Abstract
Understanding how parental distance affects offspring fitness, i.e., the effects of inbreeding and outbreeding in natural populations, is a major goal in evolutionary biology. While inbreeding is often associated with fitness reduction (inbreeding depression), interpopulation outcrossing may have either positive (heterosis) or negative (outbreeding depression) effects. Within a metapopulation, all phenomena may occur with various intensities depending on the focal population (especially its effective size) and the trait studied. However, little is known about interpopulation variation at this scale. We here examine variation in inbreeding depression, heterosis, and outbreeding depression on life-history traits across a full-life cycle, within a metapopulation of the hermaphroditic snail Physa acuta. We show that all three phenomena can co-occur at this scale, although they are not always expressed on the same traits. A large variation in inbreeding depression, heterosis, and outbreeding depression is observed among local populations. We provide evidence that, as expected from theory, small and isolated populations enjoy higher heterosis upon outcrossing than do large, open populations. These results emphasize the need for an integrated theory accounting for the effects of both deleterious mutations and genetic incompatibilities within metapopulations and to take into account the variability of the focal population to understand the genetic consequences of inbreeding and outbreeding at this scale.
THE importance of inbreeding and outbreeding on fitness has been recognized for a long time (Darwin 1876; Dobzhansky 1936; Wright 1937; Crow 1948) and has more recently reclaimed importance on theoretical (Lynch 1991; Schierup and Christiansen 1996; Merilä and Sheldon 1999; Bierne et al. 2002; Edmands and Timmerman 2003; Shpak 2005) and conservational (Frankham 1999; Quilichini et al. 2001; Marr et al. 2002; Tallmon et al. 2004; Aspi et al. 2006; Willi et al. 2007) grounds. It is widely assumed that there is some optimal genetic distance (or degree of relatedness) between parents, above and below which offspring fitness decreases (Price and Waser 1979; Waser 1993). However, little can be predicted about this optimum, essentially because the two sides are explained by different theories and assumed to result from different categories of mutations.
Within populations, mating between relatives (inbreeding) is often associated with reduction in performance when compared to outbreeding, a phenomenon known as inbreeding depression (Charlesworth and Charlesworth 1987, 1999). Inbreeding depression has been observed in most plant and animal species (Husband and Schemske 1996; Crnokrak and Roff 1999; Keller and Waller 2002) and has been usually attributed to the expression of recessive lethal and sublethal mutations maintained in appreciable frequency in large outbred populations (Mukai et al. 1974; Deng et al. 1998; Keller and Waller 2002). To a lesser extent, overdominance (Charlesworth and Charlesworth 1999; Li et al. 2001) and epistasis (Lynch 1991; Lynch and Walsh 1998) may also contribute to the low performance in inbred individuals.
The same category of mutations is thought to be responsible for heterosis, i.e., the higher fitness in interpopulation hybrids compared to offspring of within-population crosses (Lynch and Walsh 1998). Heterosis seems to be mainly produced by the presence of complementary sets of deleterious recessive alleles within both parental populations and the masking of their effects in the F1 heterozygotes (Lynch and Walsh 1998; Remington and O'Malley 2000). Such alleles generally have small effects on fitness; hence they can approach fixation by random genetic drift in some isolated populations (Kimura et al. 1963; Lynch 1991; Whitlock et al. 2000). As for inbreeding depression, overdominance and epistatic interactions may also be involved in heterosis (Lynch 1991; Lynch and Walsh 1998).
However, hybridization between divergent populations may eventually result in a loss in fitness, a phenomenon known as outbreeding depression (Lynch 1991; Edmands 1999), considered as a first step in the way to speciation. Such a decline in fitness may be apparent in F1 hybrids, which have a complete haploid set of genes from each parent, but it is usually considered more likely to be observed in F2 and later generations, when deleterious interactions among recessive alleles at different loci get exposed to selection due to recombination (Lynch 1991; Edmands 1999, 2007). On the other hand, outbreeding depression can appear in the F1 as a result of the disruption of local adaptation (gene × environment interactions), underdominance, or epistatic interactions such as the breakup of favorable additive × additive epistatic effects (Burton 1990; Lynch 1991; Edmands 1999), all of which can be subsumed under the term “genetic incompatibilities.” Unlike the alleles responsible for inbreeding depression and heterosis (which are unconditionally deleterious), alleles creating genetic incompatibilities become deleterious only when they interact with a foreign genetic background. In among-population crosses, either heterosis or outbreeding depression, depending on the pairs of populations considered, has been observed in natural populations of plants (Waser and Price 1994; Trame et al. 1995; Byers 1998; Fenster and Galloway 2000; Waser et al. 2000; Pélabon et al. 2005; Willi and Van Buskirk 2005) and animals (Burton 1990; Edmands 1999; Gharrett et al. 1999; Aspi 2000; Andersen et al. 2002; Gilk et al. 2004).
Many parameters determine the expected levels of inbreeding depression, heterosis, and outbreeding depression in natural populations, including effective population size, selection coefficients, dominance coefficients, migration rates, and mutation rates (Whitlock et al. 2000). Among these, the effective population size is of great importance since genetic drift will be greatest when the effective size of local populations is smallest (Hartl and Clark 1997), which increases the likelihood of fixation of alleles (Kimura et al. 1963; Bataillon and Kirkpatrick 2000). For this reason, one expects smaller populations to exhibit reduced inbreeding depression compared to larger ones (Bataillon and Kirkpatrick 2000; Glémin 2003). On the other hand, since genetic drift and mutation cause allele frequencies to vary among local populations, smaller population size leads to a higher level of genetic differentiation among populations and hence increases heterosis caused by deleterious recessives (Whitlock et al. 2000). For the same reason, small population sizes may increase outbreeding depression due to genetic incompatibilities. Consider, for instance, an underdominant locus with alleles A and a, with fitness AA > aa > Aa. In this case, outbreeding depression is due to the increased heterozygosity in interpopulation F1 compared to within-population crosses. Natural selection favors the fixation of either A or a alleles within demes (while migration among demes tends to restore intermediate frequencies). In small demes, the effect of random genetic drift adds up to that of selection and favors even more rapid divergence and fixation of alternative alleles. Thus, interpopulation hybridization should lead to stronger outbreeding depression in small demes than in larger ones. This principle remains valid with other types of genetic incompatibilities than underdominant loci (e.g., epistasis). However, because heterosis and outbreeding depression both increase when population size is small, they may partly cancel each other, and thus the overall effect of population size is not easy to predict.
The expected effects of decreasing selection intensity are similar, to a large extent, to those of decreasing population size. This is because in both cases selection becomes weaker relative to genetic drift. Therefore, the consequences of inbreeding and outbreeding may not be the same depending on the trait under consideration and are likely to vary among traits expressed at different life stages. For example, selection is expected to be weaker on late-expressed traits (Hamilton 1966; Rose 1991). Another possible origin of variation among traits is that some traits are more mutable than others: for example, many semilethal mutations on basic functions will not be expressed further than early life stages because mutant individuals die early. Early-expressed traits therefore appear as larger “mutational targets” than late-expressed traits.
Most studies of the effects of inbreeding and outbreeding (or, more generally, parental divergence) on offspring fitness in natural populations are performed by crossing a set of individuals from a single population to increasingly distant mates. However, given the intrinsically stochastic nature of genetic drift and the process of fixation of deleterious mutations, the variation in the effective size, in the degree of isolation, and in the intensity of selection among populations, it is not clear to which extent the observed patterns (e.g., heterosis or outbreeding depression at a certain distance) are general or specific to the focal population and traits. For example, even in a metapopulation where on average there is neither heterosis nor outbreeding depression, different populations may have different average breeding values for a given trait. Such traits will tend to increase upon outcrossing (heterosis) if the focal population is below average and will tend to decrease (outbreeding depression) if it is above average. We therefore need more studies to understand the amount of variation of the effects of inbreeding and outbreeding among demes in a metapopulation.
In this article we examine how the effects of inbreeding and outbreeding vary (and covary) among traits expressed at different life stages, and among populations, in the hermaphroditic self-fertile freshwater snail Physa acuta. These populations are taken within a few kilometers in a single metapopulation, harboring different local habitats characterized by different expected population sizes and degrees of isolation. We use controlled crosses and molecular analyses to ask several questions: (i) Do inbreeding depression, heterosis, and/or outbreeding depression occur simultaneously at the same geographic scale?; (ii) If so, do they affect in the same way different traits expressed early and late in the life-cycle?; and (iii) Do local populations differ in the amount of inbreeding depression and heterosis or outbreeding depression, and are these differences related to local population characteristics represented by the habitat type?
MATERIALS AND METHODS
Studied species and sampled populations:
The studied species, P. acuta, is a simultaneously hermaphroditic freshwater snail exhibiting both high outcrossing rates (80–100%, Jarne et al. 2000; Henry et al. 2005) and high inbreeding depression (78–90%, Jarne et al. 2000; Escobar et al. 2007). It is an oviparous species laying egg capsules, typically containing a few tens of eggs. Hatching occurs within 6–7 days after egg laying, and sexual maturity is attained within 6–8 weeks at 22°−24° in laboratory conditions.
Ten populations of P. acuta were sampled within 25 km around Montpellier, southern France, in October–November 2005 (Table 1). Five of these were from isolated ponds and 5 from rivers. Pond populations are assumed to have lower effective size and to be more isolated from the rest of the metapopulation than are river populations. A previous study performed on 24 French populations of P. acuta has provided support for this assumption by finding more variation at microsatellite loci (number of alleles and gene diversity) in rivers than in ponds and higher Fst among ponds than among rivers (Bousset et al. 2004). This is in line with the fact that rivers are open habitats harboring large-sized populations that experience few bottlenecks, whereas ponds are closed habitats with small-sized populations subject to frequent bottlenecks (Bousset et al. 2004; Henry et al. 2005).
TABLE 1.
