Abstract
Multiple theoretical studies have focused on the concerted evolution of the tandemly repeated rRNA genes of eukaryotes; however, these studies did not consider the transposable elements that interrupt the rRNA genes in many organisms. For example, in insects, R1 and R2 have been stable components of the rDNA locus for hundreds of millions of years, suggesting either that they have minimal effects on fitness or that they are unable to be eliminated. We constructed a simulation model of recombination and retrotransposition within the rDNA locus that addresses the population dynamics and fitness consequences associated with R1 and R2 insertions. The simulations suggest that even without R1 and R2 retrotransposition the frequent sister chromatid exchanges postulated from various empirical studies will, in combination with selection, generate rDNA loci that are much larger than those needed for transcription. These large loci enable the host to tolerate high levels of R1 and R2 insertions with little fitness consequences. Changes in retrotransposition rates are likely to be accommodated by adjustments in sister chromatid exchange (SCE) rate, rather than by direct selection on the number of uninserted rDNA units. These simulations suggest that the rDNA locus serves as an ideal niche for the long-term survival of transposable elements.
THE ribosomal RNA genes (rDNA) of eukaryotes are organized as hundreds to thousands of tandem units. Each unit is composed of an 18S, a 5.8S, and a 28S rRNA gene. The number of these units can vary considerably within a species with most organisms encoding many more units than are needed to be transcribed at any one time (Conconi et al. 1992a,b; Dammann et al. 1995). The level of nucleotide divergence between rDNA units within species has been estimated to be <0.1% (Ganley and Kobayashi 2007; Stage and Eickbush 2007). The ability of the rDNA units to change their sequence over time, yet retain high identity within species has been termed concerted evolution (reviewed in Eickbush and Eickbush 2007). Many theoretical studies that focused on the concerted evolution of the rDNA locus have revealed that unequal crossovers and gene conversions can readily account for the high levels of sequence identity between repeats (Ohta 1980; Ohta and Dover 1983; Nagylaki 1984; Walsh 1987; Stephan 1989). These studies have also shown that either stabilizing selection or loop deletion is needed to prevent the number of repeats from becoming unbounded (Walsh 1987; Lyckegaard and Clark 1991). Finally, empirical data have suggested that sister chromatid exchange is the most frequent type of recombination within rDNA loci (Petes 1980; Seperack et al. 1988; Schlotterer and Tautz 1994).
In many groups of animals a significant fraction of the rDNA units are interrupted by transposable elements highly specialized for insertion into conserved sites within the rRNA genes (Eickbush and Eickbush 2007). rDNA units inserted by these elements are nonfunctional, suggesting selective pressure against these insertions, yet the rDNA-specific transposable elements are highly successful. For example, the R1 and R2 retrotransposons of arthropods are present in all lineage of arthropods tested to date with many species containing multiple families of R1 and/or R2 (Jakubczak et al. 1991; Burke et al. 1998). The wide distribution of these elements appears to be explained by their vertical transmission since the origin of this phylum (Burke et al. 1998; Malik et al. 1999; Gentile et al. 2001). R1 and R2 are dynamic components of the rDNA locus with insertion frequencies that can exceed 50% of the units (Jakubczak et al. 1992). Studies of R1 and R2 elements in Drosophila have shown that the collections of R1 and R2 within the rDNA loci turn over rapidly at the population level (Jakubczak et al. 1992; Perez-Gonzalez and Eickbush 2001). While the rates of insertion and deletion have been estimated (Perez-Gonzalez and Eickbush 2002; Zhang et al. 2008), no studies have attempted to address the impact of these elements on the host.
In this article we present simulation studies of the rDNA loci in Drosophila that incorporate as variable parameters R1 and R2 retrotransposition rates, various types and rates of recombination, and natural selection. These studies provide insights into why the rDNA loci of most organisms contain many more rDNA units than are needed for transcription, how the levels of R1 and R2 insertions in the rDNA locus are affected by the rates of recombination and retrotransposition, and what possible fitness consequences are associated with the presence of R1 and R2.
METHODS
The simulation model was implemented in C language, compiled using gcc (version 4.0.1, Apple Computer), and run on Mac Pro computers. The simulation program is available upon request. To minimize the effect of genetic drift, for every set of parameters the simulations were repeated 50 times and the results averaged. The standard errors are presented in Figures 3, 5, and 6 but are frequently so small as to be covered by the symbol used to present the data point. The ranges of value used for the various parameters in the simulations are shown in Table 1.
