ad450sd)d | 100 | 0 | 0 | e |
| che-3(e1124) | 100 | 0 | 0 | e |
| osm-6(p811) | 100 | 0 | 0 | e |
| Double mutants with egl-4 | ||||
| che-3(e1124); egl-4(n479) | 82 | 18 | 0 | <0.001f |
| che-3(e1124); egl-4(ad450sd) | 100 | 0 | 0 | e |
| Double mutants with kin-29 | ||||
| che-3(e1124); kin-29(oy38) | 5 | 85 | 10 | <0.001g |
| che-3(e1124); kin-29(oy39) | 90 | 10 | 0 | <0.001h |
| osm-6(p811); kin-29(oy38) | 0 | 14 | 86 | <0.001g |
| osm-6(p811); kin-29(oy39) | 0 | 99 | 1 | <0.001h |
| lim-4∷Kv1.2 expression | ||||
| Ex[lim-4∷Kv1.2]i | 100 | 0 | 0 | e |
| kin-29(oy38); Ex[lim-4p∷Kv1.2]i | 0 | 11 | 89 | <0.001g |
| kin-29(oy39); Ex[lim-4p∷Kv1.2]i | 1 | 87 | 12 | <0.001h |
| egl-4(n479); kin-29(oy38); Ex[lim-4p∷Kv1.2]i | 0 | 0 | 100 | g |
| Triple mutants with kin-29 and egl-4 | ||||
| osm-6(p811); egl-4(n479); kin-29(oy38) | 0 | 0 | 100 | g |
| Double mutants with unc-13, unc-31 and unc-104 | ||||
| unc-13(e51) | 100 | 0 | 0 | e |
| unc-13(e51); kin-29(oy39) | 0 | 0 | 100 | h |
| unc-104(e1265) | 100 | 0 | 0 | e |
| unc-104(e1265); kin-29(oy39) | 0 | 0 | 100 | h |
| unc-31(e169) | 100 | 0 | 0 | e |
| unc-31(e169); kin-29(oy38) | 0 | 0 | 100 | g |
n = 80–200.
Adult animals grown at 20° were examined in all cases. All strains contain stably integrated copies of str-1p∷gfp fusion genes.
Expression of str-1p∷gfp was examined at ×50 magnification. Expression levels were defined as in Table 1.
P-values were determined as in Table 1.
Compared to wild type.
Compared to egl-4(n479).
Compared to kin-29(oy38).
Compared to kin-29(oy39).
Data shown are from one or two independent transgenic lines.
TABLE 5.
Mutations in mef-2 and hda-4 suppress the str-1p∷gfp expression phenotypes of egl-4 and kin-2 mutants
| % expressing str-1p∷gfp in at least one AWB neuron at the indicated levelb
|
||||
|---|---|---|---|---|
| Straina | Strong | Reduced | Weak/none | P-valuesc |
| Wild typed | 100 | 0 | 0 | |
| egl-4(n479)d | 0 | 92 | 8 | <0.001e |
| egl-4(ad450sd)d | 100 | 0 | 0 | e |
| daf-11(sa195)d | 0 | 0 | 100 | <0.001e |
| odr-1(n1936)d | 0 | 53 | 47 | <0.001e |
| gsa-1(ce94gf)d | 0 | 31 | 69 | <0.001e |
| kin-2(ce179)d | 0 | 3 | 97 | <0.001e |
| mef-2(gv1) | 100 | 0 | 0 | e |
| hda-4(oy57) | 100 | 0 | 0 | e |
| Suppression of mutations in the kin-2 pathway | ||||
| mef-2(gv1); gsa-1(ce94gf) | 97 | 3 | 0 | <0.001f |
| mef-2(gv1); kin-2(ce179) | 94 | 6 | 0 | <0.001g |
| hda-4(oy57) kin-2(ce179) | 95 | 5 | 0 | <0.001g |
| Suppression of mutations in the egl-4 pathway | ||||
| mef-2(gv1); egl-4(n479) | 97 | 3 | 0 | <0.001h |
| hda-4(oy57); egl-4(n479) | 88 | 12 | 0 | <0.001h |
| mef-2(gv1); egl-4(ad450sd) | 100 | 0 | 0 | e |
| mef-2(gv1); daf-11(sa195) | 98 | 2 | 0 | <0.001i |
| mef-2(gv1); odr-1(n1936) | 100 | 0 | 0 | <0.001j |
n > 80.
Animals were grown at 20° with the exception of daf-11 mutant animals, which were grown at 15°. All strains carry stably integrated copies of str-1p∷gfp fusion genes.
Expression was examined in adult animals at ×50 magnification. Expression levels were defined as in Table 1.
P-values were determined as in Table 1.
Compared to wild type.
Compared to gsa-1(ce94gf).
Compared to kin-2(ce179).
Compared to egl-4(n479).
Compared to daf-11(sa195).
Compared to odr-1(n1936).
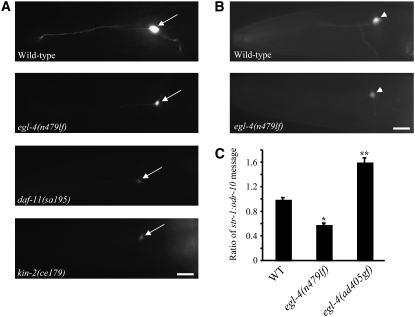
Figure 1.—
cGMP signaling and EGL-4 regulate chemoreceptor gene expression. (A) The expression of str-1p∷gfp in an AWB neuron of adult animals of the indicated genotypes. Arrows indicate the AWB cell body. Images are lateral views; anterior is at left. Images were acquired using identical exposure times at ×400 magnification. Bar, 15 μm. All strains contain stably integrated copies of a str-1p∷gfp transgene. (B) Expression of str-3p∷gfp in an ASI neuron in wild-type (top) and egl-4(n479) (bottom) mutant animals. The ASI cell body is indicated by an arrowhead. Images are lateral views; anterior is at left. Images were acquired using identical exposure times at ×400 magnification. Bar, 15 μm. Strains contain stably integrated copies of a str-3p∷gfp transgene. (C) Levels of endogenous str-1 messages are downregulated in egl-4(lf) mutants and upregulated in egl-4(gf) mutants. Shown is the ratio of endogenous str-1 message to endogenous odr-10 (Sengupta et al. 1996) message as quantified by qRT–PCR in animals of the indicated genotypes. Expression of odr-10 is unaffected in egl-4 mutants (Daniels et al. 2000). The mean of the ratios from two independent experiments is shown. Error bars denote the SEM. * and ** indicate values that are different from that of wild type at P < 0.05 and P < 0.01, respectively.
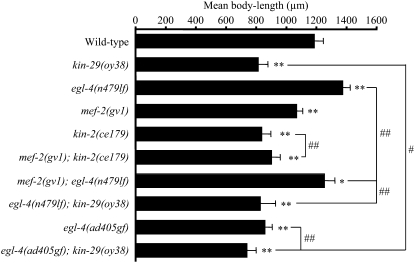
Figure 2.—
Mutations in mef-2 partly suppress the egl-4(lf) defects in the regulation of body size. Shown is the mean body length of adult animals of the indicated genotypes. The body lengths of at least 20 animals were measured for each strain. * and ** indicate values that are different at P < 0.05 and P < 0.01, respectively, from that of wild type; # and ## indicate values that are different at P < 0.05 and P < 0.01, respectively, between the values compared by brackets. Statistical analyses were performed using one-way ANOVA and adjusted for multiple comparisons using Tukey's HSD post-hoc test. Error bars are the SD. All strains contain integrated copies of a str-1p∷gfp fusion gene.
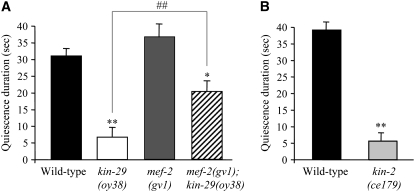
Figure 3.—
kin-29 and kin-2 mutants exhibit defects in food-induced quiescence behavior. (A and B) Quiescence duration of nonfasted animals of the indicated genotypes after feeding with HB101 bacteria for 6 hr. Quiescence is defined as cessation of both locomotion and pharyngeal pumping. n = 20 each in two independent assays. Values are the mean ± SEM. * and ** indicate values that are different at P < 0.05 and P < 0.01, respectively, from that of wild type; ## indicates values that are different at P < 0.01 between the values compared by brackets in A. Statistical analyses were performed using one-way ANOVA and adjusted for multiple comparisons in A using Tukey's HSD post-hoc test. All strains contain integrated copies of a str-1p∷gfp fusion gene.