Origin, coordinates, and neutral-genetic variation parameters for the studied populations
| Habitat | Population | Coordinates | N alleles | Allelic richness | Hexp | Hobs | Fis | s | |
|---|---|---|---|---|---|---|---|---|---|
| River | Buz | 43.46 N | 3.59 E | 4.0 ± 1.6 | 3.53 ± 1.32 | 0.56 ± 0.18 | 0.51 ± 0.24 | 0.086 ± 0.097* | 0.004, NS |
| River | Lam | 43.47 N | 3.43 E | 4.1 ± 2.0 | 3.59 ± 1.46 | 0.59 ± 0.14 | 0.53 ± 0.12 | 0.075 ± 0.054, NS | 0.005, NS |
| River | Lez | 43.43 N | 3.49 E | 6.2 ± 2.7 | 3.99 ± 1.40 | 0.60 ± 0.22 | 0.50 ± 0.23 | 0.139 ± 0.062*** | 0.009, NS |
| River | Mos | 43.40 N | 3.46 E | 6.2 ± 2.9 | 4.42 ± 1.84 | 0.64 ± 0.17 | 0.55 ± 0.23 | 0.111 ± 0.084** | 0.03, NS |
| River | Sal | 43.41 N | 3.55 E | 5.6 ± 2.8 | 4.43 ± 2.02 | 0.58 ± 0.26 | 0.53 ± 0.21 | 0.087 ± 0.061* | 0.08, NS |
| Pond | Vio 01 | 43.44 N | 3.42 E | 5.2 ± 2.3 | 4.07 ± 1.82 | 0.60 ± 0.21 | 0.44 ± 0.28 | 0.203 ± 0.090*** | 0.30** |
| Pond | Vio 02 | 43.45 N | 3.43 E | 2.9 ± 0.8 | 2.28 ± 1.13 | 0.54 ± 0.17 | 0.39 ± 0.22 | 0.246 ± 0.100** | 0.26, NS |
| Pond | Vio 07 | 43.45 N | 3.46 E | 5.3 ± 2.3 | 4.00 ± 1.49 | 0.55 ± 0.21 | 0.49 ± 0.20 | 0.087 ± 0.076* | 0.008, NS |
| Pond | Vio 11 | 43.46 N | 3.47 E | 3.4 ± 1.2 | 2.94 ± 0.89 | 0.48 ± 0.21 | 0.54 ± 0.25 | −0.119 ± 0.058, NS | 0.03, NS |
| Pond | Vio 12 | 43.46 N | 3.48 E | 3.6 ± 2.1 | 3.01 ± 1.84 | 0.42 ± 0.23 | 0.37 ± 0.21 | 0.130 ± 0.072* | 0.20, NS |
| All rivers | 5.2 ± 1.1 | 3.99 ± 0.43 | 0.59 ± 0.03 | 0.52 ± 0.02 | 0.11 ± 0.03 | 0.002 | |||
| All ponds | 4.1 ± 1.1 | 3.26 ± 0.76 | 0.52 ± 0.07 | 0.45 ± 0.07 | 0.14 ± 0.17 | 0.17 | |||
N alleles, number of alleles; Hexp, gene diversity; Hobs, observed heterozygosity; s, selfing rate. *P < 0.05; **P < 0.01; ***P < 0.001; NS, P > 0.05.
Experimental setting:
Wild mature individuals (G0) were sampled in each population (100–200 individuals) and brought alive into the laboratory. A subsample from each population (∼30 individuals) was preserved in 70% ethanol for subsequent molecular analyses. In the laboratory, G0 individuals spent 1 week in high-density conditions (∼20–30 individuals per 3-liter aquariums) to ensure cross-fertilization. Behavioral and molecular evidence suggests that self-fertilization is very unlikely whenever mates are abundant (Jarne et al. 2000; Bousset et al. 2004; Henry et al. 2005; David et al. 2007). After this, 24–90 individuals per population were isolated in 75-ml plastic boxes and reproduction was checked every 2 days. Additional snails, not related to experimental families, were kept as large stock colonies (one colony per population) in optimal conditions to serve as mating partners later.
After some mortality occurred, 13–40 families per population were created by isolating egg capsules from G0 parents in 75-ml boxes. The G1 offspring of a given clutch were raised together for 2 weeks. At 15 days, density was standardized (five individuals per box) and at 22 days, prior to sexual maturity, G1 offspring of each family were isolated (one individual per box). This experimental protocol avoids mating among relatives while reducing the number of boxes to handle. At 30 days, offspring from each family were randomly split into three treatments: selfing and intra- and interpopulation outcrossing.
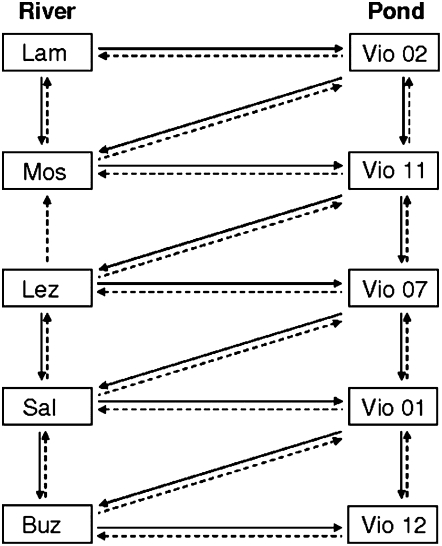
Selfers (N = 503) were kept isolated throughout the experiment until they reproduced. Intrapopulation outcrossers (N = 401) were given adult mates, randomly chosen from the stock colony corresponding to their own population. Interpopulation outcrossers (N = 872) were given adult mates, randomly chosen from a stock colony corresponding to a different population. For this treatment, each population was crossed with at least two different populations, one pond and one river, following the crossing scheme shown in Figure 1. Four types of crosses (female × male) were therefore created: pond × pond, pond × river, river × pond, and river × river. In both intra- and interpopulation outcrossing treatments, each G1 individual was mated with at least three different randomly chosen adult partners by periods of 48 hr each. To identify partners, we marked them with a dot of varnish on the shell (Henry and Jarne 2007). Eggs obtained during mating periods were discarded because of uncertainty in maternity. Only eggs obtained after the removal of partners were conserved for subsequent measures. Reproduction in all treatments was checked three times a week.
Figure 1.—
Crossing scheme for interpopulation hybridizations. Each population was crossed with at least one river and one pond population. Arrows are from females to males. Thirty-three types of interpopulation crosses were performed (note that the female Mos × male Lez cross could not be performed).
Snails were maintained at 25° under a 12 hr:12 hr light:dark photoperiod and fed with boiled lettuce. Food and water were changed twice a week.
Traits measured:
G2 eggs (mean ± SD: 78 ± 37 eggs per mother) were counted and isolated in 75-ml boxes. An estimate of early survival was obtained as the number of living G2 individuals 15 days after the egg laying divided by the number of eggs in the clutch (hereafter, 0- to 15-day survival). As in the previous generation, density was adjusted at 15 days (five individuals per box) and individuals were isolated at 22 days. Survival between 15 and 22 days was computed. Size (height from the base to the apex of the shell) was measured in G2 individuals at isolation (22 days) by means of digital pictures (Nikon coolpix 5900) analyzed with the ImageJ 1.34s software (Rasband 2007). At 45 days, virgin G2 individuals were pair mated with a single adult partner from the stock colony corresponding to the original maternal population during 24 hr. As above, partners were marked with a dot of varnish on the shell and clutches laid throughout the mating period were discarded. At 49 days, fecundity was measured as the number of eggs laid after the removal of the partner (i.e., through 72 hr). Additionally, we computed 22- to 49-day survival.
At 50 days, 1–26 G2 individuals (mean ± SD: 6 ± 4 individuals) per family and population were individually marked using a numbered, colored plastic tag for honeybees (Ickowicz Apiculture, Bollène, France) and a dot of varnish on the shell (Henry and Jarne 2007). This double marking provided unique combinations for >1200 snails. Tagged individuals (N = 1253) were randomized and grouped in 3-liter plastic aquariums at constant density (100 individuals per aquarium). Density was adjusted within aquariums every week, using when necessary unmarked nonexperimental individuals randomly taken from stock colonies. Individuals were randomized across aquaria every week to avoid common-environment effects. Mortality within aquaria was checked three times a week. Late survival of marked snails was then quantified as the age at death minus 49 days. This measure is thought to represent the reproductive life span (RLS) because the first reproduction occurred at 46–49 days for most G2 individuals (72%), constrained by the fact that we provided the mates only at this time. Every 2 weeks, marked G2 snails were isolated in 75-ml boxes during 72 hr to quantify their fecundity. At that moment, snails were measured by means of digital pictures, as described above.
Selfed and intra- and interpopulation outcrossed 0- to 15-day survival was recorded from 60,396 G2 eggs; 15- to 22- and 22- to 49-day survivals were estimated from 5837 and 2322 individuals, respectively, and RLS was estimated in 1253 individuals. Fecundity was estimated from 1801 individuals. Size was measured in 5150 individuals at 22 days and in 1117 individuals at 150 days.
Molecular analyses:
DNA was extracted using the DNeasy blood and tissue kit (QIAGEN, Valencia, CA) from 100 mg of ∼30 G0 individuals per population (N = 276), maintained in 70% ethylic alcohol, and eluted in 200 μl of sterilized water.
Ten loci were used for the PCR amplification: Pac1, Pac2, and Pac5 (Sourrouille et al. 2003); locus 27, locus 32-B, and locus 59-B (Monsutti and Perrin 1999); and Pasu1-02, Pasu1-09, Pasu1-11, and Pasu1-12 (J. Goudet, personal communication). The loci used were multiplexed in three subsets for genotype analysis: (i) Pac1, Pac2, Pac5, Pasu1-12, and locus 27; (ii) locus 32-B and locus 59-B; and (iii) Pasu1-02, Pasu1-09, and Pasu1-11. PCRs were conducted in a Mastercycler thermocycler (Eppendorf) using 10.0-μl final volumes, including 0.2 μm of each primer and 10 ng of genomic DNA, using the QIAGEN multiplex PCR kit.
PCR conditions were as follows: a 15-min activation of the HotStartTaq DNA polymerase at 95°; 30 cycles including 30 sec initial denaturation at 94°, 90 sec annealing, and 60 sec extension at 72°; followed by a 30-min final extension at 60°. For genotyping, 3.0 μl of diluted multiplexed amplicon were pooled with 13.0 μl of deionized formamide and 0.2 μl GeneScan-500 LIZ size standard and analyzed on an ABI Prism 3100 genetic analyzer.
Statistical analyses:
G2 individuals are characterized by their treatment (selfing and intra- and interpopulation outcrossing), maternal habitat (pond, river), maternal population (nested within maternal habitat), and family (nested within maternal population). In addition to the above factors, interpopulation-hybrid characterization includes the paternal habitat (pond, river) and paternal population (nested within paternal habitat).
Estimates of survival (0–15, 15–22, and 22–49 days, and RLS) were combined into a single cumulative measure (reproductive life expectancy, RLE) representing the expected number of days during which an individual is able to lay eggs. RLE was calculated as the product of survival rates from 0 to 49 days and RLS. Fecundity (number of eggs per day of reproductive life) was estimated by dividing the total number of eggs a G2 individual laid during all 72-hr isolation periods before dying and the number of days in isolation. Finally, the expected lifetime reproductive success was calculated as the product of RLE and fecundity.
Deviance analyses were performed on survival and fecundity measures with R 2.4.0 (R Development Core Team 2006). Survival up to 49 days was analyzed by fitting generalized linear models (GLM), using a binomial error distribution (logit-link function). Data on RLS (ages at death minus 49 days, because all individuals were 49 days old when measures started) were analyzed using a GLM with Gamma error distribution (inverse-link function), which is convenient to analyze survival data without censoring (Crawley 2005). Fecundity per day of reproductive life was analyzed by a GLM using Poisson error distribution (log-link function).