Figure 3.—
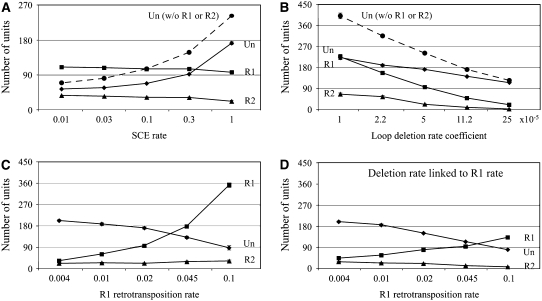
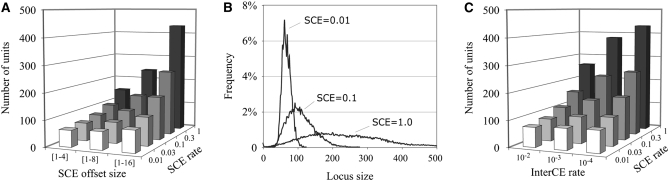
Effects of R1 and R2 retrotransposition on the rDNA locus. (A) The numbers of uninserted (Un), R1-inserted (R1, includes double-inserted), and R2-inserted (R2, includes double-inserted) rDNA units are plotted as a function of SCE rate (InterCE = 1 × 10−4, offset = 1–8, loop-deletion coefficient = 5 × 10−5, R1 rate = 0.02, R2 rate = 0.005). Dashed line, number of units (uninserted) under identical parameters but without R1 and R2. (B) The number of inserted and uninserted units as a function of the loop deletion rate (InterCE = 1 × 10−4, offset = 1–8, SCE = 1.0, R1 rate = 0.02, R2 rate = 0.005). Dashed line, number of units (uninserted) under identical parameters but without R1 and R2. (C) Mean number of inserted and uninserted units as a function of R1 retrotransposition rate (InterCE = 1 × 10−4, offset = 1–8, SCE = 1.0, loop-deletion coefficient = 5 × 10−5, R2 rate = 0.005). (D) Mean number of inserted and uninserted units when the deletion rate was also a function of the R1 retrotransposition rate. All parameters were as in C except that the deletion rate was also a function of R1 retrotransposition (see methods).
Figure 5.—
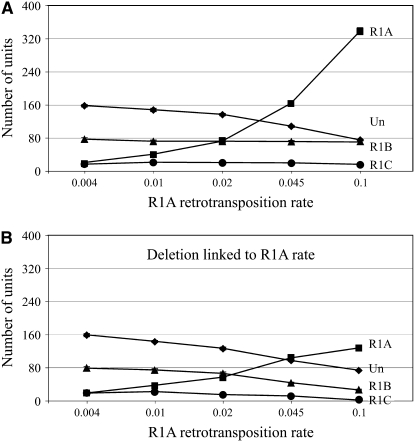
Competition among R1 families. The numbers of inserted (R1) and uninserted (UN) units were plotted as a function of the R1A family retrotransposition rate. All parameters were as in Figure 3C, except R2 was not added, R1B rate = 0.02, and R1C rate = 0.005. (A) The loop-deletion coefficient = 5 × 10−5. (B) The deletion rate was determined as in Figure 3D.
Figure 6.—
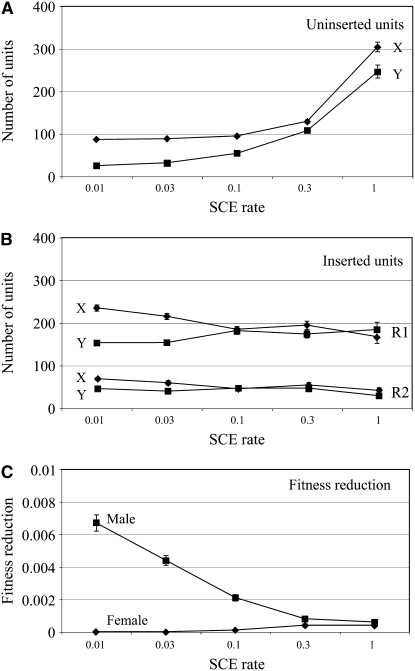
Simulations involving rDNA loci on both the X and the Y chromosomes. All parameters were as in Figure 3A with the number of inserted and uninserted units separately plotted for the X and Y loci. (A) Mean number of uninserted units as a function of SCE rates. (B) Mean number of R1- and R2-inserted units as a function of SCE rates. (C) Fitness reduction of males and females as a function of SCE rates.
TABLE 1.
Parameter values used in the simulations
| Parameter | Range tested | Empirical estimates |
|---|---|---|
| SCE ratea | 0.01–1.0 | No estimate; 0.2 in yeast (Petes 1980), 0.04 in Daphnia (McTaggart et al. 2007) |
| InterCE ratea | 10−2−10−4 | 10−4 (Williams et al. 1989) |
| SCE, InterCE offset | 1–16 units | No estimate |
| Selection, w1 | 10–90 units | 40 units (Hawley and Marcus 1989; Jakubczak et al. 1992) |
| Selection, w2 | 60–100 units | No estimate |
| Loop deletion coefficienta | 1–25 (× 10−5) | No estimate |
| Deletion offset | 1–15 units | No estimate |
| Retrotransposition ratesa | 0.004–0.1 | 0.0015–0.13 (Perez-Gonzalez and Eickbush 2002; Zhang et al. 2008) |
Events per rDNA locus per generation.