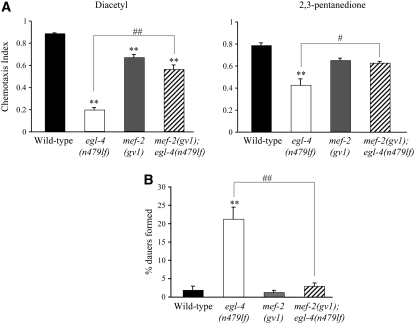
Figure 4.—
Mutations in mef-2 suppress the chemosensory behavioral and dauer formation defects of egl-4(lf) mutants. (A) Responses of adult animals of the indicated genotypes to a point source of 1 μl of a 1:1000 dilution of diacetyl or 1 μl of a 1:10,000 dilution of 2,3-pentanedione. Each data point is the average of two independent assays in duplicate with ∼100 animals in each assay. (B) Percentage of dauers formed upon exposure of animals of the indicated genotypes to 5 μl of crude pheromone extract. n = 300; two independent experiments. For both A and B, error bars equal the SEM. ** indicates values that are different at P < 0.01 from that of wild type; # and ## indicate values that are different at P < 0.05 and P < 0.01, respectively, between the values compared by brackets. Statistical analyses were performed using one-way ANOVA and Tukey's HSD post-hoc test for multiple comparisons. All strains contain integrated copies of a str-1p∷gfp fusion gene.
RESULTS
EGL-4 regulates chemoreceptor gene expression:
Since both EGL-4 and KIN-29 act in sensory neurons to regulate multiple aspects of C. elegans physiology and behavior (Fujiwara et al. 2002; Lanjuin and Sengupta 2002; Raizen et al. 2008; You et al. 2008), we investigated whether, similar to KIN-29, EGL-4 also regulates the expression of CR genes. Expression of gfp driven under the regulatory sequences of the candidate CR genes str-1, sra-6, and srh-234 is strongly downregulated in the AWB, ASH, and ADL neurons, respectively, in kin-29 mutants, whereas the expression of str-3p∷gfp in the ASI neurons is weakly downregulated (Lanjuin and Sengupta 2002). The expression of srd-23, srsx-3, and sru-38 CRp∷gfp fusion genes is unaffected in kin-29 mutants (van der Linden et al. 2007). We found that expression of str-1p∷gfp and str-3p∷gfp fusion genes was also downregulated in egl-4(n479) [henceforth referred to as egl-4(lf)] mutants in the AWB and ASI neurons, respectively (Figure 1, A and B; Table 1; supplemental Table 1), although expression of additional CRp∷gfp fusion genes was unaltered (supplemental Table 1). Expression of egl-4 in the AWB neurons under the lim-4 promoter (Sagasti et al. 1999) rescued the altered str-1p∷gfp expression phenotype of egl-4(lf) mutants (Table 1), indicating that EGL-4 acts cell autonomously to regulate CR gene expression.
egl-4(ad450sd) mutants exhibit phenotypes consistent with a gain-of-function (gf) mutation in egl-4 (Raizen et al. 2006). str-1p∷gfp expression appeared to be unaffected in egl-4(ad450sd) [henceforth referred to as egl-4(gf)] mutants (Table 1). Since it is possible that we were unable to readily quantify upregulated gfp expression perhaps due to overexpression from multiple copies of the stably integrated str-1p∷gfp transgene, we further examined the levels of endogenous str-1 message using qRT–PCR. Levels of endogenous str-1 message were significantly downregulated in egl-4(lf) mutants and were upregulated in egl-4(gf) mutants (Figure 1C). Taken together, these results imply that EGL-4 and KIN-29 regulate partly overlapping sets of CR genes.
Chemoreceptor gene expression is regulated by cGMP:
EGL-4 has been shown to act in a pathway with receptor guanylyl cyclases in mediating olfactory responses and food-induced quiescence behaviors (L'Etoile et al. 2002; You et al. 2008). Since the AWB olfactory neurons express the daf-11 and odr-1 guanylyl cyclases (Birnby et al. 2000; L'Etoile et al. 2002), we examined whether mutations in these genes also affect expression of str-1p∷gfp. We found that expression of str-1p∷gfp was downregulated in both daf-11 and odr-1 mutants (Figure 1A; Table 1), with daf-11 mutants exhibiting a more severe phenotype than either egl-4(lf) or odr-1 mutants.
Mutations in daf-11 and odr-1 are predicted to decrease cGMP levels in the AWB neurons. Addition of the exogenous membrane-permeable cGMP analog 8-Br-cGMP has been shown to bypass the requirement for daf-11 and odr-1 in dauer formation and in the maintenance of ciliary morphology (Schackwitz et al. 1996; Mukhopadhyay et al. 2008). Addition of 8-Br-cGMP was sufficient to partly bypass the daf-11 mutant phenotype in the regulation of str-1p∷gfp expression and enhanced expression in a wild-type background (Table 1 and supplemental Figure 1). Thus, cGMP signaling regulates str-1p∷gfp expression.
We next determined whether DAF-11/ODR-1 act via EGL-4 to regulate CR gene expression. Although we were unable to examine egl-4(lf); daf-11 double mutants since these animals constitutively entered the dauer stage even under permissive conditions, egl-4(lf); odr-1 double mutants exhibited CR gene expression phenotypes similar to those of egl-4(lf) mutants alone (Table 1). Moreover, egl-4(gf) fully suppressed the CR gene expression phenotype of both daf-11 and odr-1 mutants (Table 1). egl-4(gf) was previously shown to also suppress the quiescence phenotypes of daf-11 mutants (You et al. 2008). These results suggest that DAF-11 and ODR-1 act nonredundantly to regulate EGL-4 in the regulation of CR gene expression, but that DAF-11 may also regulate additional targets.
PKA inhibits KIN-29 to regulate chemoreceptor gene expression:
Although EGL-4 activity is cGMP dependent, cGMP levels have not been reported to regulate SIK activity. However, cAMP signaling has been shown to act via cAMP-dependent PKAs to regulate SIKs. In hepatocytes and muscle cells, elevated cAMP signaling, for example, during fasting, results in increased SIK expression (Takemori et al. 2002; Koo et al. 2005; Berdeaux et al. 2007). In addition, PKA-mediated phosphorylation of SIK results in cytoplasmic translocation of SIK and inhibition of activity (Takemori et al. 2002; Okamoto et al. 2004; Berdeaux et al. 2007; Wang et al. 2008). Thus, PKA acts at both the transcriptional and post-translational levels to regulate SIK function.
Since KIN-29 encodes a member of the SIK family, we examined whether PKA regulates KIN-29, and hence str-1p∷gfp gene expression. Animals carrying null mutations in the kin-1 catalytic and kin-2 regulatory subunits of PKA die as embryos; we therefore used the kin-2(ce179) allele, which acts recessively but results in a holoenzyme that is hypersensitive to cAMP levels for PKA activation (Schade et al. 2005; Charlie et al. 2006). str-1p∷gfp expression was severely decreased in kin-2(ce179) mutants (Table 2 and Figure 1A) and could be rescued by expression of kin-2 in the AWB neurons (Table 2). PKA activity is regulated by the GSA-1 Gαs subunit and the ACY-1 adenylyl cyclase in the regulation of neurotransmitter release and locomotion (Reynolds et al. 2005; Schade et al. 2005). gf mutations in both gsa-1 and acy-1 also downregulated str-1p∷gfp expression, and gsa-1(gf); kin-2 double mutants exhibited str-1p∷gfp expression phenotypes similar to those of kin-2 mutants alone (Table 2). These results suggest that increased cAMP signaling acts via PKA to downregulate str-1p∷gfp expression.