Deviance analyses were performed in two subsets: (i) selfed and intrapopulation outcrossed performance and (ii) intra- and interpopulation outcrossed performance, to test for inbreeding depression and heterosis (or outbreeding depression), respectively. In both cases analyses started with a complete model including all main effects: treatment, maternal habitat, maternal population (nested within maternal habitat), family (nested within maternal population), and all interactions. Then, terms were sequentially deleted, starting from higher-order interactions. To account for overdispersion, deviance ratios were computed to determine whether dropping a term from the model significantly reduced the explained deviance. These ratios are approximately F-distributed and are equivalent to F-values in ordinary ANOVA (Crawley 2005).
Size measures were analyzed separately. We fitted individual Bertalanffy's growth curves, using
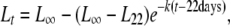 |
(1) |
where L∞ represents the mean maximum size, L22 represents the size at 22 days (i.e., our first measure), and k is a relative growth rate. Only individuals with four or more size measures were taken into account for this analysis. Growth parameters were estimated using maximum-likelihood nonlinear fits performed with Mathematica 5.1 (Wolfram Research).
We computed size at 150 days (L150) using Equation 1. Both L22 and L150 were analyzed with mixed-model ANOVA using the same factors as above. Treatment and habitat were analyzed as fixed effects, whereas population, family, and all interactions containing these two factors were considered as random effects.
Survival, fecundity, and size traits were log-transformed to express values in both a relative and an additive scale. The difference in the logarithm of fitness traits is a natural and statistically well-behaved measure of inbreeding depression and heterosis, in contrast to the traditional ratio measures (e.g., Johnston and Schoen 1994). Log-inbreeding depression was estimated for each character, family, population, and maternal habitat using the estimator
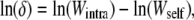 |
(2) |
where Wintra corresponds to the mean fitness of intrapopulation-outcrossed offspring and Wself holds for the mean fitness of selfed offspring. Note that Equation 2 is not a simple logarithmic transformation of the classic estimator of inbreeding depression, but a measure of the inbreeding load (Charlesworth and Charlesworth 1987; Willis 1999). Likewise, log heterosis was estimated using
 |
(3) |
where Winter holds for the mean fitness of interpopulation hybrids. Note that log-transformed estimates can be back transformed to recover classic estimates using 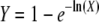 ; Y represents either inbreeding depression (δ) or heterosis (H). For survival traits, in some cases W = 0 (no egg or individual survived) and logarithmic transformation was impossible. To avoid biasing the data set by dropping these values, we used the transformation ln(0.01 + W) instead of ln(W) (Escobar et al. 2007).
; Y represents either inbreeding depression (δ) or heterosis (H). For survival traits, in some cases W = 0 (no egg or individual survived) and logarithmic transformation was impossible. To avoid biasing the data set by dropping these values, we used the transformation ln(0.01 + W) instead of ln(W) (Escobar et al. 2007).
Microsatellite data served to estimate neutral variation within populations and between habitat types, fixation indexes (within and among populations and between habitat types, namely Fis, Fst, and Fct, respectively), and intrapopulation selfing rates. Molecular data were analyzed with Arlequin 3.11 (Excoffier et al. 2005), FSTAT 2.9.3 (Goudet 1995), and RMES (David et al. 2007). The RMES software estimates selfing rates on the basis of the distribution of multilocus heterozygosity; these estimates, unlike those derived from Fis, have been shown to be insensitive to technical artifacts such as null alleles (David et al. 2007). Differences in allelic richness and expected heterozygosity between ponds and rivers were calculated, and significance of the test was assessed using a scheme of 1000 permutations (keeping the number of samples in each habitat type constant) with FSTAT 2.9.3 (Goudet 1995).
Components of phenotypic variation for life-history traits were estimated among and within populations, as well as between habitats, using data from the intrapopulation outcrossing treatment. ANOVA were computed to estimate variance components. Analyses included habitat, population, and family as random effects. Survival and fecundity measures were log-transformed, whereas size measures were not. Variance components served to calculate quantitative indexes of phenotypic divergence (Spitze 1993) among populations within habitats (Qst) and between habitats (Qct), using
 |
(4) |
and
 |
(5) |
We additionally performed deviance analyses to distinguish between additive and nonadditive effects on hybrid fitness, as well as between symmetric and asymmetric nonadditive effects, using data from both intra- and interpopulation outcrossing treatments. We constructed an additive design matrix containing the number of genes (0, 1, or 2) contributed by each of the 10 populations to each cross (for example, in the cross female A × male B, A and B contribute one gene each; while in the intrapopulation cross A × A, population A contributes two genes and population B contributes none). We then fitted GLMs on (i) the above-described matrix of additive effects (in this way fitted values of interpopulation hybrids are forced to equal the midparent value); (ii) the identity of both parental populations, irrespective of which population acted as mother or father, i.e., A × B = B × A (one value is fitted for each cross except that reciprocal crosses are forced equal); and (iii) maternal × paternal interaction in which we did distinguish reciprocal crosses (i.e., A × B ≠ B × A). The first fit indicates additive effects on hybrid fitness, the second model includes symmetric nonadditive effects, and the third term indicates asymmetric nonadditive effects (e.g., maternal effects). Significance of the terms was obtained by means of deviance ratios, as described above.
Finally, three types of correlation analyses were performed: (i) between inbreeding depression, heterosis, and neutral diversity parameters (allelic richness and gene diversity); (ii) between population genetic divergence (calculated as 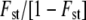 ; Rousset 1997) and interpopulation hybrid performance and heterosis; and (iii) between inbreeding depression and heterosis for all measured traits. Tests for associations were performed using Pearson's r and Spearman's ρ with JMP 7.0 (SAS Institute, Cary, NC).
; Rousset 1997) and interpopulation hybrid performance and heterosis; and (iii) between inbreeding depression and heterosis for all measured traits. Tests for associations were performed using Pearson's r and Spearman's ρ with JMP 7.0 (SAS Institute, Cary, NC).
RESULTS
Timing of inbreeding depression, heterosis, and outbreeding depression:
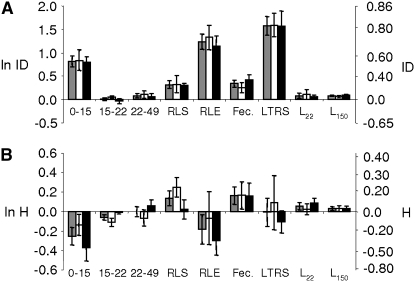
The magnitude of inbreeding depression, heterosis, and outbreeding depression markedly changed across the life cycle. We detected strong inbreeding depression early in the life cycle (mean value of 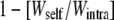 ± SEM over populations [range]: 0- to 15-day survival, 0.56 ± 0.06 [0.15–0.77]), no inbreeding depression at middle-age stages (15–22 days, 0.01 ± 0.03 [−0.28–0.12]; 22–49 days, 0.08 ± 0.04 [−0.13–0.30]), and significant inbreeding depression for adult survival (RLS: 0.27 ± 0.05 [0.01–0.64]) (Table 2; Figure 2A). When combining survival at all stages (RLE), we detected a very strong inbreeding depression (0.71 ± 0.05 [0.38–0.87]), mostly due to the high value at 0–15 days (Figure 2A). Inbreeding depression on fecundity was also significant (0.29 ± 0.06 [−0.01–0.52]) (Table 2; Figure 2A). Our cumulative fitness measure (the lifetime reproductive success), representing the number of eggs an individual lays across a full-life cycle, exhibited a very strong inbreeding depression (0.79 ± 0.05 [0.45–0.91]) (Figure 2A). Size measures exhibited slight, though significant inbreeding depression (22 days, 0.08 ± 0.03 [−0.07–0.24]; 150 days, 0.09 ± 0.02 [0.02–0.16]) (Table 2; Figure 2A).
± SEM over populations [range]: 0- to 15-day survival, 0.56 ± 0.06 [0.15–0.77]), no inbreeding depression at middle-age stages (15–22 days, 0.01 ± 0.03 [−0.28–0.12]; 22–49 days, 0.08 ± 0.04 [−0.13–0.30]), and significant inbreeding depression for adult survival (RLS: 0.27 ± 0.05 [0.01–0.64]) (Table 2; Figure 2A). When combining survival at all stages (RLE), we detected a very strong inbreeding depression (0.71 ± 0.05 [0.38–0.87]), mostly due to the high value at 0–15 days (Figure 2A). Inbreeding depression on fecundity was also significant (0.29 ± 0.06 [−0.01–0.52]) (Table 2; Figure 2A). Our cumulative fitness measure (the lifetime reproductive success), representing the number of eggs an individual lays across a full-life cycle, exhibited a very strong inbreeding depression (0.79 ± 0.05 [0.45–0.91]) (Figure 2A). Size measures exhibited slight, though significant inbreeding depression (22 days, 0.08 ± 0.03 [−0.07–0.24]; 150 days, 0.09 ± 0.02 [0.02–0.16]) (Table 2; Figure 2A).
TABLE 2.
Inbreeding depression for survival, fecundity and size measures
| 0- to 15-day survival
|
15- to 22-day survival
|
22- to 49-day survival
|
RLS
|
Fecundity
|
Size 22 days
|
Size 150 days
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source | Dev | F | Dev | F | Dev | F | Dev | F | Dev | F | MSQ | F | MSQ | F |
| Treatment | 2584.0 | 144.72*** | 1.9 | 0.89, NS | 3.7 | 2.27, NS | 10.9 | 34.58*** | 168.8 | 36.26*** | 0.101 | 6.37* | 0.033 | 22.01*** |
| (1; 902) | (1; 628) | (1; 594) | (1; 590) | (1; 590) | (1; 70) | (1; 10) | ||||||||
| Population | 1147.0 | 7.61*** | 148.4 | 8.55*** | 101.6 | 7.53*** | 18.4 | 7.12*** | 118.4 | 2.91*** | 0.062 | 3.47** | 0.101 | 4.63* |
| (9; 893) | (9; 619) | (9; 585) | (9; 581) | (9; 581) | (8; 62) | (7; 10) | ||||||||
| Habitat × treatment | 54.0 | 3.23, NS | 2.9 | 1.51, NS | 2.2 | 1.44, NS | 0.2 | 0.63, NS | 15.1 | 3.35, NS | 0.024 | 1.52, NS | 0.001 | 0.07, NS |
| (1; 892) | (1; 618) | (1; 584) | (1; 580) | (1; 580) | (1; 70) | (1; 10) | ||||||||
| Pop. × treatment | 679.0 | 5.27*** | 20.4 | 1.33, NS | 15.7 | 1.31, NS | 4.2 | 1.83, NS | 45.9 | 1.28, NS | 0.009 | 0.48, NS | 0.015 | 1.06, NS |
| (8; 884) | (8; 610) | (8; 576) | (8; 572) | (8; 572) | (8; 62) | (7; 55) | ||||||||
| Family | 6550.8 | 3.69*** | 413.9 | 1.65*** | 266.0 | 1.27* | 37.9 | 0.96, NS | 630.9 | 1.02, NS | 0.022 | 0.97, NS | 0.022 | 1.51, NS |
| (166; 718) | (152; 458) | (150; 426) | (138; 434) | (138; 434) | (55; 55) | (44; 44) | ||||||||
| Family × treatment | 1678.9 | 2.23*** | 114.5 | 1.23, NS | 94.9 | 1.28, NS | 15.2 | 1.20, NS | 254.0 | 1.30, NS | 0.022 | 4.49*** | 0.015 | 1.16, NS |
| (80; 638) | (58; 400) | (55; 371) | (45; 389) | (45; 389) | (55; 1298) | (44; 182) | ||||||||
Dev, deviance explained by the factor considered (i.e., change in deviance between models with and without that factor); MSQ, mean square; RLS, reproductive life span. The treatment factor has two modalities: selfed and intrapopulation outcrossed. Numerator and denominator degrees of freedom are given in parentheses with the corresponding F-values. ***P < 0.001; **P < 0.01; *P < 0.05; NS, P > 0.05. Note that for size at 150 days, there was no selfed progeny from population Vio 12. Therefore, the degrees of freedom associated with population are 7 instead of 8.