Recombination:
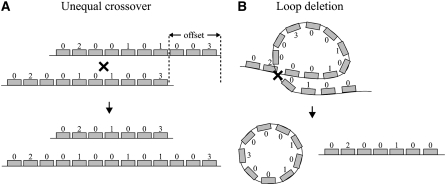
An rDNA locus was modeled by a string of symbols with uninserted units represented by 0, R1-inserted by 1, R2-inserted by 2, and double-inserted by 3 (Figure 1A). To simulate crossover events, two rDNA arrays were aligned by their centers, shifted relative to each other by an offset randomly determined between 1 and the maximum offset size, the two arrays were cut at a random location in the overlap region, and the ends of the two arrays exchanged (Figure 1A). Sister chromatid exchange (SCE) involved crossovers between two copies of an rDNA locus after DNA replication. Interchromosomal exchange (InterCE) involved exchanges between two X loci or the X and Y loci. A direct estimate of the SCE rate exists only for yeast (Petes 1980) and Daphnia (McTaggart et al. 2007); however, it has been suggested to be more frequent than InterCE in Drosophila melanogaster (Schlotterer and Tautz 1994). The InterCE rate has been estimated in D. melanogaster at 10−4 events per chromosome per generation for both X–X and X–Y chromosome exchanges (Williams et al. 1989). Estimates of the offset sizes involved in these exchanges are also unavailable. We have therefore used three ranges in our simulations: maximum of 4 units, maximum of 8 units, and maximum of 16 units.
Figure 1.—
Diagram of the simulations of sister chromatid exchange (SCE) and loop deletions. (A) The rDNA loci were modeled as strings of symbols with uninserted units represented by 0, R1-inserted by 1, R2-inserted by 2, and double-inserted by 3. The outcome of recombination was determined by two parameters: the offset size (an integer between 1 and 4, 1 and 8, or 1 and 16 in the various simulations) and the crossover location (random in all simulations). (B) Loop deletions occur when part of a chromatid forms a loop and a crossover generates an extrachromosomal circle that is lost.
Natural selection:
Fitness (ω) was simulated as a ramp function of the number of uninserted units (ηu) according to the following:
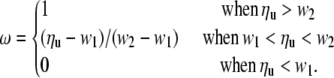 |
The number of gametes generated by each individual was calculated as ω times the maximum number of gametes. The maximum number of gametes was set at six for all simulations shown; however, increasing this number had no detectable effect on the outcomes of the simulations (data not shown). Because the number of gametes had to be an integer, fitness was a step function rather than linear when w1 < ηu < w2. Female fitness was determined by the number of uninserted units on both X rDNA loci. In the initial simulations males were assigned maximum fitness. When the Y locus was added, male fitness was determined by the number of uninserted units on both the X and the Y loci. The mean rDNA locus size for D. melanogaster has been estimated between 165 and 230 units (Hawley and Marcus 1989; Lyckegaard and Clark 1991; Jakubczak et al. 1992); however, only 40–50 units appear sufficient to avoid the bobbed phenotype, a phenotype that would be strongly selected against (Hawley and Marcus 1989). Fitness reduction was calculated as 1 − ωbar, where ωbar was the average fitness of the population.
Deletion:
There were two forms of deletions: intrachromosomal recombination (loop deletions) (Figure 1B) and deletions triggered by retrotransposition. The loop deletion rate (δloop) was set to be proportional to locus size (i) by the coefficient (k1). The retrotransposition-triggered deletion rate (δretro) was set to be proportional to R1 retrotransposition rate (χ) by the coefficient (k2, arbitrarily set at 0.6). In most simulations only loop deletion was included. When both forms of deletions were included, the total deletion rate was the sum of the two:
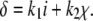 |
While direct estimates of the size of loop deletions and retrotransposition-induced deletions are not available, they typically involve multiple R1 or R2 copies (Perez-Gonzalez and Eickbush 2002; Zhang et al. 2008); thus we have used the larger range (1–15 units) for all simulations. The positions of the deletions were randomly selected within the locus.
R1 and R2 retrotransposition:
For R1 and R2 retrotransposition, an uninserted unit or unit containing the other insertion was randomly selected and converted into an inserted or double-inserted unit. Retrotransposition rates were independent of the number of elements in the locus. R1 and R2 retrotransposition rates for multiple laboratory stocks of D. melanogaster and D. simulans have been found to vary over a wide range (Perez-Gonzalez and Eickbush 2002; Zhang and Eickbush 2005; Zhang et al. 2008).
RESULTS
The simulated diploid populations comprised 2000 females and 2000 males formed by the random selection of gametes from the previous generation. Fitness was determined by the total number of uninserted rDNA units present in the rDNA loci on one pair of homologous chromosomes and was expressed as the number of gametes produced per individual. At each generation gametes experienced crossovers, loop deletions, and retrotransposition events with predetermined probabilities (see methods). Gene conversions were not part of the simulations as they would seldom change the number of rDNA units or the number of R1 and R2. At the start of each simulation, the population was seeded with rDNA loci commonly found in D. melanogaster, namely 210 units in size with R1 and R2 insertion levels at 0.34 and 0.12, respectively (Jakubczak et al. 1992; Averbeck and Eickbush 2005). The final locus compositions (i.e., size and fraction of R1 and R2 inserted) after 5000 generations were independent of the starting composition (data not shown). The rDNA loci in species of the melanogaster species subgroup are located either on the X chromosome alone or on both the X and Y chromosomes (Roy et al. 2005). In D. melanogaster the loci are located on both the X and Y chromosomes. Our initial simulations followed only the rDNA locus on the X chromosomes; subsequent simulations included the Y-linked rDNA locus.