We next determined whether KIN-2 and KIN-29 act in a linear or parallel pathway to regulate str-1p∷gfp gene expression. Since the tight linkage of kin-2, kin-29, and kyIs104 (stably integrated str-1p∷gfp transgene) on LG X prevented the construction of a kin-2 kin-29 kyIs104 strain, we instead examined gsa-1(gf); kin-29 mutants. The str-1p∷gfp expression phenotype of these double-mutant animals was similar to that of kin-29 mutants alone (Table 2), suggesting that KIN-29 acts downstream of, and is inhibited by, cAMP signaling. Analysis of the KIN-29 protein sequence revealed eight predicted PKA phosphorylation sites, three of which are highly conserved in the related SIK1, SIK2, and/or SIK3 kinases (supplemental Figure 2A). Of these, the S517 residue in KIN-29 is analogous to the PKA target sites S577 and S587 in SIK1 and SIK2, respectively (Takemori et al. 2002) (supplemental Figure 2A). Mutating Ser577 to Ala results in nuclear localization of SIK1 (Takemori et al. 2002). However, a GFP-tagged KIN-29[S517A] protein retained cytoplasmic localization in all cell types (supplemental Figure 3) and rescued the kin-29 mutant phenotype of reduced str-1p∷gfp expression (Table 2). Thus, elevated cAMP levels may act via PKA to directly or indirectly target multiple sites on KIN-29 and inhibit KIN-29 function.
EGL-4 acts downstream of, or partly in parallel to, KIN-29 to regulate chemoreceptor gene expression:
Since lf mutations in both kin-29 and egl-4 downregulate str-1p∷gfp expression, we next determined whether EGL-4 and KIN-29 act in parallel or linear pathways to regulate gene expression. Double mutants carrying putative null alleles of both kin-29 and egl-4 or daf-11 exhibited str-1p∷gfp gene expression phenotypes similar to those of kin-29 mutants alone (Table 3). egl-4(lf); kin-2 mutant animals were unhealthy and exhibited pale bodies, uncoordinated locomotion, and slow growth (data not shown). These animals also exhibited markedly low levels of str-1p∷gfp expression similar to those of kin-29 mutants. However, egl-4(gf) partly suppressed the CR gene expression defects of kin-29(oy38) mutants and fully suppressed the CR gene expression phenotypes of kin-2(ce179) mutants (Table 3). These observations are consistent with the notion that EGL-4 acts downstream of, or partly in parallel to, KIN-2/KIN-29 to regulate CR gene expression.
Reduction of sensory activity suppresses the chemoreceptor gene expression defects of kin-29 and egl-4 mutants:
Chemosensory neurons in C. elegans exhibit ciliated sensory endings, which are essential for correct sensation and transduction of environmental cues (Ward et al. 1975; Ware et al. 1975; Perkins et al. 1986). Mutations in genes such as che-3, encoding a dynein motor, and osm-6, encoding a conserved protein required to build and maintain cilia, result in markedly defective ciliary structures and altered development and behavior (Perkins et al. 1986; Collet et al. 1998; Wicks et al. 2000; Mukhopadhyay et al. 2007). In previous work, it has been suggested that sensory inputs inhibit EGL-4 activity in the regulation of body size and locomotor activity (Fujiwara et al. 2002). To determine whether this is also the case in the regulation of CR gene expression, we examined str-1p∷gfp expression in che-3; egl-4(lf) and che-3; egl-4(gf) mutants. Surprisingly, however, che-3 mutations suppressed the str-1p∷gfp expression defects of egl-4(lf) mutants (Table 4). str-1p∷gfp expression was unaffected in che-3; egl-4(gf) mutants and was similar to the expression levels in either single mutant alone (Table 4). These results exclude the possibility that sensory inputs act solely via inhibition of EGL-4 to regulate CR gene expression.
Our genetic epistasis experiments suggest that EGL-4 and KIN-29 may act in partly parallel pathways to regulate CR gene expression. A simple model based on the above results might suggest that sensory inputs inhibit both EGL-4 and KIN-29, such that upregulation of the KIN-29-regulated pathway in che-3; egl-4(lf) animals is sufficient to bypass the egl-4(lf) phenotype. If this is the case, reduction of sensory input would be predicted to also partly bypass the CR expression phenotype of kin-29 mutants due to activation of the EGL-4-regulated pathway. As shown in Table 4, mutations in che-3 and osm-6 also partly suppressed the kin-29 mutant phenotypes in the regulation of CR gene expression. Mutations in the fkh-2 forkhead domain transcription factor gene result in defects in AWB dendritic extension and ciliary morphology (Mukhopadhyay et al. 2007). We observed a significant correlation between the upregulation of str-1∷gfp expression and defective dendritic and/or ciliary structures in the AWB neurons of fkh-2(ok683) kin-29(oy39) double mutants (95% of AWB neurons with defective dendritic/ciliary structure expressed str-1∷gfp at wild-type levels, as compared to 3% of AWB neurons with wild-type morphology; n = 49).
To further confirm that the observed suppression was due to reduced neuronal activity as a consequence of altered sensory inputs, we examined str-1p∷gfp expression in animals expressing the rat delayed rectifying voltage-gated potassium channel Kv1.2 (Stuhmer et al. 1989; Grissmer et al. 1994). It has previously been shown that expression of Kv1.1 or Kv1.2 results in reduction of activity of a subset of sensory neurons and downregulation of expression of a subset of KIN-29-independent CR genes in the ASI chemosensory neurons (Peckol et al. 1999, 2001). We expressed a rat Kv1.2 cDNA in the AWB sensory neurons using the lim-4 promoter, which drives expression in the AWB olfactory and additional inter- and motorneurons (Sagasti et al. 1999). Transgenic animals expressing this fusion gene phenocopied lim-4 mutants in their locomotory behavior (Sagasti et al. 1999) (data not shown), suggesting that expression of Kv1.2 results in decreased inter- and/or motorneuron function. Although no effect was observed on str-1 expression upon expression of Kv1.2 in a wild-type background, we observed partial suppression of the downregulated str-1p∷gfp expression phenotype in kin-29 mutants (Table 4). These results suggest that reduction of sensory inputs partly bypasses the requirement of either the KIN-29 or the EGL-4 pathway in the regulation of CR gene expression.
If loss of sensory inputs activates both the EGL-4 and KIN-29 pathways to upregulate str-1p∷gfp expression, we would predict that loss of both egl-4 and kin-29 gene functions would fully abolish the sensory-input-mediated upregulation of gene expression. Indeed, we found this to be the case (Table 4). Thus, str-1p∷gfp expression was strongly downregulated in osm-6; egl-4(lf); kin-29 triple mutants or upon expression of lim-4p∷Kv1.2 in egl-4(lf); kin-29 double mutants. These results further suggest that reduction of sensory activity bypasses kin-29(lf) or egl-4(lf) mutations by activating the EGL-4 or KIN-29 pathways, respectively, in the regulation of str-1p∷gfp expression. No suppression was observed in animals mutant for the genes unc-13, unc-31, and unc-104 (Table 4), which are required for neurotransmitter release, dense core vesicle fusion, and trafficking of synaptic vesicles (Hall and Hedgecock 1991; Maruyama and Brenner 1991; Ahmed et al. 1992; Avery et al. 1993; Ailion et al. 1999; Richmond et al. 1999), implying that inputs from other neurons may not play a role in the regulation of CR gene expression.
Mutations in the hda-4 class II HDAC and the mef-2 transcription factor genes suppress the egl-4(lf) and kin-2 phenotypes of reduced chemoreceptor gene expression:
We next turned to identification of molecules that act downstream of EGL-4 and KIN-29 in the regulation of CR gene expression. We previously showed that lf mutations in hda-4 and mef-2 suppress all examined phenotypes of kin-29(lf) mutants, including altered CR gene expression, body size, and dauer formation (van der Linden et al. 2007). If cAMP signaling inhibits KIN-29, we would predict that lf mutations in mef-2 and hda-4 would also bypass the decreased str-1p∷gfp expression phenotypes of gsa-1(gf) and kin-2 mutants. As shown in Table 5, mutations in both mef-2 and hda-4 efficiently suppressed the CR gene expression phenotypes of both gsa-1(gf) and kin-2 mutants. mef-2(gv1) and hda-4(oy57) also fully suppressed the egl-4(lf) phenotype of downregulated str-1p∷gfp gene expression, but had no effect in the egl-4(ad450sd) mutant background (Table 5). Moreover, mutations in mef-2 suppressed the gene regulation defects of daf-11 and odr-1 mutants (Table 5). These results indicate that both KIN-29 and EGL-4 act via MEF-2 and HDA-4 to regulate the expression of str-1p∷gfp.