Figure 2.—
Inbreeding depression and heterosis for life-history traits. Left y-axes, the log-inbreeding depression (ln ID in A), and the log heterosis (ln H in B); right y-axes, classic inbreeding depression (A) and heterosis (B). Shaded bars, mean values over the two habitat types; open bars, means for ponds; and solid bars, means for rivers. Error bars were calculated as SEM over populations. RLS, reproductive life span; RLE, reproductive life expectancy; Fec., fecundity; LTRS, lifetime reproductive success; L22, size at 22 days; L150, size at 150 days. Note that ln ID and ln H (left y-axes) do not represent a simple logarithmic transformation of values presented in the right y-axes (see materials and methods for details).
On the other hand, significantly negative heterosis (i.e., outbreeding depression) for survival was detected in the two earliest age classes (mean value of 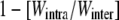 ± SEM over populations [range]: 0–15 days, −0.29 ± 0.12 [−1.03–0.18]; 15–22 days, −0.06 ± 0.03 [−0.32–0.02]), no heterosis at 22–49 days (−0.004 ± 0.06 [−0.45–0.19]) and significant and positive heterosis in adult survival (RLS: 0.13 ± 0.07 [−0.17–0.34]) (Table 3; Figure 2B). Combining all survival estimates, we detected negative heterosis on RLE (−0.20 ± 0.22 [−1.96–0.39]) (Figure 2B). In contrast, significant positive heterosis was detected in fecundity (0.15 ± 0.08 [−0.29–0.47]). Remarkably, when combining RLE and fecundity, we found neither heterosis nor outbreeding depression on the lifetime reproductive success (−0.02 ± 0.18 [−1.41–0.59]) (Figure 2B). Size at 22 days exhibited no significant heterosis (0.05 ± 0.04 [−0.12–0.19]), and the heterosis observed on size at 150 days was very low though significant (0.03 ± 0.02 [–0.04–0.11]) (Table 3; Figure 2B).
± SEM over populations [range]: 0–15 days, −0.29 ± 0.12 [−1.03–0.18]; 15–22 days, −0.06 ± 0.03 [−0.32–0.02]), no heterosis at 22–49 days (−0.004 ± 0.06 [−0.45–0.19]) and significant and positive heterosis in adult survival (RLS: 0.13 ± 0.07 [−0.17–0.34]) (Table 3; Figure 2B). Combining all survival estimates, we detected negative heterosis on RLE (−0.20 ± 0.22 [−1.96–0.39]) (Figure 2B). In contrast, significant positive heterosis was detected in fecundity (0.15 ± 0.08 [−0.29–0.47]). Remarkably, when combining RLE and fecundity, we found neither heterosis nor outbreeding depression on the lifetime reproductive success (−0.02 ± 0.18 [−1.41–0.59]) (Figure 2B). Size at 22 days exhibited no significant heterosis (0.05 ± 0.04 [−0.12–0.19]), and the heterosis observed on size at 150 days was very low though significant (0.03 ± 0.02 [–0.04–0.11]) (Table 3; Figure 2B).
TABLE 3.
Heterosis for survival, fecundity, and size measures
| 0- to 15-day survival
|
15- to 22-day survival
|
22- to 49-day survival
|
RLS
|
Fecundity
|
Size 22 days
|
Size 150 days
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source | Dev | F | Dev | F | Dev | F | Dev | F | Dev | F | MSQ | F | MSQ | F |
| Habitat | 46 | 2.26, NS | 2.6 | 1.16, NS | 8.1 | 5.22* | 5.1 | 20.09*** | 2.7 | 0.72, NS | 0.033 | 1.42, NS | 0.054 | 0.56, NS |
| (1; 1271) | (1; 992) | (1; 942) | (1; 993) | (1; 991) | (1; 74) | (1; 17) | ||||||||
| Population | 1891 | 12.42*** | 237.1 | 14.73*** | 158.7 | 14.21*** | 11.5 | 5.86*** | 497.9 | 19.24*** | 0.043 | 1.62, NS | 0.167 | 6.02* |
| (8; 1263) | (8; 984) | (8; 934) | (8; 985) | (8; 983) | (8; 68) | (8; 7) | ||||||||
| Treatment | 356.0 | 18.98*** | 21.2 | 10.64** | 0.5 | 0.36, NS | 7.4 | 31.17*** | 24.7 | 7.69** | 0.053 | 2.12, NS | 0.150 | 6.70* |
| (1; 1262) | (1; 983) | (1; 933) | (1; 984) | (1; 982) | (1; 73) | (1; 17) | ||||||||
| Habitat × treatment | 106.0 | 5.67* | 7.1 | 3.57, NS | 0.6 | 0.43, NS | 3.8 | 16.01*** | 0.2 | 0.06, NS | 0.020 | 0.81, NS | 0.032 | 1.43, NS |
| (1; 1261) | (1; 982) | (1; 932) | (1; 983) | (1; 981) | (1; 73) | (1; 17) | ||||||||
| Pop. × treatment | 612.0 | 4.18*** | 14.0 | 0.88, NS | 15.0 | 1.34, NS | 5.3 | 2.85** | 153.6 | 6.22*** | 0.012 | 0.43, NS | 0.028 | 1.71, NS |
| (8; 1253) | (8; 974) | (8; 924) | (8; 975) | (8; 973) | (8; 68) | (8; 48) | ||||||||
| Family | 7317.0 | 2.52*** | 579.6 | 1.76*** | 291.1 | 1.14, NS | 47.5 | 1.18*** | 723.6 | 1.40** | 0.032 | 0.92, NS | 0.016 | 0.95, NS |
| (196; 1057) | (190; 784) | (188; 736) | (180; 795) | (180; 793) | (62; 62) | (44; 44) | ||||||||
| Family × treatment | 2891.0 | 3.06*** | 96.9 | 0.90, NS | 113.6 | 1.40* | 10.2 | 0.97, NS | 161.6 | 1.21, NS | 0.035 | 5.64*** | 0.017 | 1.27, NS |
| (73; 984) | (62; 722) | (62; 674) | (47; 748) | (47; 746) | (62; 1807) | (44; 288) | ||||||||
Definitions are the same as in Table 2. Treatment now includes two modalities: within-population outcrossing and interpopulation outcrossing. Population refers to the maternal populations (common to both treatments).
Neutral and phenotypic divergence among populations:
We first analyzed the neutral genetic variation within populations and we found that rivers exhibited more neutral variation than ponds (Table 1): differences in gene diversity were significantly greater in rivers than in ponds (one-sided P-value = 0.006) and nearly significant for the allelic richness (one-sided P-value = 0.061). The multilocus selfing rate was significant in one pond population (Vio 01, s = 0.30), whereas other pond populations and all river populations exhibited selfing rates not significantly different from zero (Table 1). Analysis of genetic and geographical divergence among populations did not provide evidence for isolation by distance (Mantel test, Z = 81.74, r = −0.18, P = 0.73). Genetic divergence (Fst) among all populations was 0.12 ± 0.01. It was stronger among pond populations (0.20 ± 0.07) than among river populations (0.08 ± 0.02). The genetic differentiation between the two habitat types (pond vs. river) was quite low, though significant (AMOVA Fct: 0.02 ± 0.03, P < 0.05) (Table 4).
TABLE 4.
Genetic variances associated with the maternal habitat type on measured life-history traits
| Habitat | Variance component | 0- to 15-day survival | 15- to 22-day survival | 22- to 49-day survival | RLS | Fecundity | Size 22 days | Size 150 days | Mean Fst | Mean Qst | AMOVA Fct |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pond | Interpopulation | 0.050 | 0.024 | 0.634 | 1.905 | 5.141 | 0.0008 | 0.005 | |||
| Intrapopulation | 1.334 | 0.075 | 0.334 | 42.748 | 3.838 | 0.002 | 0.001 | ||||
| Qst | 0.018 | 0.138 | 0.487 | 0.022 | 0.401 | 0.159 | 0.689 | 0.20 ± 0.07 | 0.27 ± 0.26 | ||
| River | Interpopulation | 0.194 | 0.142 | 0.323 | 34.348 | 0.453 | 0.0003 | 0.003 | |||
| Intrapopulation | 0.296 | 0.129 | 0.283 | 89.322 | 0.735 | 0.002 | 0.003 | ||||
| Qst | 0.247 | 0.355 | 0.363 | 0.161 | 0.236 | 0.064 | 0.335 | 0.08 ± 0.02 | 0.25 ± 0.11 | ||
| Qct | 0.020 | −0.060 | −0.030 | 0.410 | −0.100 | −0.020 | −0.110 | 0.02 ± 0.03 |
Variance components were calculated using the intrapopulation outcrossing treatment alone. Survival and fecundity variances were calculated after logarithmic transformation, whereas sizes were not. RLS, reproductive life span.
We then analyzed population variance for life-history traits using the intrapopulation outcrossing treatment alone. We found that the habitat type significantly affected both early survival and RLS: rivers exhibited on average slightly greater 0- to 15-day survival than ponds (0.58 ± 0.07 and 0.49 ± 0.06, respectively) and much longer RLS (51.03 ± 3.19 and 37.57 ± 1.24 days, respectively) (Table 5). Among-population variation (within habitats) was significant for all measured traits, whereas within-population (among-family) variation was significant only for 0- to 15- and 15- to 22-day survival and the two size measures (Table 5). Among- and within-population variances for life-history traits served to calculate the quantitative index of population divergence (Qst) between habitat types and to compare this measure with the among-population fixation index on neutral markers (Fst). On the one hand, rivers had mean Qst almost always greater than mean Fst (the only exception corresponds to size at 22 days). On the other hand, ponds exhibited mean Qst lower than mean Fst for 0- to 15- and 15- to 22-day survival, RLS, and size at 22 days and Qst > Fst for 22- to 49-day survival, fecundity, and size at 150 days (Table 4). Finally, Fct was almost always >Qct, with the exception of RLS, where Qct ≫ Fct (Table 4), consistent with the large difference observed between the two habitats for this trait.