Effects of unequal crossovers on the size of the rDNA locus:
SCE between the two copies of an rDNA locus after DNA replication has been suggested as the most frequent recombination event within the rDNA loci (Petes 1980; Seperack et al. 1988; Schlotterer and Tautz 1994; Gonzalez and Sylvester 2001; McTaggart et al. 2007). To evaluate the effect of these exchanges we varied both the SCE rate (0.01–1 per locus per generation) and the offset size (random between 1 and 4, 1 and 8, or 1 and 16 units) while low rates of loop deletion (Walsh 1987; Lyckegaard and Clark 1991) were used to keep the locus size from becoming unbounded (R1 and R2 were not included in these first studies). As shown in Figure 2A at low SCE rates and offset sizes the average number of rDNA units in most loci was only somewhat larger than that needed for maximum fitness (w2 = 100 units per fly or 50 units per locus). As the SCE rate and offset size increased, locus size also increased such that at the highest rate and offset the mean locus size was over eight times that needed for maximum fitness. Varying w2 over the range of 60–200 units led to a corresponding change in locus size at low SCE but minimally affected locus size at high SCE values (data not shown). Varying w1 over the range 10–90 units or changing the fitness function between w2 and w1 had minimal effects on mean locus size (data not shown).
Figure 2.—
Effects of SCE and InterCE rates on rDNA locus size (R1 and R2 not present). (A) Mean rDNA locus size as a function of offset size and SCE rate (w1 = 50, w2 = 100, InterCE = 1 × 10−4, loop-deletion coefficient = 5 × 10−5) (see methods). (B) Distribution of locus sizes in the population at various SCE rates (offset = 1–8). All other parameters were as in A. (C) Mean locus size as a function of InterCE and SCE rates (w1 = 50, w2 = 100, offset = 1–16, loop-deletion coefficient = 5 × 10−5).
SCE events by themselves do not result in a net increase or decrease in the number of rDNA units in a population. How, therefore, can higher SCE rates increase the locus size of the population? Because each SCE event yields one larger and one smaller rDNA locus, high SCE rates generate larger variations in locus size for the population (Figure 2B). Natural selection will eliminate the smallest loci from the population, thereby increasing the mean locus size. Less frequent SCE generates lower variation and results in locus sizes closer to the size required by natural selection.
While SCE increased the mean size of the rDNA locus, InterCE, or unequal crossovers between the two X chromosomes, had the opposite effect. In Figure 2C, the SCE rate was again varied from 0.01 to 1 and the InterCE rate from 10−4 to 10−2 per locus per generation (no effect was observed below 10−4). Mean locus size decreased as the InterCE rate was increased because it permitted recombination between large and small loci in the population, thereby reducing size variation. The recombination rate between the two X-linked rDNA loci in D. melanogaster has been estimated at 10−4 per locus per generation (Williams et al. 1989), the rate used in all subsequent simulations.
Effects of R1 and R2:
Figure 3 summarizes the effects of R1 and R2 elements on the rDNA locus. Figure 3A shows the composition of the locus at varying SCE rates and constant R1 and R2 retrotransposition rates (arbitrarily set at 0.02 and 0.005 retrotranspositions per locus per generation, respectively). For comparison, the number of rDNA units in the absence of R1 and R2 is also shown at each SCE rate (Figure 3A, dashed line). As the SCE rate increased the number of uninserted units increased (as in Figure 2A); however, the number of R1 and R2 inserted units decreased slightly. As a consequence at the lowest SCE rate the locus was nearly three times larger in the presence of R1 and R2 than without (220 units vs. 75 units), while at the highest SCE rate the locus was only 20% larger in the presence of R1 and R2 (300 units vs. 250 units). The stability of R1 and R2 numbers relative to the SCE rate is because while higher SCE rates increase the variation in the number of both inserted and uninserted units present in the population, natural selection eliminates only those chromosomes with low numbers of uninserted units. Therefore, only uninserted units are driven to higher mean values by the SCE rate.
Shown in Figure 3B are the effects of loop-deletion rate on rDNA loci with and without R1 and R2 insertions (SCE = 1). Because a larger locus would be expected to undergo more loop deletions, in our simulations the deletion rates were set proportional to locus size (see methods). At the lowest deletion rate the rDNA loci were large (∼500 units) with high levels of R1 and R2 insertions. A 25-fold increase in the deletion rate decreased the number of uninserted units 2-fold, while R1-inserted units decreased 11-fold and R2-inserted units were eliminated. While loop deletions equally removed both inserted and uninserted units from the loci, the greater effect on inserted units was because natural selection retained the number of uninserted units in the population.
In Figure 3C, R1 retrotransposition rates were tested over the range 0.004–0.1 per locus per generation, while the R2 rate was held constant at 0.005. A 25-fold increase in R1 retrotransposition increased the number of R1-inserted units 8-fold, decreased the number of uninserted units ∼2-fold, but only minimally affected the number of R2-inserted units. Thus higher retrotransposition rates by one element (R1 or R2) significantly reduce the number of uninserted units but not the number of the other element. Part of the stability of both R1 and R2 in a locus is because both elements can be inserted into the same unit. However, as described below, even without this ability multiple families of the same element can readily coexist in the same rDNA locus.
Recently, high retrotransposition rates within laboratory stocks of D. simulans were correlated with deletions involving multiple rDNA units per event (Zhang et al. 2008). The increased deletion was suggested to result from chromosome breakage induced by the retrotransposition process itself. Simulations were conducted in which the deletion rate was divided into two components: loop deletion based on locus size and induced deletions based on retrotransposition rate. These rates were arbitrarily adjusted in Figure 3D so that the total deletion rate at the lowest R1 retrotransposition rate was similar to the deletion rate used in Figure 3C (see methods). Linking the deletion rate to retrotransposition prevented the number of R1 elements from increasing as dramatically and decreased the number of R2 elements, while selection maintained the number of uninserted units. As would be expected, the extent of this effect was dependent upon the proportional constant between retrotransposition and deletion.