EGL-4 acts via both MEF-2-dependent and -independent pathways to regulate body size:
We next determined whether mef-2 mutations also suppress additional egl-4(lf) phenotypes. Both kin-29 and egl-4 mutants exhibit defects in body-size regulation although kin-29 mutants are small, whereas egl-4(lf) mutants are long (Daniels et al. 2000; Lanjuin and Sengupta 2002; Fujiwara et al. 2002; Hirose et al. 2003; Nakano et al. 2004; Maduzia et al. 2005) (Figure 2). Both kinases act in sensory neurons to regulate body size, and mutations in mef-2 and hda-4 fully suppress the body-size defects of kin-29 mutants (Fujiwara et al. 2002; Lanjuin and Sengupta 2002; van der Linden et al. 2007). As expected, kin-2(ce179) mutants were also small, and these body-size defects could be partly rescued by mef-2 mutations (Figure 2).
It has previously been shown that mutations in the daf-3 SMAD transcription factor and the DAF-5 SKI protein suppress the body-size defects of egl-4 mutants (Patterson et al. 1997; Daniels et al. 2000; da Graca et al. 2004). Mutations in mef-2 also partly suppressed the body-size defects of egl-4(lf) mutants (Figure 2). The body size of egl-4(lf); kin-29 mutants was similar to that of kin-29 mutants alone; however, the body size of egl-4(gf); kin-29 mutants was significantly smaller than that of either single mutant alone (Figure 2). These results suggest that while KIN-29 acts solely through MEF-2/HDA-4 to regulate body size, EGL-4 acts via both MEF-2-dependent and -independent (and likely DAF-3/5-dependent) pathways in the regulation of body size. However, we are unable to rule out the possibility of additive effects of egl-4 and mef-2 mutations on the regulation of body size.
The food-induced quiescence behavioral defects of kin-29, but not of egl-4 mutants, are partly suppressed by mutations in mef-2:
When grown on high-quality food (Shtonda and Avery 2006), at any given time, over 90% of wild-type animals are found to be in a quiescent state where they cease locomotion and terminate feeding (You et al. 2008). These food-induced quiescence behaviors have been suggested to be related to satiety behaviors in mammals (Schwartz et al. 2000; Woods et al. 2000). egl-4(lf) mutants exhibit defects in food-induced quiescence behaviors such that these animals continuously move and feed when grown on high-quality food (You et al. 2008). kin-29 mutants were also found to be defective in food-induced quiescence behavior (Figure 3A). However, the quiescence defects of kin-29 and egl-4 mutants were qualitatively distinct. While egl-4(lf) mutants continued to move and feed, kin-29 mutants often ceased locomotion but continued to pump when grown on high-quality food. kin-2(ce179) mutants exhibited quiescence defects similar to those of kin-29 mutants in that kin-2 mutant animals also ceased to move but continued to pump on high-quality food (Figure 3, A and B), as might be predicted if PKA inhibits KIN-29 activity. The quiescence defect of kin-29 mutants was partly suppressed by mutations in mef-2 (Figure 3A), although mef-2 mutations failed to suppress the quiescence defects of egl-4(lf) animals (data not shown). Thus, both EGL-4 and KIN-29 are defective in a food-induced behavior, but use distinct pathways to regulate this behavioral phenotype.
Mutations in mef-2 suppress the chemosensory behavior and dauer formation defects of egl-4 mutants:
Wild-type animals are attracted toward a point source of the volatile odorants diacetyl and 2,3-pentanedione, which are sensed by the AWA and AWC olfactory neuron types, respectively (Bargmann et al. 1993; Chou et al. 2001). egl-4(lf) mutants exhibit defects in their responses to both these odorants, although they retain responses to other odorants sensed by the AWA and AWC neurons (Daniels et al. 2000) (Figure 4A). Mutations in daf-3 suppress the chemosensory defects of egl-4(lf) mutants to both diacetyl and 2,3-pentanedione, while mutations in daf-5 suppress only the behavioral defect toward diacetyl (Daniels et al. 2000). Mutations in mef-2 resulted in weak but significant defects in the responses to diacetyl, but suppressed the olfactory behavioral defects of egl-4(lf) mutants to both diacetyl and 2,3-pentanedione (Figure 4A). Thus, EGL-4 acts via both DAF-3 and MEF-2 to regulate chemosensory behaviors.
In addition to exhibiting chemosensory behavioral defects, egl-4(lf) mutants also exhibit defects in dauer formation (Golden and Riddle 1984b; Daniels et al. 2000). C. elegans produces dauer pheromone, a complex mixture of small molecules throughout its life cycle (Golden and Riddle 1982; Jeong et al. 2005; Butcher et al. 2007). High concentrations of pheromone signal adverse conditions and trigger entry into the dauer stage. egl-4(lf) mutants are hypersensitive to dauer pheromone, such that they form dauers at concentrations that are not sufficient to promote dauer formation in wild-type animals (Golden and Riddle 1984b; Daniels et al. 2000) (Figure 4B). Mutations in either daf-3 or daf-5 fully suppress dauer formation by egl-4(lf) mutants (Daniels et al. 2000). Similarly, mutations in mef-2 also suppressed the increased sensitivity of egl-4(lf) mutants to dauer pheromone (Figure 4B), indicating that EGL-4 acts via DAF-3, DAF-5, and MEF-2 to regulate dauer formation.
Mutations in predicted PKG/PKA phosphorylation sites result in cytoplasmic localization of HDA-4:
In mammalian cells, phosphorylation of class II HDACs at a residue analogous to the S198 residue in HDA-4 (as well as at other residues) results in translocation of HDACs from the nucleus to the cytoplasm (McKinsey et al. 2000a,b; Kao et al. 2001; Chawla et al. 2003; Linseman et al. 2003). Sequestering of HDACs in the cytoplasm alleviates the gene repression functions of these proteins. However, in C. elegans, phosphorylation of HDA-4 at the S198 residue, likely by KIN-29, does not alter the subcellular localization of HDA-4, although the gene repression functions of the protein are abrogated (van der Linden et al. 2007). Thus, a mutant HDA-4[S198A] protein remains nuclear localized and mediates constitutive repression of str-1p∷gfp expression (van der Linden et al. 2007).
We examined whether the localization of functional GFP-tagged HDA-4 was affected in kin-2(ce179) or egl-4(lf) mutant backgrounds. Localization of HDA-4 was unaltered in kin-2, kin-29, che-3, egl-4(lf), and egl-4(lf); kin-29 mutant backgrounds (Figure 5, A–E; van der Linden et al. 2007), suggesting that lf of egl-4 or kin-29 alone, or together, or activation of PKA function does not affect HDA-4 localization. We could not directly examine the requirement of PKA in modulating HDA-4 function due to the lethality of kin-1 and kin-2 null alleles. Interestingly, however, HDA-4 was cytoplasmically localized in a subset of neurons in che-3; egl-4 mutants, although HDA-4 localization remained nuclear in non-neuronal cell types (Figure 5F and data not shown). One interpretation of these results is that PKA activity is downregulated in a subset of neurons in che-3 mutants and that downregulation of both PKA and PKG activity results in cytoplasmic translocation of HDA-4.
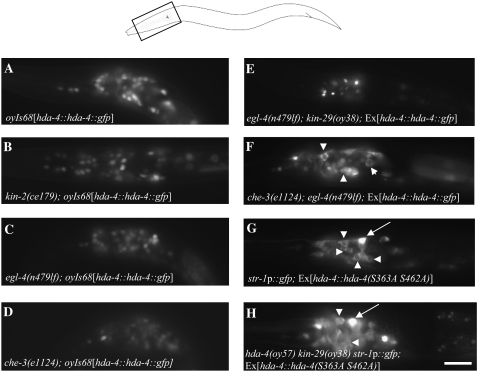
Figure 5.—
Localization of HDA-4 may be regulated by both PKA- and PKG-mediated phosphorylation. (A–F) The expression of stably integrated (A–D) or extrachromosomal copies (E and F) of a functional full-length gfp-tagged hda-4 transgene (van der Linden et al. 2007) in the indicated genetic backgrounds. Note cytoplasmic localization in a subset of cells in F (arrowheads). (G and H) A GFP-tagged HDA-4[S363A S462A] protein is localized to the cytoplasm (arrowheads) in the indicated genetic backgrounds. Arrows point to str-1p∷gfp expression in an AWB neuron. (A–H) Images of L1/L2 larval stage animals were acquired using identical exposure times at ×400 magnification. The area imaged in A–H is indicated by a box in the cartoon at top. Bar, 20 μm.