TABLE 5.
Deviance analyses for the intrapopulation outcrossing treatment alone
| 0- to 15-day survival
|
15- to 22-day survival
|
22- to 49-day survival
|
RLS
|
Fecundity
|
Size 22 days
|
Size 150 days
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source | Dev | F | Dev | F | Dev | F | Dev | F | Dev | F | MSQ | F | MSQ | F |
| Habitat | 152.4 | 7.60** | 0.23 | 0.12, NS | 3.74 | 2.32, NS | 7.9 | 34.73*** | 1.0 | 0.23, NS | 0.033 | 1.59, NS | 0.013 | 0.13, NS |
| (1; 399) | (1; 343) | (1; 330) | (1; 332) | (1; 332) | (1; 130) | (1; 8) | ||||||||
| Population | 962.4 | 6.69*** | 132.0 | 10.12*** | 76.5 | 6.76*** | 4.1 | 2.32* | 132.1 | 4.03*** | 0.079 | 3.43** | 0.112 | 6.35*** |
| (8; 391) | (8; 335) | (8; 322) | (8; 324) | (8; 324) | (8; 123) | (8; 130) | ||||||||
| Family | 4242.8 | 3.43*** | 272.1 | 1.93*** | 184.5 | 1.27, NS | 23.9 | 1.09, NS | 423.5 | 1.02, NS | 0.028 | 5.70*** | 0.019 | 1.48* |
| (120; 271) | (114; 221) | (112; 210) | (102; 222) | (102; 222) | (112; 1337) | (98; 195) | ||||||||
Dev, deviance explained by the factor considered (i.e., change in deviance between models with and without that factor); MSQ, mean square; RLS, reproductive life span. Numerator and denominator degrees of freedom are given in parentheses with the corresponding F-values. ***P < 0.001; **P < 0.01; *P < 0.05; NS, P > 0.05.
Population variability for inbreeding depression, heterosis, and outbreeding depression:
Among-population variation in inbreeding depression was significant only for 0- to 15-day survival (population-by-treatment interaction in Table 2). However, this variation was not related with the maternal habitat (mean δ ± SEM ponds, 0.56 ± 0.11; rivers, 0.55 ± 0.05; Table 2; Figure 2A). In all other traits inbreeding depression neither varied between habitats nor varied among populations within habitats (Table 2; Figure 2A). Importantly, almost all populations exhibited inbreeding depressions on the lifetime reproductive success >0.5, the threshold above which cross-fertilization is assumed to be favored in most theoretical models. The only exception corresponds to the Buz population (0.45) (Table 6). This result is in line with the fact that P. acuta is a preferentially cross-fertilizing species.
TABLE 6.
Log-inbreeding depression and log heterosis on measured life-history traits per population
| Population | 0- to 15-day survival | 15- to 22-day survival | 22- to 49-day survival | RLS | RLE | Fecundity | LTRS | Size 22 days | Size 150 days |
|---|---|---|---|---|---|---|---|---|---|
| Buz | 0.5378 | −0.2453 | −0.1184 | 0.3086 | 0.4826 | 0.1220 | 0.6047 | 0.0417 | 0.1061 |
| (−0.4282) | (0.0068) | (0.1806) | (−0.1341) | (−0.3750) | (0.3550) | (−0.0199) | (−0.0334) | (0.0624) | |
| Lam | 0.9963 | −0.0097 | 0.2470 | 0.2319 | 1.4654 | 0.5617 | 2.0271 | −0.0108 | 0.1137 |
| (−0.4405) | (−0.0323) | (−0.0804) | (−0.0297) | (−0.5828) | (0.4686) | (−0.1142) | (0.1791) | (0.0958) | |
| Lez | 1.1252 | 0.0633 | 0.0720 | 0.4380 | 1.6985 | 0.7451 | 2.4436 | 0.2248 | 0.0441 |
| (−0.4119) | (−0.0530) | (0.0604) | (0.4018) | (−0.0027) | (−0.2522) | (−0.2548) | (−0.0221) | (−0.0171) | |
| Mos | 0.8016 | −0.0139 | 0.1431 | 0.2828 | 1.2136 | 0.4534 | 1.6671 | 0.0083 | 0.1456 |
| (−0.7071) | (0.0174) | (0.2075) | (−0.1541) | (−0.6363) | (0.2508) | (−0.3855) | (0.1511) | (0.0161) | |
| Sal | 0.5352 | 0.0511 | −0.0254 | 0.2949 | 0.8559 | 0.2379 | 1.0937 | 0.0298 | 0.0765 |
| (0.1327) | (−0.0094) | (−0.0650) | (0.0261) | (0.0844) | (−0.0183) | (0.0661) | (0.1600) | (0.0006) | |
| Vio 01 | 1.4636 | 0.0817 | 0.0302 | 0.2534 | 1.8289 | 0.2338 | 2.0627 | 0.2461 | 0.1047 |
| (−0.4186) | (−0.0712) | (0.0555) | (0.3395) | (−0.0949) | (−0.2320) | (−0.3269) | (−0.0819) | (−0.0405) | |
| Vio 02 | 1.0051 | 0.0402 | 0.1730 | 0.0110 | 1.2293 | 0.5188 | 1.7481 | 0.4001 | 0.0738 |
| (−0.1125) | (−0.0634) | (0.0979) | (0.3225) | (0.2445) | (0.6434) | (0.8880) | (0.0497) | (0.1189) | |
| Vio 07 | 0.1637 | 0.0309 | 0.3504 | 0.0984 | 0.6434 | 0.0583 | 0.7017 | 0.1627 | −0.0241 |
| (0.2041) | (−0.0444) | (−0.0915) | (0.4229) | (0.4912) | (0.0753) | (0.5665) | (0.0372) | (0.0211) | |
| Vio 11 | 0.5757 | −0.0174 | 0.1097 | 0.2661 | 0.9341 | 0.4977 | 1.4317 | −0.0200 | 0.1106 |
| (−0.0725) | (−0.0938) | (−0.0279) | (0.3049) | (0.1106) | (0.1459) | (0.2565) | (−0.1173) | (−0.0244) | |
| Vio 12 | 0.9936 | 0.1271 | −0.1263 | 1.0220 | 2.0164 | −0.0121 | 2.0044 | −0.2351 | NA |
| (−0.2943 | (−0.2801) | (−0.3726) | (−0.1367) | (−1.0837) | (0.2031) | (−0.8806) | (0.2150) | (0.0774) |
Log-inbreeding depression is provided outside and log heterosis within brackets. Note that for size at 150 days there was no surviving selfed progeny from population Vio 12; therefore the inbreeding depression could not be estimated (NA).
For each population i, the effect of outbreeding can be measured, keeping maternal effects constant, by using the relative difference (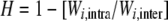 ) between the performance of offspring from crosses within population i (Wi,intra) and that of interpopulation hybrids whose mothers belong to population i and fathers belong to another population (Wi,inter). This relative difference significantly varied among populations for early survival (0–15 days), RLS, and fecundity (maternal population-by-treatment interaction; Table 3). Importantly, this variation was significantly related to the maternal habitat type for two of these three characters (maternal habitat-by-treatment interaction; Table 3): 0- to 15-day survival and RLS. Early survival decreased more upon interpopulation crossing when the mother population was in a river than when it was in a pond (H = −0.45 ± 0.18 in rivers and −0.15 ± 0.12 in ponds). RLS increased more upon outbreeding when the mother population was in a pond than when it was in a river (H = 0.22 ± 0.09 and 0.02 ± 0.09, respectively). These interactions were still significant when the two types of interpopulation crosses (with a pond and with a river paternal population) were distinguished in the analysis (data not shown). All other fitness traits showed similar effects of outcrossing between ponds and rivers (Figure 2B). Analysis of the lifetime reproductive success (LTRS) showed that rivers tended to exhibit negative heterosis (i.e., outbreeding depression) on the cumulative fitness measure, whereas ponds tended to have positive heterosis (Figure 2B; Table 6). However, this trend was not significant due to large variation among populations within each habitat.
) between the performance of offspring from crosses within population i (Wi,intra) and that of interpopulation hybrids whose mothers belong to population i and fathers belong to another population (Wi,inter). This relative difference significantly varied among populations for early survival (0–15 days), RLS, and fecundity (maternal population-by-treatment interaction; Table 3). Importantly, this variation was significantly related to the maternal habitat type for two of these three characters (maternal habitat-by-treatment interaction; Table 3): 0- to 15-day survival and RLS. Early survival decreased more upon interpopulation crossing when the mother population was in a river than when it was in a pond (H = −0.45 ± 0.18 in rivers and −0.15 ± 0.12 in ponds). RLS increased more upon outbreeding when the mother population was in a pond than when it was in a river (H = 0.22 ± 0.09 and 0.02 ± 0.09, respectively). These interactions were still significant when the two types of interpopulation crosses (with a pond and with a river paternal population) were distinguished in the analysis (data not shown). All other fitness traits showed similar effects of outcrossing between ponds and rivers (Figure 2B). Analysis of the lifetime reproductive success (LTRS) showed that rivers tended to exhibit negative heterosis (i.e., outbreeding depression) on the cumulative fitness measure, whereas ponds tended to have positive heterosis (Figure 2B; Table 6). However, this trend was not significant due to large variation among populations within each habitat.
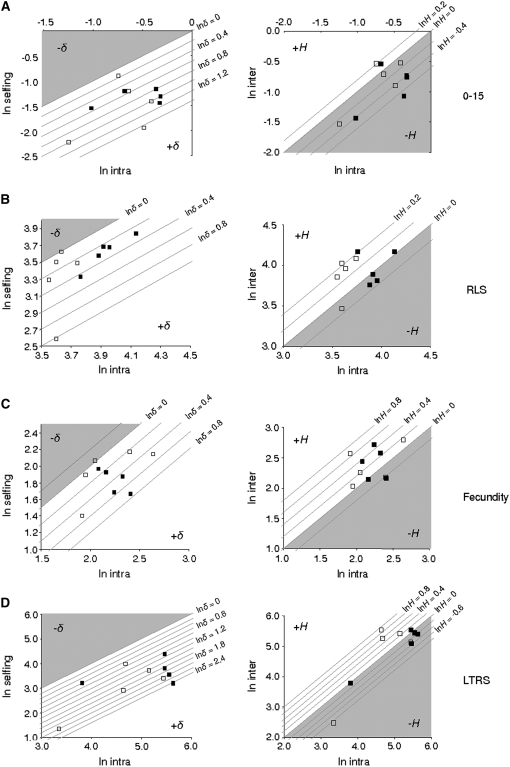
For each population, we plotted the fitness of inbred individuals and interpopulation hybrids against the fitness of within-population cross-fertilized individuals to know whether differences in inbreeding depression and heterosis among populations could be due to additive effects. Under additive effects, populations having above-average values for fitness components in the intrapopulation outcrossing treatment would tend to exhibit less heterosis (or more outbreeding depression) than populations below average. This is because (additive) breeding values of interpopulation hybrids are the average of their two parental populations and therefore lower than the best, and higher than the worst, of these two parental populations. With regard to inbreeding depression, the same principle holds for different families within a population: families with above-average breeding values will tend to show less inbreeding depression than below-average families. Because we sampled a finite number of families within each population, the average of our sample can be, by chance, either higher or lower than the true population mean, resulting in a downward or an upward bias (respectively) on our estimate of inbreeding depression and a lower apparent inbreeding depression in above-average populations. No such tendencies were, however, observed in our analyses (Figure 3 and supplemental Figure 1).