Fitness reduction under different parameters:
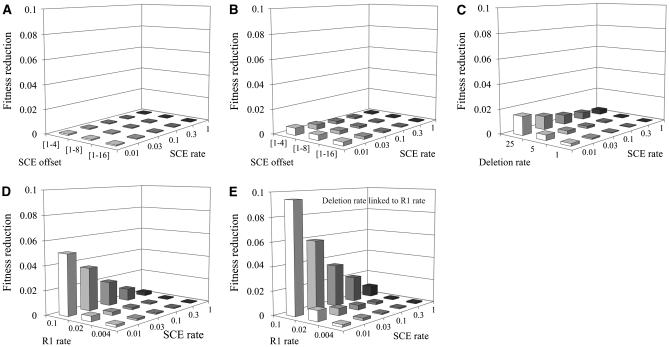
Figure 4 compares the fitness reduction, 1 – ωbar (ωbar is the mean population fitness), for all SCE, deletion, and retrotransposition parameters described in Figures 1–3. Even in the absence of R1 and R2 there is some fitness reduction because SCE occasionally generates loci that are too small (Figure 4A). In the presence of R1 and R2 (retrotransposition rates of 0.02 and 0.005, respectively) there is a further reduction in fitness, but this effect is small at high SCE rates (Figure 4B). Increasing the loop-deletion rate (with R1 and R2) reduces fitness at all SCE rates (Figure 4C). Increasing the R1 retrotransposition rate also reduces fitness, but the effect is minimal at higher SCE rates (Figure 4D). For example, at the highest R1 rate and SCE simulated, 80% of the rDNA units were inserted (see Figure 3C), but the fitness reduction was only 0.003 (2% of the organisms had less than maximum fitness). Finally, linking the deletion rate to the R1 retrotransposition rate significantly reduces fitness (Figure 4E), even though it results in many fewer inserted units (see Figure 3D).
Figure 4.—
Fitness reduction in the presence and absence of R1 and R2 elements. (A) Fitness reduction (1 – ω) as a function of SCE rates and offset size in the absence of R1 and R2 (all parameters were as in Figure 2A). (B) Fitness reduction as a function of SCE rates and offset size with R1 and R2 (R1 rate = 0.02; R2 rate = 0.005; all other parameters were as in A). (C) Fitness reduction as a function of SCE rates and deletion rate. Parameters were as in B except the offset size was held at 1–8 and the loop-deletion coefficient was varied. (D) Fitness reduction as a function of SCE rate and R1 retrotransposition rate. Parameters were as in B except the offset = 1–8 and the loop-deletion coefficient = 5 × 10−5. (E) Fitness reduction as a function of SCE rate and R1 retrotransposition rate with the deletion rate linked to the retrotransposition rate. All parameters were as in D except the deletion rate was determined as in Figure 3D.
The simulations in Figures 3 and 4 suggest that high SCE rates allow most individuals in a population to maintain fitness even at high rates of R1 and R2 retrotransposition. Increasing the deletion rate, either in response to R1 and R2 or as a byproduct of their activity, reduces the total number of insertions but at the cost of significant fitness reduction.
Competition among different R1 subfamilies:
Multiple families of R1 and/or R2 are frequently detected within the same species and appear stable over long periods of evolution (Burke et al. 1998; Gentile et al. 2001; Kojima and Fujiwara 2005). Because the insertion of an R1 (or R2) element disrupts the target site for the insertion of a second R1 (or R2) element, multiple families are in direct competition for uninserted units. To study this competition, three R1 families were simulated in a population (Figure 5A). The retrotransposition rate of one family (R1A) was varied (0.004–0.1), while the rates of the other two families (R1B and R1C) were held constant (0.02 and 0.005). As the number of R1A elements increased with its retrotransposition rate, the number of uninserted units decreased twofold, while the number of R1B and R1C elements decreased only slightly. This stability is because the level of each family is a function of its own retrotransposition rate and the deletion rate. When the deletion rate was also linked to an increasing R1A retrotransposition rate, the R1B and R1C families were greatly reduced (Figure 5B). These results suggest multiple families can coexist within a rDNA locus when each family is retrotransposing independently and the combined retrotransposition activity of all families does not lead to a high deletion rate.
Comparison of the X- and Y-rDNA loci:
All simulations to this point followed only the X chromosome rDNA loci. In these simulations the X loci in males underwent SCE, deletion, and retrotransposition rates as in females but were not subject to selection because males were assigned maximum fitness. In Figure 6, simulations were conducted with both the X and the Y rDNA loci with parameters similar to that in Figure 3A. Male fitness was determined by the total number of uninserted units on the X and Y loci. InterCE between the X and Y loci was set at 10−4, the rate determined from direct observation (Williams et al. 1989); however, again this low rate had little effect on locus size (data not shown).