PKA and PKG have been shown to target similar consensus sites (Kennelly and Krebs 1991). We examined the HDA-4 protein sequence and identified the S363 and S462 residues as potential PKG/PKA phosphorylation sites. The S462 residue is highly conserved in mammalian class II HDACs (supplemental Figure 2B). A GFP-tagged HDA-4[S363A S462A] mutant protein localized to the cytoplasm in wild-type animals and did not affect str-1p∷gfp expression (Figure 5G). Moreover, this mutant HDA-4 protein only partly abolished suppression of the str-1p∷gfp expression phenotype when expressed in hda-4 kin-29 mutants (Figure 5H; 32% of hda-4 kin-29 double mutants expressing this mutant protein retained wild-type levels of str-1p∷gfp expression; n = 73). These results suggest that PKA- and PKG-mediated phosphorylation of HDA-4 at S363/462 may be required for nuclear localization of HDA-4. In addition, KIN-29 phosphorylates HDA-4 at S198 to alleviate its gene repression properties.
DISCUSSION
We have described a complex mechanism of regulation of CR gene expression in C. elegans. In the AWB olfactory neuron type, levels of expression of the str-1p∷gfp CR gene are regulated via integration of signals from multiple pathways. Results described here suggest that increased cAMP levels downregulate str-1p∷gfp expression via inhibition of the KIN-29 SIK, whereas increased cGMP levels promote str-1p∷gfp expression via activation of the EGL-4 PKG (Figure 6A). These antagonistic effects on CR gene expression may allow for more precise regulation of CR expression levels than would be possible via a single pathway alone.
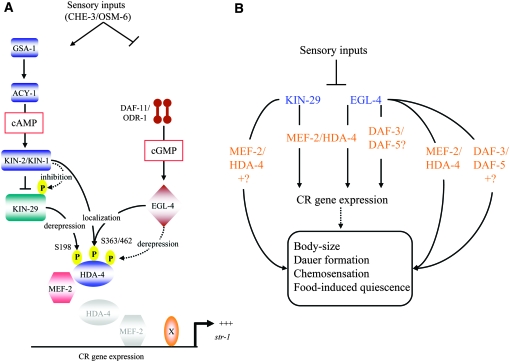
Figure 6.—
Model of KIN-29- and EGL-4-regulated pathways in the modulation of CR gene expression and sensory behaviors. (A) KIN-29 and EGL-4 act in partly parallel pathways to regulate str-1p∷gfp expression. Increased activity of the GSA-1 Gαs and ACY-1 adenylyl cyclase result in increased intracellular cAMP levels and activation of PKA. PKA inhibits KIN-29 activity via direct or indirect phosphorylation. EGL-4 activity is regulated by cGMP levels generated by the DAF-11 and ODR-1 receptor guanylyl cyclases. Sensory inputs downregulate DAF-11/ODR-1, but may upregulate ACY-1 activity. KIN-29 and EGL-4 may phosphorylate HDA-4 at the S198 and other residue(s) to alleviate the gene repression properties of HDA-4. Nuclear localization of HDA-4 requires phosphorylation at the S363/S462 residues by PKA and EGL-4. Proposed phosphorylation is indicated by dashed arrows. See discussion for additional details. (B) KIN-29 acts primarily via MEF-2 and HDA-4 to regulate CR gene expression, body size, dauer formation, and food-induced quiescence behaviors (van der Linden et al. 2007 and this work). EGL-4 acts via MEF-2/HDA-4 to regulate CR gene expression and via MEF-2 and the DAF-3 SMAD and DAF-5 SKI proteins to regulate additional sensory behaviors (Daniels et al. 2000).
Sensory inputs inhibit both KIN-29 and EGL-4:
On the basis of our genetic experiments, we suggest that, under normal growth conditions, activity of both the EGL-4 and KIN-29 the pathways is required for expression of str-1p∷gfp. Thus, str-1p∷gfp expression levels are decreased in either egl-4(lf) or kin-29 single mutants. However, upon loss of sensory signaling, such as in che-3 or osm-6 mutants, activity of both the EGL-4 and the KIN-29 pathways may be upregulated, such that one pathway would be able to bypass the requirement for the second pathway in the regulation of str-1p∷gfp expression (Figure 6A).
Sensory inputs have previously been suggested to inhibit EGL-4 activity via downregulation of cGMP levels (Fujiwara et al. 2002). In the vertebrate visual transduction cascade, light activates a cGMP phosphodiesterase, thereby decreasing intracellular cGMP levels leading to hyperpolarization (Arshavsky et al. 2002). Similarly, it has recently been shown that the AWC olfactory neurons in C. elegans are hyperpolarized in the presence of odorants, likely via downregulation of cGMP levels (Chalasani et al. 2007). Surprisingly, however, our experiments also suggest that sensory inputs inhibit KIN-29, perhaps via upregulation of cAMP signaling (Figure 6A). Although we have not formally ruled out the possibility that sensory inputs act via cAMP-independent pathways to regulate KIN-29, cAMP signaling has also been shown to regulate SIKs in mammalian cells (Takemori et al. 2002; Okamoto et al. 2004; Koo et al. 2005; Berdeaux et al. 2007; Wang et al. 2008). Our observations imply that chemosensory cues may have opposite effects on different signaling pathways in individual chemosensory neurons in C. elegans. This is not unprecedented, since it has been shown that different odors can activate or inhibit individual olfactory receptor neurons in Drosophila, Manduca, and rats and that, in Drosophila, a given odor can elicit either excitatory or inhibitory responses via different receptors (Duchamp-Viret et al. 1999; Shields and Hildebrand 2000; De Bruyne et al. 2001; Hallem et al. 2004). Since each chemosensory neuron in C. elegans expresses multiple CR genes (Troemel et al. 1995), sensory inputs may act via different CR subsets to differentially regulate KIN-29 and EGL-4, thereby allowing for fine tuning of gene expression in response to distinct environmental conditions.
The functions of KIN-29 and EGL-4 are mediated via multiple targets and mechanisms:
Both KIN-29 and EGL-4 appear to act primarily via MEF-2 and HDA-4 to regulate str-1p∷gfp expression. We showed previously that MEF-2 is not required for activation of str-1p∷gfp expression in the AWB neurons (van der Linden et al. 2007). Instead, in the absence of phosphorylation of HDA-4 at the S198 residue, a MEF-2/HDA-4 complex interacts directly with cis-regulatory sequences upstream of str-1 to repress gene expression (van der Linden et al. 2007). KIN-29 is able to directly phosphorylate HDA-4 in vitro at the S198 residue (van der Linden et al. 2007), and recently, SIK1 has been shown to directly target class II HDACs in skeletal muscle (Berdeaux et al. 2007). It is possible that EGL-4 also directly targets HDA-4 at as yet unidentified residues to alleviate its gene-repressive functions.
We have also uncovered a second mode of HDA-4 regulation based on subcellular localization. Phosphorylation of the conserved putative PKA/PKG target residues S363 and S462, but not S198, appears to be necessary for nuclear localization of HDA-4, an essential requirement for its gene repression functions. S363 and S462 may be targeted redundantly by EGL-4 and PKA (Figure 6A). Although no function has yet been ascribed to the analogous residues in mammalian HDACs, the phosphorylation state of the residues analogous to S198 is the primary regulator of the subcellular localization of class II HDACs in mammals. Mammalian class II HDACs are phosphorylated by kinases such as calcium/calmodulin-dependent protein kinases, protein kinase D, and SIK1 (McKinsey et al. 2000a,b; Kao et al. 2001; Chawla et al. 2003; Linseman et al. 2003; Dequiedt et al. 2005; Parra et al. 2005; Backs et al. 2006; Matthews et al. 2006; Berdeaux et al. 2007), but unlike the case in C. elegans, phosphorylated HDACs are localized to the cytoplasm. Thus, in C. elegans, the subcellular localization and repressive functions of HDA-4 may be regulated via phosphorylation of different sites by different kinases. Multiple modes of regulation of HDA-4 function may allow for integration of complex sensory inputs at the level of regulation of HDA-4 activity and consequently for gene expression. It will be interesting to determine whether similar mechanisms operate to regulate class II HDAC functions in a cell- or tissue-specific manner in vertebrates (Martin et al. 2007).