Figure 3.—
Log-fitness traits for self-fertilized offspring (ln selfing, left) and interpopulation hybrids (ln inter, right) as a function of log-fitness traits for intrapopulation outcrossed offspring (ln intra). Only the most important fitness components are represented. (A) 0- to 15-day survival; (B) reproductive life span (RLS); (C) fecundity; (D) lifetime reproductive success (LTRS). Lines represent constant values of log-inbreeding depression (ln δ, left and log heterosis (ln H, right). Open squares, ponds; solid squares, rivers. Open areas represent positive values and shaded areas negative values of inbreeding depression and heterosis, respectively (note that negative heterosis represents outbreeding depression). Note change in scales among graphs.
Additive, nonadditive, and asymmetric effects of interpopulation hybrids:
We further analyzed data from both intra- and interpopulation outcrossing treatments to determine the relative importance of additive, nonadditive, and asymmetric effects of interpopulation hybrids. By nonadditivity, we mean that interpopulation crosses differ from the average of parental populations. By asymmetry, we mean a difference in the fitness of a cross between two populations, depending on which population assumes the maternal role. Significant nonadditive effects were present for all traits, and significant asymmetries between reciprocal crosses were detected in all traits but RLS and size at 150 days (Table 7). This is visible in Figure 4 (and supplemental Figure 2), which shows that most interpopulation crosses differ from their midparent value and often lie outside the parental range. Nonadditive effects are either positive (heterosis) or negative (outbreeding depression), depending on the trait and population. Figure 4 also illustrates that the two reciprocal crosses often yield contrasted results.
TABLE 7.
Deviance analyses testing for asymmetries between reciprocal crosses in fitness traits
| 0- to 15-day survival
|
15- to 22-day survival
|
22- to 49-day survival
|
RLS
|
Fecundity
|
Size 22 days
|
Size 150 days
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | Dev | F | Dev | F | Dev | F | Dev | F | Dev | F | Dev | F | Dev | F |
| Additive | 1662 | 9.60*** | 214 | 11.67*** | 148.7 | 11.67*** | 18.0 | 8.16*** | 396.3 | 13.18*** | 0.59 | 7.69*** | 1.34 | 9.76*** |
| (9; 1263) | (9; 984) | (9; 934) | (9; 985) | (9; 983) | (9; 1941) | (9; 386) | ||||||||
| Symmetric nonadditive | 1523 | 4.90*** | 71.3 | 2.10** | 43.5 | 1.84* | 19.0 | 4.88*** | 213.3 | 3.95*** | 1.18 | 8.73*** | 0.71 | 3.20*** |
| (17; 1246) | (17; 967) | (17; 917) | (17; 968) | (17; 966) | (17; 1924) | (16; 370) | ||||||||
| Asymmetric nonadditive | 1133 | 4.02*** | 58.2 | 1.84* | 42.0 | 1.91* | 3.7 | 1.02, NS | 182.1 | 3.74*** | 0.45 | 3.88*** | 0.21 | 1.15, NS |
| (16; 1230) | (16; 951) | (16; 901) | (16; 952) | (16; 950) | (15; 1909) | (13; 357) | ||||||||
Definitions are the same as in Table 2. Data include intra- and interpopulation crosses. The additive model constrains interpopulation crosses to have performances equal to their midparent value. The symmetric nonadditive model relaxes this constraint, but constrains reciprocal interpopulation crosses to be equal. Finally the asymmetric nonadditive model fits one value for each cross including reciprocals.
Figure 4.—
Intra- and interpopulation outcrossing performances for the most important fitness components. (A) 0- to 15-day survival; (B) reproductive life span; (C) fecundity. Solid squares and lines represent intrapopulation outcrossing; shaded squares and lines represent interpopulation outcrossing. Results are presented for each pair of populations (two shaded squares and two solid squares). For instance, the first two populations on the left (Lam and Mos) should be read as follows: the solid square on the left represents the value for the Lam × Lam cross, and the shaded square represents the value for the cross female Lam × male Mos. The solid square on the right represents the value for the Mos × Mos cross, and the shaded square the value for the female Mos × male Lam cross. In this representation, differences between parental populations are indicated by the slope of the solid line, and asymmetries between reciprocal crosses are indicated by the slope of the shaded line. Different shadings denote heterosis (interpopulation hybrids fitter than intrapopulation crosses) and outbreeding depression (vice versa) as indicated. By convention, the river is on the left and the pond on the right in all mixed pairs of populations. Note that one cross (female Mos × male Lez) could not be performed.
Correlation analyses:
Three types of association tests were performed: (i) across maternal populations, between neutral-diversity parameters and inbreeding depression or heterosis; (ii) across pairs of populations, between population-genetic divergence and interpopulation-hybrid performance; and (iii) across maternal populations, between inbreeding depression and heterosis for all measured traits. None of the three types of correlations were significant, when considering our global measure of fitness (lifetime reproductive success). The same was true for all other traits, after correction for multiple comparisons (supplemental Tables 1–3).
DISCUSSION
In this study we manipulated the relatedness among parental individuals, from selfing to interpopulation outcrossing, as well as the direction of hybridization (e.g., we distinguished between female A × male B and female B × male A crosses), and evaluated fitness effects in F1 in the form of inbreeding depression, heterosis, and outbreeding depression expressed during a full-life cycle. We further analyzed theoretical predictions concerning the effective population size, neutral genetic diversity, and genetic variance on all three parameters.
Inbreeding depression, heterosis, and outbreeding depression simultaneously occur in the F1 of a structured population:
Our results show that within a metapopulation, inbreeding depression, heterosis, and outbreeding depression are simultaneously expressed on different traits. Inbreeding depression affects basically all fitness traits and is generally high (0.79 on the cumulative fitness measure on average), which is not surprising in a predominantly outcrossing species (70–100% outcrossing from our molecular data, in agreement with previous data on the same species; Jarne et al. 2000; Escobar et al. 2007). To a certain extent heterosis, the increase in fitness traits upon interpopulation crossing, reflects the same type of mutations as inbreeding depression; i.e., it also stems from directional dominance, most likely due to a constant mutational input of deleterious, recessive alleles (Lynch 1991). An additional force is needed to generate heterosis: genetic drift, allowing allele frequencies to diverge among populations (Whitlock et al. 2000). The importance of drift in our snail metapopulation is indicated by significant and relatively high neutral microsatellite Fst (at such a small scale, all populations within 25 km), mostly in the range 10–20% (overall 12%). This degree of population structure appears sufficient for populations to fix (or nearly fix) different sets of mildly deleterious mutations, thus producing heterosis upon crossing.
The occurrence of significant outbreeding depression in the F1 of demes a few kilometers apart is more surprising. Outbreeding depression is usually considered an important issue when divergent populations, often recognized as distinct subspecies or well on the way to speciation, are brought into contact (e.g., during reintroduction programs) rather than for two demes in a local metapopulation. Moreover, outbreeding depression is considered more likely to be expressed in the F2, because different populations can more easily accumulate genetic incompatibilities (i.e., alleles with negative epistatic interaction) when the latter are completely recessive (the Dobzhansky–Muller model; Orr and Turelli 2001). Our data show that genetic incompatibilities that are not purely recessive can accumulate in metapopulations and diverge in frequency among demes by genetic drift, even without long-term complete isolation, in such a way as to generate substantial outbreeding depression in the F1. The existence of such incompatibilities adds complexity to the interpretation of the observed pattern, because interpopulation F1 will express the balance between two classes of alleles with opposite effects: deleterious recessives (creating heterosis) and genetic incompatibilities (creating outbreeding depression). Similar data have been obtained in plants from demes separated by short distances (Waser and Price 1989; Fenster 1991; Waser and Price 1994; Fischer and Matthies 1997).
Inbreeding depression, heterosis, and outbreeding depression reveal differential impacts of mutations along the life cycle:
The magnitude of inbreeding depression, heterosis, and outbreeding depression was very variable along the life cycle in P. acuta. Inbreeding depression was strongest early in life, inexistent at intermediate stages, and increased again after sexual maturity (RLS and fecundity). Such stage-specific differences in inbreeding depression have already been observed in other predominantly outcrossing organisms, especially plants (Husband and Schemske 1996; Koelewijn et al. 1999). They are thought to arise because both mutational pressure and selection intensity vary among stages. Strong early-acting inbreeding depression reflects the expression of recessive lethal and sublethal mutations affecting basic functions (Jarne and Charlesworth 1993; Husband and Schemske 1996; Remington and O'Malley 2000). In organisms that survive the juvenile stage, the inbreeding depression observed in later life stages is probably due to a more restricted set of partially recessive mildly deleterious genes with age-specific expression (e.g., genes involved in late growth or reproduction). In agreement with this idea, we found no correlation between inbreeding depression among life stages, suggesting that genes causing inbreeding depression are to some extent independent across the life cycle.
The fitness in F1 interpopulation hybrids reveals the balance between heterosis and outbreeding depression. Theory states that the fitness loss caused by genetic incompatibilities has to exceed twice the benefit from dominance for the F1 population to exhibit outbreeding depression (Lynch 1991). Our results show that outbreeding depression predominates at early stages while heterosis predominates at the reproductive stage (RLS and fecundity). This suggests that viability is relatively more affected by genetic incompatibilities while mutations affecting specifically the reproductive stage are more of the deleterious recessive type. The expression of strong incompatibilities affecting viability is surprising because, in the context of speciation, it is often assumed that hybrid fertility is compromised earlier than hybrid viability during the process of population divergence (Presgraves 2003). For example, interspecific F1's often show heterosis for survival and condition, while at least one sex is sterile (Orr and Turelli 2001). Interestingly, in our data set, late-acting heterosis and early-acting outbreeding depression cancel out when analyzing our cumulative fitness measure (the lifetime reproductive success); i.e., averaging over the entire life cycle, interpopulation F1 hybrids are as fit as offspring from local residents. As for inbreeding depression, heterosis was not correlated among life stages, suggesting independence among genes at different stages of the life cycle. Furthermore, inbreeding depression and heterosis were not correlated to each other at any stage of the life cycle, which is expected because inbreeding depression is due to mutations segregating within demes, while heterosis is more affected by mutations that reach fixation in some demes.