The trend toward greater numbers of uninserted units with increased SCE rate was observed for both the X and the Y loci (Figure 6A). However, the mean numbers of uninserted rDNA units on the X loci were higher than those on the Y loci, as well as higher than in the simulations that did not include the Y loci (compare to Figure 3A). The increase in the X chromosome loci had an interesting origin. Due to drift, the mean X and Y rDNA loci fluctuated in size within the simulated populations. While X loci were maintained by selection in females, selection on the Y loci in males can be relaxed when large X loci are present in the population. When the simulations were allowed to continue for tens of thousands of generations, this absorbing boundary resulted in the loss of the Y loci (Y-locus loss was more rapid at low SCE rates). The only way to maintain the Y loci indefinitely in the population was to require this locus for male fitness (data not shown).
The numbers of R1 and R2 inserted units in the X and Y loci were similar at high SCE rates (Figure 6B). At low SCE rates the Y loci were already starting to become smaller at the end of our 5000-generation simulations due to the X-locus compensation, and thus the numbers of R1 and R2 inserted units were also declining. As would be expected from this partial compensation by the X loci, fitness reduction at low SCE rates was much greater in males than in females (Figure 6C).
DISCUSSION
This report attempts to simulate the recombination parameters and fitness constraints that determine the size and composition of eukaryotic rDNA loci. Unequal crossovers randomly increase and decrease the size of rDNA loci. Maintenance of a minimum locus size is readily explained by the need for the synthesis of sufficient levels of rRNA. Preventing the locus size from randomly increasing to high levels has been suggested to result either from stabilizing selection (i.e., selection against large locus size) or from loop deletion (Walsh 1987; Lyckegaard and Clark 1991). We focused on loop deletion in this study because circular rDNA derived from loop-deletion events have been detected in various organisms (Cohen et al. 2003). Low levels of these events readily prevented the rDNA loci from becoming unbounded and thus eliminated the need for stabilizing selection in our simulations.
On the basis of both direct measurements in yeast and the crustacean, Daphnia (Petes 1980; McTaggart et al. 2007) and studies of inter- vs. intrachromosomal rDNA polymorphisms in flies and humans (Seperack et al. 1988; Schlotterer and Tautz 1994; Gonzalez and Sylvester 2001), SCE has been suggested as the most frequent form of unequal crossover in the rDNA locus. Our simulations suggest that this SCE rate has a significant impact on rDNA locus size. High rates of SCE increase the mean rDNA locus size by generating greater variation within the population, thereby enabling selection to be more effective. In contrast to SCE, InterCE generally reduces size variation within the population by permitting recombination between large and small loci. Thus, the net effect of InterCE is to reduce locus size. Because in most organisms the rates of InterCE are predicted to be low (Petes 1980; Williams et al. 1989), InterCE is unlikely to have a major effect on rDNA locus size.
These results help to explain the large size of the rDNA locus in eukaryotes. For example, yeast, which maintains a small streamlined genome, encodes two to three times more rDNA units than are transcribed during rapid growth (Dammann et al. 1995). Most eukaryotes transcribe an even smaller fraction of their rDNA units (Conconi et al. 1992a,b; Ye and Eickbush 2006). Our simulations suggest that if an organism maintains high SCE rates, the number of units within the rDNA loci is relatively independent of the number of units needed for maximum fitness. In other words, the size of an rDNA locus for a species may be more dependent on the SCE rate than on the number of units needed for the synthesis of rRNA. While there have been many theoretical as well as empirical studies documenting how crossovers can lead to the concerted evolution of rDNA (Ohta 1980; Petes 1980; Ohta and Dover 1983; Nagylaki 1984; Walsh 1987; Seperack et al. 1988; Stephan 1989; Lyckegaard and Clark 1991; Schlotterer and Tautz 1994), these studies did not enumerate this important aspect of rDNA loci dynamics.
We suggest that R1 and R2 are abundant and evolutionarily stable components of the rDNA locus because the large loci generated by high SCE rates will tolerate high levels of R1 and R2 insertion. Our simulations indicate that the number of R1- and R2-inserted units per locus is a balance between their retrotransposition rates and the deletion rate but is not substantially changed by the SCE rate. Thus for a population undergoing high rates of SCE, active retrotransposition will produce many inserted (inactive) units, but the increase in locus size due to SCE will minimize the effects of these insertions on locus size and fitness. Without a high SCE rate organisms must rely on the less efficient mechanism of direct selection to maintain the minimum number of uninserted units. The alternative approach to combat high rates of retrotransposition by increasing the deletion rate for the rDNA locus is also less attractive. Unless a mechanism evolves to specifically delete inserted units, an increased deletion rate will also reduce the number of uninserted units. This decrease means the organism must again rely on less efficient direct selection to maintain the minimum number of uninserted units. Thus so long as an organism is not sensitive to rDNA locus size, high levels of R1 and R2 retrotransposition rates are most readily compensated by changes in the SCE rate.