While KIN-29 appears to act primarily via MEF-2 and HDA-4 to regulate multiple behaviors and physiological processes, the case for EGL-4 is more complex (Figure 6B). EGL-4 acts via MEF-2 to regulate str-1p∷gfp expression and partly via MEF-2 as well as DAF-3 and DAF-5 to regulate body size, chemosensory behaviors, and dauer formation (Daniels et al. 2000 and this work). Moreover, while daf-3 mutations fully suppress all examined egl-4(lf) defects in chemosensory behaviors, daf-5 mutations suppress only a subset of these phenotypes (Daniels et al. 2000). One interpretation of these observations is that EGL-4 acts via distinct effectors in different cell types to regulate the expression of different gene subsets. The complexity of signaling pathways mediating EGL-4 function has been noted previously (Daniels et al. 2000; Raizen et al. 2006). An important next step is to define the complete gene sets regulated by KIN-29 and EGL-4 in sensory neurons and to identify the effectors mediating their functions.
Multiple mechanisms regulate chemoreceptor gene expression in C. elegans:
Why must CR gene expression be regulated so precisely in C. elegans? We and others have previously hypothesized that plasticity in CR gene expression contributes to behavioral plasticity in C. elegans (Peckol et al. 2001; Lanjuin and Sengupta 2002; Nolan et al. 2002; van der Linden et al. 2007). Environmental conditions and internal metabolic state may be translated into defined patterns and levels of expression of CRs and other chemosensory neuron-expressed genes. The expression of different subsets of CR genes has been shown to be regulated by population density, food availability, developmental stage of the animal, and neuronal activity (Peckol et al. 2001; Nolan et al. 2002; A. van der Linden and P. Sengupta, unpublished data). Dietary restriction and infection by pathogenic bacteria have also been shown to regulate gene expression in chemosensory neuron types (Zhang et al. 2005; Bishop and Guarente 2007), indicating that these neurons are able to monitor cellular state to alter gene expression patterns accordingly.
Although we cannot yet confirm whether altered regulation of CR gene expression is causal to the physiological phenotypes of kin-29 and egl-4 mutants, we suggest that regulation of CR gene expression may partly underlie the modulation of food-seeking and acquisition behaviors, as well as food-regulated physiological processes such as body size, quiescence, and dauer formation in C. elegans. EGL-4 and KIN-29 activity in sensory neurons may therefore integrate multiple chemosensory inputs to direct precise patterns and levels of expression of distinct, but overlapping sets of CR and other genes. We have shown that EGL-4 regulates the expression of str-1 and str-3, but not that of five additional CR genes examined. Since the C. elegans genome is predicted to encode >1500 CR genes (Troemel et al. 1995; Robertson and Thomas 2006), it is likely that EGL-4 also regulates additional CR genes. In addition to CR genes, targets of these kinase pathways may include additional chemosensory neuron-expressed signaling molecules, which may regulate the activity and neurohormone outputs of these neuron types (You et al. 2008).
The roles of PKGs and SIKs in food-related behaviors are conserved:
The roles of PKGs and SIKs in the regulation of food-related behaviors appear to be conserved across species. For example, in Drosophila and honeybees, PKG levels regulate foraging, a complex food-acquisition behavior (Osborne et al. 1997; Ben-Shahar et al. 2002). Variations in PKG levels also regulate responses to chemicals such as sucrose, food-acquisition behaviors, and experience-dependent modification of food intake (Scheiner et al. 2004; Kaun et al. 2007). Similarly, cAMP signaling via SIKs and the related AMP-activated kinase have been shown to play critical roles in the regulation of energy homeostasis, food intake, and energy expenditure in Drosophila, C. elegans, and mammals (Apfeld et al. 2004; Minokoshi et al. 2004; Screaton et al. 2004; Dowell et al. 2005; Kahn et al. 2005; Kim and Lee 2005; Koo et al. 2005; Greer et al. 2007; Wang et al. 2008). Although SIKs have been shown to act via multiple factors including CREB, TORC, MEF2, and class II HDACs (Screaton et al. 2004; Koo et al. 2005; Berdeaux et al. 2007), the regulators, effectors, and targets of PKG signaling in food-related behaviors are largely unknown. We have found that PKG acts with a SIK-signaling pathway to regulate chemoreceptor gene expression and other sensory behaviors and that PKG function is mediated via multiple downstream factors, including MEF-2. Given the high degree of conservation of signaling pathways across species, we suggest that SIKs and PKG may act similarly in other species. Identification of additional targets and regulators of both signaling pathways will lead to a better understanding of the mechanisms by which animals modulate behavior and development in response to changing external and internal conditions.
Acknowledgments
We thank Kristin Ma and Rinho Kim for expert technical assistance, the Caenorhabditis Genetics Center for strains, and Don Katz for help with statistical analyses. This work was supported by National Science Foundation grant IOS-0542372 to P.S. and National Institutes of Health grant DK074065 to L.A.
References
- Ahmed, S., I. N. Maruyama, R. Kozma, J. Lee, S. Brenner et al., 1992. The Caenorhabditis elegans unc-13 gene product is a phospholipid-dependent high-affinity phorbol ester receptor. Biochem J. 287 995–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ailion, M., T. Inoue, C. I. Weaver, R. W. Holdcraft and J. H. Thomas, 1999. Neurosecretory control of aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 96 7394–7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcedo, J., and C. Kenyon, 2004. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41 45–55. [DOI] [PubMed] [Google Scholar]
- Apfeld, J., and C. Kenyon, 1999. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402 804–809. [DOI] [PubMed] [Google Scholar]
- Apfeld, J., G. O'Connor, T. McDonagh, P. S. DiStefano and R. Curtis, 2004. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 18 3004–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshavsky, V. Y., T. D. Lamb and E. N. Pugh, Jr., 2002. G proteins and phototransduction. Annu. Rev. Physiol. 64 153–187. [DOI] [PubMed] [Google Scholar]
- Avery, L. A., C. I. Bargmann and H. R. Horvitz, 1993. The Caenorhabditis elegans unc-31 gene affects multiple nervous system-controlled functions. Genetics 134 435–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backs, J., K. Song, S. Bezprozvannaya, S. Chang and E. N. Olson, 2006. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Invest. 116 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann, C. I., 2006. Chemosensation in C. elegans, edited by The C. Elegans Research Community. WormBook, http://www.wormbook.org.