Genetic divergence among populations and habitat types:
As expected, and in agreement with previous studies (Bousset et al. 2004; Henry et al. 2005), rivers exhibited more neutral genetic variation and less genetic divergence (Fst) than ponds. River populations also outperformed pond populations with respect to early (0–15 days) and late (RLS) survival (intrapopulation outcrossing treatment). This suggests that small population size and isolation enhance genetic drift within populations, which in turn has parallel effects on neutral variation (less variation within and more variation among demes) and on nonneutral variation (larger standing genetic load).
Comparisons between phenotypic (Qst) and molecular (Fst) divergence are usually interpreted as characterizing the type of selection (spatially heterogeneous selection if Qst > Fst and homogeneous selection when Fst > Qst; McKay and Latta 2002; Goudet and Büchi 2006). However, this interpretation should be viewed with caution here because the traits involved (survival and fecundity) are likely to be positively selected in all populations rather than selected for intermediate optima, and we did not detect correlations with other traits that could be indicative of trade-offs. Although heterogeneous selection cannot be excluded, the fact that the habitat type (river vs. pond) explains more variance in RLS than in molecular traits (Qct > Fct), as mentioned above, likely reflects a differential impact of drift (fixation of mildly deleterious alleles by drift being more frequent in small isolated populations) rather than different selection regimes. Similarly, the larger Qst for most traits in the pond group of populations, relative to the river group, probably reflects a larger influence of drift in pond populations (so that even selected traits behave more like neutral ones in ponds). At any rate, one should remain cautious in comparing Fst and Qst here, both because of the imprecision in Qst estimates (based on relatively few populations) and because of dominance, the presence of deleterious recessives, and epistasis, all of which can affect classical predictions (Whitlock 1999; López-Fanjul et al. 2003; Goudet and Büchi 2006).
Because the mean and variance in mutation frequency across demes depend on genetic drift, inbreeding depression, heterosis, and outbreeding depression are all expected to vary stochastically among populations. This is basically what we found. However, variation in inbreeding depression seems limited to the early survival stage, and basically all populations show strong inbreeding depression. On the other hand, the balance between outbreeding depression and heterosis seems more variable, with positive values for some population pairs and negative values for others, for both early and late traits. More specific predictions can be made on interpopulation variation assuming that local populations vary with respect to effective size and isolation. In small and isolated populations, deleterious allele frequencies should more easily drift away from the deterministic equilibrium and from the metapopulation mean (up to local fixation) (Kimura et al. 1963; Bataillon and Kirkpatrick 2000; Glémin et al. 2001). They should therefore exhibit less inbreeding depression and more heterosis than larger ones. However, considering that heterosis is expected to arise from milder mutations than inbreeding depression on average, the sensitivity to population sizes should differ: populations must be very small to lose a substantial part of their inbreeding depression, while even moderately small effective sizes allow heterosis to increase (Kirkpatrick and Jarne 2000; Glémin et al. 2001; Glémin 2003). Habitat (pond vs. river), our proxy for population size, did not explain interpopulation variation in inbreeding depression in our data set; however, it did have a significant effect on heterosis for RLS in the expected direction; i.e., ponds benefited more from interpopulation outcrossing than did rivers. This suggests that the range of variation in effective size among the studied populations was sufficient to generate differential rates of fixation of mildly deleterious recessive alleles, but populations were still large enough to avoid the fixation of stronger mutations and thus retain their inbreeding depression. Interpreting why the effect of interpopulation outcrossing on early survival was less detrimental to ponds than to rivers is more complicated. Assuming that this effect represents the sum of a negative term (outbreeding depression) and a positive one (heterosis), this observation could be explained (as for RLS) by the increase in heterosis expected in small populations. However, small population size, by enhancing the influence of genetic drift and genetic divergence among populations, should in principle increase both heterosis and outbreeding depression, and the effect could therefore go both ways. Unfortunately, we lack precise theoretical expectations to predict whether both outbreeding depression and heterosis could respond to the same range of variation in population size.
Additive and nonadditive effects on variation in heterosis and outbreeding depression:
The variation in heterosis or outbreeding depression among populations could be due in part to additive effects, i.e., differences in the average breeding value among populations for a given trait. For example, although alleles with purely additive effects generate neither heterosis nor outbreeding depression on average, they contribute to the variance among populations because populations with below-average breeding values (e.g., pond populations) will tend to gain fitness, while populations with above-average breeding value (e.g., river populations) will tend to lose fitness, when mixing their genes with genes from other populations. If additive effects were mainly responsible for differences in heterosis/outbreeding depression among populations, we expect (i) that these differences would be determined mainly by the performances in the within-population crosses, (ii) that the highest heterosis should be observed in ponds, when crossing them with a river population, though not (or less) when crossing them with another pond population. We have no evidence for either i or ii. The interaction between maternal habitat type and cross type (i.e., the variation in heterosis among maternal habitat types) remains significant once the paternal habitat is controlled for in the model. Moreover, very significant nonadditive interactions among the paternal and maternal populations are observed, and heterosis occurs in pond × pond crosses just as it does in pond × river crosses. Interactions in a given population pair seem highly idiosyncratic, with some heterotic crosses and some crosses showing outbreeding depression, sometimes involving the same maternal populations (Figure 4 and supplemental Figure 2). In addition, many of the crosses show asymmetry between reciprocal crosses, which is globally significant for all fitness traits but RLS. Asymmetries can result from several mechanisms, including maternal effects (Roach and Wulff 1987), nucleocytoplasmic gene interactions (Galloway and Fenster 1999; Pélabon et al. 2005), and locally adapted alleles interacting differently with native and foreign genetic backgrounds (Wright 1931; Wade 1992).
With respect to early survival, the observed asymmetry is certainly strongly influenced by maternal effects. Even if eggs were separated from their mother immediately after oviposition, maternal effects can be mediated through egg cytoplasm (e.g., amount of nutritive reserves) and such mechanisms may dominate all other sources of asymmetry in early-expressed characters (Roach and Wulff 1987). Conversely, maternal effects are probably limited when evaluating adult traits, such as fecundity. In this case, hypotheses invoking genetic background, such as nucleocytoplasmic incompatibilities, commonly observed for fecundity traits (reviewed in Budar et al. 2003), are probably involved. Further research is needed to determine whether the genetic divergence between P. acuta populations involves other mechanisms than nuclear genes.
In conclusion, we provide evidence for the simultaneous expression of inbreeding depression, heterosis, and outbreeding depression in first-generation offspring, at the scale of local demes within a metapopulation. We show that the relative importance of the three phenomena varies along the life cycle, i.e., among different fitness traits, which are exposed to different impacts of both mutation pressure and selection. Specifically, inbreeding depression and outbreeding depression are more intense in early-expressed traits while heterosis is moderate and more visible during the reproductive stage. In addition, we provide evidence that the effects of interpopulation crossing largely vary depending on the focal maternal populations. A large part of this variation is not additive and is influenced by maternal or cytoplasmic effects, reflecting unpredictable interactions between genetic backgrounds that have diverged through stochastic processes. However, as predicted by theory, habitat type, through its effect on effective population size and/or immigration rates, explains a part of the variation in heterosis among populations.
Acknowledgments
We acknowledge P. Jarne and D. Promislow for comments on the manuscript; J. Goudet for unpublished microsatellite primers; and V. Sarda, G. Hinet, J. Terraube, and E. Dincuff for help in the laboratory. J.S.E. was supported by the Programme Alban (grant E04D045840CO). Funds for this research were given by the Centre National pour la Recherche Scientifique.
References
- Andersen, D. H., C. Pertoldi, V. Scali and V. Loeschcke, 2002. Intraspecific hybridization, developmental stability and fitness in Drosophila mercatorum. Evol. Ecol. Res. 4 603–621. [Google Scholar]
- Aspi, J., 2000. Inbreeding and outbreeding depression in male courtship song characters in Drosophila montana. Heredity 84 273–282. [DOI] [PubMed] [Google Scholar]
- Aspi, J., E. Roininen, M. Ruokonen, I. Kojola and C. Vila, 2006. Genetic diversity, population structure, effective population size and demographic history of the Finnish wolf population. Mol. Ecol. 15 1561–1576. [DOI] [PubMed] [Google Scholar]
- Bataillon, T., and M. Kirkpatrick, 2000. Inbreeding depression due to mildly deleterious mutations in finite populations: size does matter. Genet. Res. 75 75–81. [DOI] [PubMed] [Google Scholar]
- Bierne, N., T. Lenormand, F. Bonhomme and P. David, 2002. Deleterious mutations in a hybrid zone: Can mutational load decrease the barrier to gene flow? Genet. Res. 80 197–204. [DOI] [PubMed] [Google Scholar]
- Bousset, L., P.-Y. Henry, P. Sourrouille and P. Jarne, 2004. Population biology of the invasive freshwater snail Physa acuta approached through genetic markers, ecological characterization and demography. Mol. Ecol. 13 2023–2036. [DOI] [PubMed] [Google Scholar]
- Budar, F., P. Touzet and R. De Paepe, 2003. The nucleo-mitochondrial conflict in cytoplasmic male sterilities revisited. Genetica 117 3–16. [DOI] [PubMed] [Google Scholar]
- Burton, R. S., 1990. Hybrid breakdown in developmental time in the copepod Tigriopus californicus. Evolution 44 1814–1822. [DOI] [PubMed] [Google Scholar]
- Byers, D. L., 1998. Effect of cross proximity on progeny fitness in a rare and a common species of Eupatorium (Asteraceae). Am. J. Bot. 85 644–653. [PubMed] [Google Scholar]
- Charlesworth, B., and D. Charlesworth, 1999. The genetic basis of inbreeding depression. Genet. Res. 74 329–340. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., and B. Charlesworth, 1987. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Evol. Syst. 18 237–268. [Google Scholar]
- Crawley, M. J., 2005. Statistics: An Introduction Using R. Wiley Publishers, London.
- Crnokrak, P., and D. A. Roff, 1999. Inbreeding depression in the wild. Heredity 83 260–270. [DOI] [PubMed] [Google Scholar]
- Crow, J. F., 1948. Alternative hypotheses of hybrid vigor. Genetics 33 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, C., 1876. The Effects of Cross and Self Fertilisation in the Vegetable Kingdom. John Murray, London.