D. melanogaster is a good example of a species with high levels of R1 and R2 insertions. A survey of geographical lines revealed broad ranges but a mean rDNA locus size of 230 units with 40% inserted with R1 and 15% inserted with R2 (Jakubczak et al. 1992). Our simulations suggest retrotransposition rates of 0.02 for R1 and 0.006 for R2, a SCE rate of 0.4, an offset size of 1–8 units, and a deletion coefficient of 5 × 10−5 can generate rDNA loci typically seen in the geographical survey. Of course compensating adjustments particularly in offset size and SCE rates, as well as the deletion and retrotransposition rates, can also generate loci with similar properties. Our only estimates of retrotransposition rates in D. melanogaster are from stocks maintained for >350 generations in the laboratory (Perez-Gonzalez and Eickbush 2002). The R1 rate was estimated at 0.1, and consistent with our simulations the elements were rapidly accumulating in the rDNA loci, while the R2 rate was estimated at 0.003 with elements gradually being lost from the stocks. While the purpose of this study was not to define precise recombinational parameters for D. melanogaster, the range of values used appear to readily explain the empirical parameters. We suggest further analysis of the empirical population data, particularly locus size and the rates at which R1 and R2 are gained and lost, will enable more accurate estimates of the various parameters that affect the rDNA locus.
Our simulations also help to explain why many species maintain multiple families of R1 and/or R2 (Burke et al. 1998; Kojima and Fujiwara 2005). In the best-studied examples, multiple R1 lineages were found to coexist in the rDNA loci of many Drosophila lineages (Gentile et al. 2001). Although these families are competing for the same insertion sites, our simulations suggest the abundance of each family is not adversely affected by an increase in the retrotransposition rate of another family.
Another prediction from our simulations relates to the presence of the rDNA loci on the X and Y chromosomes in D. melanogaster and other Drosophila species. Due to random fluctuations in the mean number of units in these loci, larger numbers of uninserted units on the X chromosome are predicted to compensate for the number of units needed on the Y chromosome. Eventually the continued reductions in selective constraints for the Y locus caused by X-locus compensation can result in the elimination of the Y rDNA locus. In the only study conducted to date on the size of the rDNA in a natural population of D. melanogaster, the mean Y-locus size was 60% that of the X locus (Lyckegaard and Clark 1991). Other studies have suggested reduced selection constraints for the size of the Y chromosome rDNA locus compared to the X locus (Frankham et al. 1980; Williams et al. 1987; Clark et al. 1990). Remarkably the rDNA units on the Y chromosome in D. simulans and D. sechellia have been lost (Roy et al. 2005). The only component of the locus that remains on the Y in these two species is tandem copies of the intergenic spacer shown to be required for X–Y pairing (McKee et al. 1992). Thus our simulations provide support for the speculations that the major factor maintaining the Y chromosome rDNA locus in D. melanogaster is a need for X–Y pairing.
Finally, there have been many attempts to model the evolution of mobile elements that insert throughout the genome (for recent reports see Le Rouzic et al. 2007; Dolgin and Charlesworth 2008). Modeling of R1 and R2 would appear more straightforward because all insertions have a uniform effect on the host, ectopic recombination is not detrimental for the rDNA locus, and recombination eventually removes old, dysfunctional copies of R1 and R2 while continually providing new sites for insertion. Indeed, this uniformity of insertion site and predicable effects of insertions argues that the rDNA locus is a reliable niche for an element to hide within the genome and may explain why R1 and R2 are more evolutionarily stable in a lineage than transposable elements that insert throughout the genome (Burke et al. 1998; Malik et al. 1999; Gentile et al. 2001). Meanwhile from the perspective of the organism, the major challenge brought on by R1 and R2 is not to retain sufficient numbers of uninserted rDNA units, but to be able to identify uninserted units for transcription or their RNA for processing into ribosomal subunits. This discrimination is presumably the key component that limits the retrotransposition activity of R1 and R2 and represents fertile ground for future studies.
Acknowledgments
We thank members of the laboratory for discussions and comments on the manuscript and Jim Fry and Allen Orr for their advice on various aspects of the simulation approach. This work was supported by a National Institutes of Health grant (GM42790) and a National Science Foundation grant (MCB-0544071).
References
- Averbeck, K. T., and T. H. Eickbush, 2005. Monitoring the mode and tempo of concerted evolution in the Drosophila melanogaster rDNA locus. Genetics 171 1837–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, W. D., H. S. Malik, W. C. Lathe, III and T. H. Eickbush, 1998. Are retrotransposons long-term hitchhikers? Nature 392 141–142. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., F. M. Szumski and E. M. Lyckegaard, 1990. Population genetics of the Y chromosome of Drosophila melanogaster: rDNA variation and phenotypic correlates. Genet. Res. 58 7–13. [DOI] [PubMed] [Google Scholar]
- Cohen, S., K. Yacobi and D. Segal, 2003. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 13 1133–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi, A., R. M. Widmer, T. Koller and J. M. Sogo, 1992. a Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57 753–761. [DOI] [PubMed] [Google Scholar]