- Bargmann, C. I., E. Hartwieg and H. R. Horvitz, 1993. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74 515–527. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar, Y., A. Robichon, M. B. Sokolowski and G. E. Robinson, 2002. Influence of gene action across different time scales on behavior. Science 296 741–744. [DOI] [PubMed] [Google Scholar]
- Berdeaux, R., N. Goebel, L. Banaszynski, H. Takemori, T. Wandless et al., 2007. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat. Med. 13 597–603. [DOI] [PubMed] [Google Scholar]
- Birnby, D. A., E. A. Link, J. J. Vowels, H. Tian, P. L. Colacurcio et al., 2000. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics 155 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, N. A., and L. Guarente, 2007. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447 545–549. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher, R. A., M. Fujita, F. C. Schroeder and J. Clardy, 2007. Small molecule signaling of dauer formation in C. elegans. Nat. Chem. Biol. 3 420–422. [DOI] [PubMed] [Google Scholar]
- Chalasani, S. H., N. Chronis, M. Tsunozaki, J. M. Gray, D. Ramot et al., 2007. Dissecting a neural circuit for food-seeking behavior in Caenorhabditis elegans. Nature 450 63–70. [DOI] [PubMed] [Google Scholar]
- Charlie, N. K., A. M. Thomure, M. A. Schade and K. G. Miller, 2006. The Dunce cAMP phosphodiesterase PDE-4 negatively regulates G α(s)-dependent and G α(s)-independent cAMP pools in the Caenorhabditis elegans synaptic signaling network. Genetics 173 111–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla, S., P. Vanhoutte, F. J. Arnold, C. L. Huang and H. Bading, 2003. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 85 151–159. [DOI] [PubMed] [Google Scholar]
- Chou, J. H., C. I. Bargmann and P. Sengupta, 2001. The Caenorhabditis elegans odr-2 gene encodes a novel Ly-6-related protein required for olfaction. Genetics 157 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet, J., C. A. Spike, E. A. Lundquist, J. E. Shaw and R. K. Herman, 1998. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics 148 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo, M. E., A. Brown, S. Mukhopadhyay, C. Gabel, A. E. Lanjuin et al., 2004. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr. Biol. 14 2245–2251. [DOI] [PubMed] [Google Scholar]
- da Graca, L. S., K. K. Zimmerman, M. C. Mitchell, M. Kozhan-Gorodetska, K. Sekiewicz et al., 2004. DAF-5 is a Ski oncoprotein homolog that functions in a neuronal TGF beta pathway to regulate C. elegans dauer development. Development 131 435–446. [DOI] [PubMed] [Google Scholar]
- Daniels, S. A., M. Ailion, J. H. Thomas and P. Sengupta, 2000. egl-4 acts through a transforming growth factor-beta/SMAD pathway in Caenorhabditis elegans to regulate multiple neuronal circuits in response to sensory cues. Genetics 156 123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne, M., K. Foster and J. R. Carlson, 2001. Odor coding in the Drosophila antenna. Neuron 30 537–552. [DOI] [PubMed] [Google Scholar]
- Dentin, R., Y. Liu, S. H. Koo, S. Hedrick, T. Vargas et al., 2007. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449 366–369. [DOI] [PubMed] [Google Scholar]
- Dequiedt, F., J. Van Lint, E. Lecomte, V. Van Duppen, T. Seufferlein et al., 2005. Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J. Exp. Med. 201 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell, P., Z. Hu and M. D. Lane, 2005. Monitoring energy balance: metabolites of fatty acid synthesis as hypothalamic sensors. Annu. Rev. Biochem. 74 515–534. [DOI] [PubMed] [Google Scholar]
- Duchamp-Viret, P., M. A. Chaput and A. Duchamp, 1999. Odor response properties of rat olfactory receptor neurons. Science 284 2171–2174. [DOI] [PubMed] [Google Scholar]
- Dulac, C., and A. T. Torello, 2003. Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat. Rev. Neurosci. 4 551–562. [DOI] [PubMed] [Google Scholar]
- Dwyer, N. D., E. R. Troemel, P. Sengupta and C. I. Bargmann, 1998. Odorant receptor localization to olfactory cilia is mediated by ODR-4, a novel membrane-asociated protein. Cell 93 455–466. [DOI] [PubMed] [Google Scholar]
- Fujiwara, M., P. Sengupta and S. L. McIntire, 2002. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron 36 1091–1102. [DOI] [PubMed] [Google Scholar]
- Golden, J. W., and D. L. Riddle, 1982. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218 578–580. [DOI] [PubMed] [Google Scholar]
- Golden, J. W., and D. L. Riddle, 1984. a The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev. Biol. 102 368–378. [DOI] [PubMed] [Google Scholar]
- Golden, J. W., and D. L. Riddle, 1984. b A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218 578–580. [DOI] [PubMed] [Google Scholar]
- Gray, J. M., J. J. Hill and C. I. Bargmann, 2005. A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 102 3184–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer, E. L., D. Dowlatshahi, M. R. Banko, J. Villen, K. Hoang et al., 2007. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 17 1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissmer, S., A. N. Nguyen, J. Aiyar, D. C. Hanson, R. J. Mather et al., 1994. Pharmacological characterization of five cloned voltage-gated K+ channels, types Kv1.1, 1.2, 1.3, 1.5, and 3.1, stably expressed in mammalian cell lines. Mol. Pharmacol. 45 1227–1234. [PubMed] [Google Scholar]
- Gruninger, T. R., D. G. Gualbarto and L. R. Garcia, 2008. Sensory perception of food and insulin-like signals influence seizure susceptibility. PLoS Genet. 4 e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, D. H., and E. M. Hedgecock, 1991. Kinesin-related gene unc-104 is required for axonal transport of synpatic vesicles in C. elegans. Cell 65 837–847. [DOI] [PubMed] [Google Scholar]
- Hallem, E. A., M. G. Ho and J. R. Carlson, 2004. The molecular basis of odor coding in the Drosophila antenna. Cell 117 965–979. [DOI] [PubMed] [Google Scholar]
- Hirose, T., Y. Nakano, Y. Nagamatsu, T. Misumi, H. Ohta et al., 2003. Cyclic GMP-dependent protein kinase EGL-4 controls body size and lifespan in C. elegans. Development 130 1089–1099. [DOI] [PubMed] [Google Scholar]
- Jackson, D. E., and F. L. Ratnieks, 2006. Communication in ants. Curr. Biol. 16 R570–R574. [DOI] [PubMed] [Google Scholar]
- Jeong, P. Y., M. Jung, Y. H. Yim, H. Kim, M. Park et al., 2005. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature 433 541–545. [DOI] [PubMed] [Google Scholar]
- Kahn, B. B., T. Alquier, D. Carling and D. G. Hardie, 2005. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 1 15–25. [DOI] [PubMed] [Google Scholar]
- Kao, H. Y., A. Verdel, C. C. Tsai, C. Simon, H. Juguilon et al., 2001. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276 47496–47507. [DOI] [PubMed] [Google Scholar]
- Kaun, K. R., C. A. Riedl, M. Chakaborty-Chatterjee, A. T. Belay, S. J. Douglas et al., 2007. Natural variation in food acquisition mediated via a Drosophila cGMP-dependent protein kinase. J. Exp. Biol. 210 3547–3558. [DOI] [PubMed] [Google Scholar]
- Kennelly, P. J., and E. G. Krebs, 1991. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 266 15555–15558. [PubMed] [Google Scholar]
- Kim, M. S., and K. U. Lee, 2005. Role of hypothalamic 5′-AMP-activated protein kinase in the regulation of food intake and energy homeostasis. J. Mol. Med. 83 514–520. [DOI] [PubMed] [Google Scholar]
- Koo, S. H., L. Flechner, L. Qi, X. Zhang, R. A. Screaton et al., 2005. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437 1109–1111. [DOI] [PubMed] [Google Scholar]
- Lanjuin, A., and P. Sengupta, 2002. Regulation of chemosensory receptor expression and sensory signaling by the KIN-29 Ser/Thr kinase. Neuron 33 369–381. [DOI] [PubMed] [Google Scholar]
- L'Etoile, N. D., C. M. Coburn, J. Eastham, A. Kistler, G. Gallegos et al., 2002. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron 36 1079–1089. [DOI] [PubMed] [Google Scholar]
- Libert, S., J. Zwiener, X. Chu, W. Vanvoorhies, G. Roman et al., 2007. Regulation of Drosophila life span by olfaction and food-derived odors. Science 315 1133–1137. [DOI] [PubMed] [Google Scholar]
- Linseman, D. A., C. M. Bartley, S. S. Le, T. A. Laessig, R. J. Bouchard et al., 2003. Inactivation of the myocyte enhancer factor-2 repressor histone deacetylase-5 by endogenous Ca(2+) //calmodulin-dependent kinase II promotes depolarization-mediated cerebellar granule neuron survival. J. Biol. Chem. 278 41472–41481. [DOI] [PubMed] [Google Scholar]
- Maduzia, L. L., A. F. Roberts, H. Wang, X. Lin, L. J. Chin et al., 2005. C. elegans serine-threonine kinase KIN-29 modulates TGFbeta signaling and regulates body size formation. BMC Dev. Biol. 5 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak, H. Y., L. S. Nelson, M. Basson, C. D. Johnson and G. Ruvkun, 2006. Polygenic control of Caenorhabditis elegans fat storage. Nat. Genet. 38 363–368. [DOI] [PubMed] [Google Scholar]
- Martin, M., R. Kettmann and F. Dequiedt, 2007. Class IIa histone deacetylases: regulating the regulators. Oncogene 26 5450–5467. [DOI] [PubMed] [Google Scholar]