- David, P., B. Pujol, F. Viard, V. Castella and J. Goudet, 2007. Reliable selfing rate estimates from imperfect population genetic data. Mol. Ecol. 16 2474–2487. [DOI] [PubMed] [Google Scholar]
- Deng, H. W., Y. X. Fu and M. Lynch, 1998. Inferring the major genomic mode of dominance and overdominance. Genetica 102/103 559–567. [PubMed] [Google Scholar]
- Dobzhansky, T., 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmands, S., 1999. Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution 53 1757–1768. [DOI] [PubMed] [Google Scholar]
- Edmands, S., 2007. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 16 463–475. [DOI] [PubMed] [Google Scholar]
- Edmands, S., and C. C. Timmerman, 2003. Modeling factors affecting the severity of outbreeding depression. Conserv. Biol. 17 883–892. [Google Scholar]
- Escobar, J. S., G. Epinat, V. Sarda and P. David, 2007. No correlation between inbreeding depression and delayed selfing in the freshwater snail Physa acuta. Evolution 61 2655–2670. [DOI] [PubMed] [Google Scholar]
- Excoffier, L., G. Laval and S. Schneider, 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1 47–50. [PMC free article] [PubMed] [Google Scholar]
- Fenster, C. B., 1991. Effect of male pollen donor and female seed parent on allocation resources to developing seeds and fruit in Chamaecrista fasciculata (Leguminosae). Am. J. Bot. 78 12–23. [Google Scholar]
- Fenster, C. B., and L. F. Galloway, 2000. Population differentiation in an annual legume: genetic architecture. Evolution 54 1157–1172. [DOI] [PubMed] [Google Scholar]
- Fischer, M., and D. Matthies, 1997. Mating structure and inbreeding and outbreeding depression in the rare plant Gentianella germanica (Gentianaceae). Am. J. Bot. 84 1685–1692. [PubMed] [Google Scholar]
- Frankham, R., 1999. Quantitative genetics in conservation biology. Genet. Res. 74 237–244. [DOI] [PubMed] [Google Scholar]
- Galloway, L. F., and C. B. Fenster, 1999. The effect of nuclear and cytoplasmic genes on fitness and local adaptation in an annual legume, Chamaecrista fasciculata. Evolution 53 1734–1743. [DOI] [PubMed] [Google Scholar]
- Gharrett, A. J., W. W. Smoker, R. R. Reisenbichler and S. G. Taylor, 1999. Outbreeding depression in hybrids between odd- and even-broodyear pink salmon. Aquaculture 173 117–129. [Google Scholar]
- Gilk, S. E., I. A. Wang, C. L. Hoover, W. W. Smoker, S. G. Taylor et al., 2004. Outbreeding depression in hybrids between spatially separated pink salmon, Oncorhynchus gorbuscha, populations: marine survival, homing ability, and variability in family size. Environ. Biol. Fish. 69 287–297. [Google Scholar]
- Glémin, S., 2003. How are deleterious mutations purged? Drift versus nonrandom mating. Evolution 57 2678–2687. [DOI] [PubMed] [Google Scholar]
- Glémin, S., T. Bataillon, J. Ronfort, A. Mignot and I. Olivieri, 2001. Inbreeding depression in small populations of self-incompatible plants. Genetics 159 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet, J., 1995. FSTAT (Version 1.2): a computer program to calculate F-statistics. J. Hered. 86 485–486. [Google Scholar]
- Goudet, J., and L. Büchi, 2006. The effects of dominance, regular inbreeding and sampling design on QST, an estimator of population differentiation for quantitative traits. Genetics 172 1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, W. D., 1966. The moulding of senescence by natural selection. J. Theor. Biol. 12 12–45. [DOI] [PubMed] [Google Scholar]
- Hartl, D. L., and A. G. Clark, 1997. Principles of Population Genetics. Sinauer Associates, Sunderland, MA.
- Henry, P. Y., and P. Jarne, 2007. Marking hard-shelled gastropods: tag loss, impact on life-history traits, and perspectives in biology. Invertebr. Biol. 126 138–153. [Google Scholar]
- Henry, P. Y., L. Bousset, P. Sourrouille and P. Jarne, 2005. Partial selfing, ecological disturbance and reproductive assurance in an invasive freshwater snail. Heredity 95 428–436. [DOI] [PubMed] [Google Scholar]
- Husband, B., and D. Schemske, 1996. Evolution of the magnitude and timing of inbreeding depression in plants. Evolution 50 54–70. [DOI] [PubMed] [Google Scholar]
- Jarne, P., and D. Charlesworth, 1993. The evolution of the selfing rate in functionally hermaphrodite plants and animals. Annu. Rev. Ecol. Evol. Syst. 24 441–466. [Google Scholar]
- Jarne, P., M. A. Perdieu, A. F. Pernot and P. David, 2000. The influence of self-fertilization and grouping on fitness attributes in the freshwater snail Physa acuta: population and individual inbreeding depression. J. Evol. Biol. 13 645–655. [Google Scholar]
- Johnston, M. O., and D. J. Schoen, 1994. On the measurement of inbreeding depression. Evolution 48 1735–1741. [DOI] [PubMed] [Google Scholar]
- Keller, L. F., and D. M. Waller, 2002. Inbreeding effects in wild populations. Trends Ecol. Evol. 17 230–241. [Google Scholar]
- Kimura, M., T. Maruyama and J. F. Crow, 1963. The mutation load in small populations. Genetics 48 1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick, M., and P. Jarne, 2000. The effect of a bottleneck on inbreeding depression and the genetic load. Am. Nat. 155 154–167. [DOI] [PubMed] [Google Scholar]
- Koelewijn, H. P., V. Koski and O. Savolainen, 1999. Magnitude and timing of inbreeding depression in Scots pine (Pinus sylvestris L.). Evolution 53 758–768. [DOI] [PubMed] [Google Scholar]
- Li, Z. K., L. J. Luo, H. W. Mei, D. L. Wang, Q. Y. Shu et al., 2001. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics 158 1737–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Fanjul, C., A. Fernández and M. A. Toro, 2003. The effect of neutral nonadditive gene action on the quantitative index of population divergence. Genetics 164 1627–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., 1991. The genetic interpretation of inbreeding depression and outbreeding depression. Evolution 45 622–629. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Marr, A. B., L. F. Keller and P. Arcese, 2002. Heterosis and outbreeding depression in descendants of natural immigrants to an inbred population of song sparrows (Melospiza melodia). Evolution 56 131–142. [DOI] [PubMed] [Google Scholar]
- McKay, J. K., and R. G. Latta, 2002. Adaptive population divergence: markers, QTL and traits. Trends Ecol. Evol. 17 285–291. [Google Scholar]
- Merilä, J., and B. C. Sheldon, 1999. Genetic architecture of fitness and nonfitness traits: empirical patterns and development of ideas. Heredity 83 103–109. [DOI] [PubMed] [Google Scholar]
- Monsutti, A., and N. Perrin, 1999. Dinucleotide microsatellite loci reveal a high selfing rate in the freshwater snail Physa acuta. Mol. Ecol. 8 1075–1092.10434426 [Google Scholar]
- Mukai, T., T. K. Watanabe and O. Yamaguchi, 1974. Genetic structure of natural populations of Drosophila melanogaster. 12. Linkage disequilibrium in a large local population. Genetics 77 771–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and M. Turelli, 2001. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55 1085–1094. [DOI] [PubMed] [Google Scholar]
- Pélabon, C., M. L. Carlson, T. F. Hansen and W. S. Armbruster, 2005. Effects of crossing distance on offspring fitness and developmental stability in Dalechampia scandens (Euphorbiaceae). Am. J. Bot. 92 842–851. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163 955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, M. V., and N. M. Waser, 1979. Pollen dispersal and optimal outcrossing in Delphinium nelsoni. Nature 277 294–297. [Google Scholar]
- Quilichini, A., M. Debussche and J. D. Thompson, 2001. Evidence for local outbreeding depression in the Mediterranean island endemic Anchusa crispa Viv. (Boraginaceae). Heredity 87 190–197. [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna.
- Rasband, W., 2007. ImageJ 1.34s. Image processing and analysis in Java. National Institutes of Health, Bethesda, MD.
- Remington, D. L., and D. M. O'Malley, 2000. Whole-genome characterization of embryonic stage inbreeding depression in a selfed loblolly pine family. Genetics 155 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach, D. A., and R. D. Wulff, 1987. Maternal effects in plants. Annu. Rev. Ecol. Evol. Syst. 18 209–235. [Google Scholar]
- Rose, M. R., 1991. Evolutionary Biology of Aging. Oxford University Press, New York.
- Rousset, F., 1997. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup, M. H., and F. B. Christiansen, 1996. Inbreeding depression and outbreeding depression in plants. Heredity 77 461–468. [Google Scholar]
- Shpak, M., 2005. The role of deleterious mutations in allopatric speciation. Evolution 59 1389–1399. [PubMed] [Google Scholar]
- Sourrouille, P., C. Debain and P. Jarne, 2003. Microsatellite variation in the freshwater snail Physa acuta. Mol. Ecol. Notes 3 21–23. [Google Scholar]
- Spitze, K., 1993. Population structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics 135 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallmon, D. A., G. Luikart and R. S. Waples, 2004. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 19 489–496. [DOI] [PubMed] [Google Scholar]
- Trame, A. M., A. J. Coddington and K. N. Paige, 1995. Field and genetic-studies testing optimal outcrossing in Agave schottii, a long-lived clonal plant. Oecologia 104 93–100. [DOI] [PubMed] [Google Scholar]
- Wade, M. J., 1992. Sewall Wright: gene interaction and the shifting balance theory. Oxf. Surv. Evol. Biol. 8 35–62. [Google Scholar]
- Waser, N. M., 1993. Population structure, optimal outbreeding, and assortative mating in angiosperms, pp. 173–199 in The Natural History of Inbreeding and Outbreeding, edited by N. W. Thornhill. University of Chicago Press, Chicago.
- Waser, N. M., and M. V. Price, 1989. Optimal outcrossing in Ipomopsis aggregata: seed set and offspring fitness. Evolution 43 1097–1109. [DOI] [PubMed] [Google Scholar]
- Waser, N. M., and M. V. Price, 1994. Crossing-distance effects in Delphinium nelsonii, outbreeding and inbreeding depression in progeny fitness. Evolution 48 842–852. [DOI] [PubMed] [Google Scholar]
- Waser, N. M., M. V. Price and R. G. Shaw, 2000. Outbreeding depression varies among cohorts of Ipomopsis aggregata planted in nature. Evolution 54 485–491. [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C., 1999. Neutral additive variance in a metapopulation. Genet. Res. 74 215–221. [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C., P. K. Ingvarsson and T. Hatfield, 2000. Local drift load and the heterosis of interconnected populations. Heredity 84 452–457. [DOI] [PubMed] [Google Scholar]
- Willi, Y., and J. Van Buskirk, 2005. Genomic compatibility occurs over a wide range of parental genetic similarity in an outcrossing plant. Proc. R. Soc. B Biol. Sci. 272 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willi, Y., M. Van Kleunen, S. Dietrich and M. Fischer, 2007. Genetic rescue persists beyond first-generation outbreeding in small populations of a rare plant. Proc. R. Soc. B Biol. Sci. 274 2357–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, J. H., 1999. The contribution of male-sterility mutations to inbreeding depression in Mimulus guttatus. Heredity 83 337–346. [DOI] [PubMed] [Google Scholar]
- Wright, S., 1931. Evolution in Mendelian populations. Genetics 16 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1937. The distribution of gene frequencies in populations. Proc. Natl. Acad. Sci. USA 23 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]