- Conconi, A., J. M. Sogo and C. A. Ryan, 1992. b Ribosomal gene clusters are uniquely proportioned between open and closed chromatin structures in both tomato leaf cells and exponentially growing suspension cultures. Proc. Natl. Acad. Sci. USA 89 5256–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann, R., R. Lucchini, T. Koller and J. M. Sogo, 1995. Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol. Cell. Biol. 15 5294–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin, E. S., and B. Charlesworth, 2008. The effects of recombination rate on the distribution and abundance of transposable elements. Genetics 178 2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush, T. H., and D. G. Eickbush, 2007. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham, R., D. A. Briscoe and R. K. Nurthen, 1980. Unequal crossing over at the rDNA tandon as a source of quantitative genetic variation in Drosophila. Genetics 95 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley, A. R., and T. Kobayashi, 2007. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 17 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile, K. L., W. D. Burke and T. H. Eickbush, 2001. Multiple lineages of R1 retrotransposable elements can coexist in the rDNA loci of Drosophila. Mol. Biol. Evol. 16 235–245. [DOI] [PubMed] [Google Scholar]
- Gonzalez, I. L., and J. E. Sylvester, 2001. Human rDNA: evolutionary patterns within genes and tandem arrays derived from multiple chromosomes. Genomics 73 255–263. [DOI] [PubMed] [Google Scholar]
- Hawley, R. S., and C. H. Marcus, 1989. Recombinational controls of rDNA redundancy in Drosophila. Annu. Rev. Genet. 23 87–120. [DOI] [PubMed] [Google Scholar]
- Jakubczak, J., W. D. Burke and T. H. Eickbush, 1991. Retrotransposable elements R1 and R2 interrupt the rDNA genes of most insects. Proc. Natl. Acad. Sci. USA 88 3295–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczak, J., M. Zenni, R. Woodruff and T. H. Eickbush, 1992. Turnover of R1 (type I) and R2 (type II) retrotransposable elements in the ribosomal DNA of Drosophila melanogaster. Genetics 131 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, K. K., and H. Fujiwara, 2005. Long-term inheritance of the 28S rDNA-specific retrotransposon R2. Mol. Biol. Evol. 22 2157–2165. [DOI] [PubMed] [Google Scholar]
- Le Rouzic, A., T. S. Boutin and P. Capy, 2007. Long-term evolution of transposable elements. Proc. Natl. Acad. Sci. USA 104 19375–19380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyckegaard, E. M., and A. G. Clark, 1991. Evolution of ribosomal RNA gene copy number on the sex chromosomes of Drosophila melanogaster. Mol. Biol. Evol. 8 458–474. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., W. D. Burke and T. H. Eickbush, 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16 793–805. [DOI] [PubMed] [Google Scholar]
- McKee, B. D., L. Habera and J. A. Vrana, 1992. Evidence that intergenic spacer repeats of Drosophila melanogaster rRNA genes function as X-Y pairing sites in male meiosis, and a general model for achiasmatic pairing. Genetics 132 529–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart, S. J., J. L. Dudycha, A. Omilian and T. J. Crease, 2007. Rates of recombination in the ribosomal DNA of apomictically propagated Daphnia obtusa lines. Genetics 175 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki, T., 1984. Evolution of multigene families under interchromosomal gene conversion. Proc. Natl. Acad. Sci. USA 81 3796–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, T., 1980. Evolution and Variation of Multigene Families. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Ohta, T., and G. Dover, 1983. Population genetics of multigene families that are dispersed into two or more chromosomes. Proc. Natl. Acad. Sci. USA 80 4079–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gonzalez, C. E., and T. H. Eickbush, 2001. Dynamics of R1 and R2 elements in the rDNA locus of Drosophila simulans. Genetics 158 1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gonzalez, C. E., and T. H. Eickbush, 2002. Rates of R1 and R2 retrotransposition and elimination from the rDNA locus of Drosophila melanogaster. Genetics 162 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T. D., 1980. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell 19 765–774. [DOI] [PubMed] [Google Scholar]
- Roy, V., L. Monti-Dedieu, N. Chaminade, S. Siljak-Yakovlev, S. Aulard et al., 2005. Evolution of the chromosomal location of rDNA genes in two Drosophila species subgroups: ananassae and melanogaster. Heredity 94 388–395. [DOI] [PubMed] [Google Scholar]
- Schlotterer, C., and D. Tautz, 1994. Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchange drives concerted evolution. Curr. Biol. 4 777–783. [DOI] [PubMed] [Google Scholar]
- Seperack, P., M. Slatkin and N. Arnheim, 1988. Linkage disequilibrium in human ribosomal genes: implications from multigene family evolution. Genetics 119 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage, D. E., and T. H. Eickbush, 2007. Sequence variation within the rRNA gene loci of 12 Drosophila species. Genome Res. 17 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, W., 1989. Tandem-repetitive noncoding DNA: forms and forces. Mol. Biol. Evol. 6 198–212. [DOI] [PubMed] [Google Scholar]
- Walsh, J. B., 1987. Persistence of tandem arrays: implications for satellite and simple-sequence DNAs. Genetics 115 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, S. M., G. R. Furnier, E. Fuog and C. Strobeck, 1987. Evolution of the ribosomal DNA spacers of Drosophila melanogaster: different patterns of variation on the X and Y chromosomes. Genetics 116 225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, S. M., J. A. Kennison, L. G. Robbins and C. Strobeck, 1989. Reciprocal recombination and the evolution of the ribosomal gene family of Drosophila melanogaster. Genetics 122 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J., and T. H. Eickbush, 2006. Chromatin structure and transcription of the R1- and R2-inserted rRNA genes of Drosophila melanogaster. Mol. Cell. Biol. 26 8781–8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., and T. H. Eickbush, 2005. Characterization of active R2 retrotransposition in the rDNA locus of Drosophila simulans. Genetics 170 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., J. Zhou and T. H. Eickbush, 2008. Rapid R2 retrotransposition leads to the loss of previously inserted copies via large deletions of the rDNA locus. Mol. Biol. Evol. 25 229–237. [DOI] [PubMed] [Google Scholar]