- Maruyama, I. N., and S. Brenner, 1991. A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 88 5729–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, S. A., P. Liu, M. Spitaler, E. N. Olson, T. A. McKinsey et al., 2006. Essential role for protein kinase D family kinases in the regulation of class II histone deacetylases in B lymphocytes. Mol. Cell. Biol. 26 1569–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey, T. A., C. L. Zhang, J. Lu and E. N. Olson, 2000. a Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey, T. A., C. L. Zhang and E. N. Olson, 2000. b Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14–3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minokoshi, Y., T. Alquier, N. Furukawa, Y. B. Kim, A. Lee et al., 2004. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature 428 569–574. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, S., Y. Lu, H. Qin, A. Lanjuin, S. Shaham et al., 2007. Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans. EMBO J. 26 2966–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, S., Y. Lu, S. Shaham and P. Sengupta, 2008. Sensory signaling-dependent remodeling of olfactory cilia architecture in C. elegans. Dev. Cell 14 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, Y., Y. Nagamatsu and Y. Ohshima, 2004. cGMP and a germ-line signal control body size in C. elegans through cGMP-dependent protein kinase EGL-4. Genes Cells 9 773–779. [DOI] [PubMed] [Google Scholar]
- Nolan, K. M., T. R. Sarafi-Reinach, J. G. Horne, A. M. Saffer and P. Sengupta, 2002. The DAF-7 TGF-beta signaling pathway regulates chemosensory receptor gene expression in C. elegans. Genes Dev. 16 3061–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, M., H. Takemori and Y. Katoh, 2004. Salt-inducible kinase in steroidogenesis and adipogenesis. Trends Endocrinol. Metab. 15 21–26. [DOI] [PubMed] [Google Scholar]
- Osborne, K. A., A. Robichon, E. Burgess, S. Butland, R. A. Shaw et al., 1997. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science 277 834–836. [DOI] [PubMed] [Google Scholar]
- Parra, M., H. Kasler, T. A. McKinsey, E. N. Olson and E. Verdin, 2005. Protein kinase D1 phosphorylates HDAC7 and induces its nuclear export after T-cell receptor activation. J. Biol. Chem. 280 13762–13770. [DOI] [PubMed] [Google Scholar]
- Patterson, G. I., A. Koweek, A. Wong, Y. Liu and G. Ruvkun, 1997. The DAF-3 Smad protein antagonizes TGF-beta-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes Dev. 11 2679–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckol, E. L., J. A. Zallen, J. C. Yarrow and C. I. Bargmann, 1999. Sensory activity affects sensory axon development in C. elegans. Development 126 1891–1902. [DOI] [PubMed] [Google Scholar]
- Peckol, E. L., E. R. Troemel and C. I. Bargmann, 2001. Sensory experience and sensory activity regulate chemosensory receptor gene expression in C. elegans. Proc. Natl. Acad. Sci. USA 98 11032–11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, L. A., E. M. Hedgecock, J. N. Thomson and J. G. Culotti, 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117 456–487. [DOI] [PubMed] [Google Scholar]
- Raizen, D. M., K. M. Cullison, A. I. Pack and M. V. Sundaram, 2006. A novel gain-of-function mutant of the cyclic GMP-dependent protein kinase egl-4 affects multiple physiological processes in Caenorhabditis elegans. Genetics 173 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen, D. M., J. E. Zimmerman, M. H. Maycock, U. D. Ta, Y. J. You et al., 2008. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451 569–572. [DOI] [PubMed] [Google Scholar]
- Reynolds, N. K., M. A. Schade and K. G. Miller, 2005. Convergent, RIC-8-dependent Gα signaling pathways in the Caenorhabditis elegans synaptic signaling network. Genetics 169 651–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, J. E., W. S. Davis and E. M. Jorgensen, 1999. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat. Neurosci. 2 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, D. L., 1988. The dauer larva, pp. 393–412 in The Nematode Caenorhabditis elegans, edited by W. B. Wood. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Robertson, H. M., and J. H. Thomas, 2006. The putative chemoreceptor families of C. elegans, edited by The C. Elegans Research Community. WormBook, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Rolls, E. T., 2005. Taste, olfactory, and food texture processing in the brain, and the control of food intake. Physiol. Behav. 85 45–56. [DOI] [PubMed] [Google Scholar]
- Sagasti, A., O. Hobert, E. R. Troemel, G. Ruvkun and C. I. Bargmann, 1999. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 13 1794–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafi-Reinach, T. R., T. Melkman, O. Hobert and P. Sengupta, 2001. The lin-11 LIM homeobox gene specifies olfactory and chemosensory neuron fates in C. elegans. Development 128 3269–3281. [DOI] [PubMed] [Google Scholar]
- Sawin, E. R., R. Ranganathan and H. R. Horvitz, 2000. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26 619–631. [DOI] [PubMed] [Google Scholar]
- Schackwitz, W. S., T. Inoue and J. H. Thomas, 1996. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17 719–728. [DOI] [PubMed] [Google Scholar]
- Schade, M. A., N. K. Reynolds, C. M. Dollins and K. G. Miller, 2005. Mutations that rescue the paralysis of Caenorhabditis elegans ric-8 (synembryn) mutants activate the G α(s) pathway and define a third major branch of the synaptic signaling network. Genetics 169 631–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiner, R., M. B. Sokolowski and J. Erber, 2004. Activity of cGMP-dependent protein kinase (PKG) affects sucrose responsiveness and habituation in Drosophila melanogaster. Learn. Mem. 11 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, M. W., S. C. Woods, D. Porte, Jr., R. J. Seeley and D. G. Baskin, 2000. Central nervous system control of food intake. Nature 404 661–671. [DOI] [PubMed] [Google Scholar]
- Screaton, R. A., M. D. Conkright, Y. Katoh, J. L. Best, G. Canettieri et al., 2004. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119 61–74. [DOI] [PubMed] [Google Scholar]
- Sengupta, P., J. H. Chou and C. I. Bargmann, 1996. odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 84 899–909. [DOI] [PubMed] [Google Scholar]
- Shields, V. D., and J. G. Hildebrand, 2000. Responses of a population of antennal olfactory receptor cells in the female moth Manduca sexta to plant-associated volatile organic compounds. J. Comp. Physiol. 186 1135–1151. [DOI] [PubMed] [Google Scholar]
- Shtonda, B. B., and L. Avery, 2006. Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol. 209 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhmer, W., J. P. Ruppersberg, K. H. Schroter, B. Sakmann, M. Stocker et al., 1989. Molecular basis of functional diversity of voltage-gated potassium channels in mammalian brain. EMBO J. 8 3235–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemori, H., Y. Katoh, N. Horike, J. Doi and M. Okamoto, 2002. ACTH-induced nucleocytoplasmic translocation of salt-inducible kinase. Implication in the protein kinase A-activated gene transcription in mouse adrenocortical tumor cells. J. Biol. Chem. 277 42334–42343. [DOI] [PubMed] [Google Scholar]
- Trent, C., N. Tsung and H. R. Horvitz, 1983. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 104 619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel, E. R., J. H. Chou, N. D. Dwyer, H. A. Colbert and C. I. Bargmann, 1995. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83 207–218. [DOI] [PubMed] [Google Scholar]
- Troemel, E. R., B. E. Kimmel and C. I. Bargmann, 1997. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91 161–169. [DOI] [PubMed] [Google Scholar]
- van der Linden, A. M., K. M. Nolan and P. Sengupta, 2007. KIN-29 SIK regulates chemoreceptor gene expression via an MEF2 transcription factor and a class II HDAC. EMBO J. 26 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels, J. J., and J. H. Thomas, 1994. Multiple chemosensory defects in daf-11 and daf-21 mutants of Caenorhabditis elegans. Genetics 138 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner, L. E., L. A. Hardaker, S. Golik and W. R. Schafer, 2000. Effect of a neuropeptide gene on behavioral states in Caenorhabditis elegans egg-laying. Genetics 154 1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B., J. Goode, J. Best, J. Meltzer, P. E. Schilman et al., 2008. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 7 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., H. Takemori, S. K. Halder, Y. Nonaka and M. Okamoto, 1999. Cloning of a novel kinase (SIK) of the SNF1/AMPK family from high salt diet-treated rat adrenal. FEBS Lett. 453 135–139. [DOI] [PubMed] [Google Scholar]
- Ward, S., N. Thomson, J. G. White and S. Brenner, 1975. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J. Comp. Neurol. 160 313–337. [DOI] [PubMed] [Google Scholar]
- Ware, R. W., D. Clark, K. Crossland and R. L. Russell, 1975. The nerve ring of the nematode Caenorhabditis elegans: sensory input and motor output. J. Comp. Neurol. 162 71–110. [Google Scholar]
- Wicks, S. R., C. J. de Vries, H. G. van Luenen and R. H. Plasterk, 2000. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev. Biol. 221 295–307. [DOI] [PubMed] [Google Scholar]
- Woods, S. C., M. W. Schwartz, D. G. Baskin and R. J. Seeley, 2000. Food intake and the regulation of body weight. Annu. Rev. Psychol. 51 255–277. [DOI] [PubMed] [Google Scholar]
- You, Y. J., J. Kim, D. M. Raizen and L. Avery, 2008. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 7 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., H. Lu and C. I. Bargmann, 2005. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438 179–184. [DOI] [PubMed] [Google Scholar]