Abstract
We performed a forward genetic screen, using Drosophila as a surrogate mosquito, to identify host factors required for the growth of the avian malaria parasite, Plasmodium gallinaceum. We identified 18 presumed loss-of-function mutants that reduced the growth of the parasite in flies. Presumptive mutation sites were identified in 14 of the mutants on the basis of the insertion site of a transposable element. None of the identified genes have been previously implicated in innate immune responses or interactions with Plasmodium. The functions of five Anopheles gambiae homologs were tested by using RNAi to knock down gene function followed by measuring the growth of the rodent parasite, Plasmodium berghei. Loss of function of four of these genes in the mosquito affected Plasmodium growth, suggesting that Drosophila can be used effectively as a surrogate mosquito to identify relevant host factors in the mosquito.
MALARIA is transmitted by anopheline mosquitoes, resulting in 300–500 million clinical cases and >1 million deaths annually, mostly among African children. Recent advances in RNA interference (RNAi) technology make it possible to manipulate mosquito gene function in vivo and to determine the effects of mosquito gene regulation on the ability to support the growth of Plasmodium. The field now faces the problem of choosing which mosquito genes should be tested. Three methods have been used successfully to date: First, genetic mapping of vector traits led to the identification of variant alleles that affect vector competence (Kumar et al. 2003). Second, transcript analysis during Plasmodium infections revealed genes that show significant changes in transcript levels and knockdown of some of these genes affects vector capacity (Levashina et al. 2001; Dong et al. 2006). Third, our understanding of the mosquito immune system is built upon analyses that were performed on another dipteran insect, the fruit fly Drosophila melanogaster (Zdobnov et al. 2002). Direct analysis of genes shown to be important in fighting microbial infections in Drosophila have also contributed to our understanding of the interaction between the mosquito host and Plasmodium parasite (Meister et al. 2005).
The movement and development of Plasmodium in the mosquito is a dangerous and difficult journey for the microbe and results in substantial parasite losses. For every thousand Plasmodium berhgei gametocytes ingested, only two viable ookinetes will be generated, and only 2–20% of these will successfully invade the midgut and develop to mature oocysts (Alavi et al. 2003). The parasites are introduced into the mosquito via an infected blood meal. Plasmodium gametocytes complete their development in the midgut lumen, fertilization takes place, and a diploid motile form called ookinete is generated. Ookinetes invade the midgut epithelium and come in direct contact with components of the mosquito immune system present in hemolymph as they reach the midgut basal lamina. Multiple complex interactions determine whether parasites will be recognized and destroyed, or survive and transform into oocysts. During the oocyst stage, parasites divide continuously and ultimately release thousands of sporozoites into the hemolymph. Sporozoites find and invade the salivary gland, reach the salivary duct, and are injected into a new host during the following blood feeding.
We can anticipate two general types of physiological changes in the mosquito that will alter Plasmodium growth. First, the mosquito can raise an active immune response against the parasite because it does not act simply as a passive culture vessel. Alterations in innate immunity have been shown to affect parasite growth. The second concerns the metabolic needs of the parasite. Plasmodium is dependent upon the mosquito for nutrition and changes in available nutrients in the host will presumably affect parasite development in positive and negative ways. Plasmodium can tolerate great losses in the early stages of development in the mosquito because the parasite population expands during the oocyst stage. Dissecting the interaction between host and parasite can teach us about innate immune mechanisms that can counter Plasmodium and can also provide new information about the growth requirements of the parasite in the insect host.
Every genetic technique has its problems; data mining from microarrays is useful but, by design, will miss genes that are not regulated in response to infection and are not obviously involved in immunity. The tissues affected by RNAi injection into a whole mosquito are not well defined and the efficacy of each probe is different; thus a negative result for an RNAi experiment does not eliminate the possibility that a gene plays a role in a given process. RNAi experiments require a good deal of work and thus researchers think carefully about which genes should be tested first. A result of these issues is that the community tends to select genes for study that have already been implicated in fighting Plasmodium in some manner. It is possible that we are systematically missing genes that either are not transcriptionally regulated during an infection or have not been previously identified as playing a role in the innate immune response. An ideal method of overcoming this gap is to perform an unbiased genetic screen because it allows the organism to tell you what is important in the host–parasite interaction.
Given that our understanding of the mosquito's innate immune system is still largely scaffolded on the Drosophila literature, we used Drosophila as a surrogate mosquito in an unbiased forward genetic screen to find insect factors affecting Plasmodium growth. We showed previously that the fruit fly could support the growth of Plasmodium gallinaceum injected into the body cavity, supporting the development of large numbers of oocysts that complete maturation and generate infective sporozoites (Schneider and Shahabuddin 2000).
We found 18 mutations that reduce the ability of the fly to support Plasmodium growth. Fourteen of these mutations were induced by P-element insertions and these transposons identify a set of genes that had no previously identified role in immunity and are not immune regulated. One risk of this Drosophila model is that it is too far removed from the natural host–parasite pair to provide useful information; however, we find that four of the five genes that we identified through mutation in Drosophila have RNAi-induced phenotypes that alter the growth of P. berghei in Anopheles gambiae.
MATERIALS AND METHODS
Mutant lines tested in genetic screen:
A total of 1045 homozygous viable P-element insertion lines from the Bloomington Drosophila Stock Center were tested in the genetic screen. These lines were generated by many labs in different backgrounds and included several types of P elements. We also tested 407 ethyl methanesulfonate (EMS)-mutagenized fly lines. These mutagenized lines were generated on an isogenized Oregon-R background. Male flies, 3–5 days old, were fed a solution of 2.5 mm EMS (Sigma-Aldrich, St. Louis) in 1% sucrose overnight. The treated males were then mated to X^X/Y (attached-X) females. Individual male progeny where then mated to X^X/Y females to generate separate mutant lines.
Ookinete preparation:
P. gallinaceum parasites were maintained in white leghorn chickens (Charles River Laboratories SPAFAS, Wilmington, MA) by serial passage. Parasites were isolated from chicken blood by previously described methods (Schneider and Shahabuddin 2000). Approximately 10 ml of heparinized blood was obtained from infected 3-week-old white leghorn chicks by cardiac puncture. The blood was diluted in warm 1× SAB (9 mm glucose, 8 mm Tris base, 138 mm sodium chloride, pH 7.3). Blood cells were separated from serum by centrifugation at 3000 rpm for 5 min at room temperature. Blood cells were then incubated for 45 min in 15 ml exflagellation medium (8% heat-inactivated chicken serum, 0.1 mm xanthurenic acid, 0.2% sodium bicarbonate in 1× SAB). Fertilized zygotes were then separated from the blood cells; ∼15 ml blood was carefully layered on top of 15 ml Ficoll-paque (Amersham Biosciences, Uppsala, Sweden) and spun for 15 min at 3500 rpm. The zygotes were removed from the Ficoll interface and resuspended in 50 ml 1× SAB. The zygotes were then spun down at 3500 rpm and the SAB was removed. The zygotes were resuspended in 10 ml 1× M199, pH 8.1, plus penicillin/streptomycin and incubated with 0.7 ml of 1 mg/ml wheat germ agglutinin (Sigma-Aldrich) for 10 min at room temperature. The agglutinated blood cells were pelleted at 700 rpm for 30 sec. The supernatant was removed and the agglutination procedure repeated once more with an additional 0.7 ml wheat germ agglutinin. The supernatant was then spun for 10 min at 3500 rpm. The supernatant was removed and the pellet containing zygotes was then incubated overnight in 1× M199 media, pH 8.1, plus penicillin (100 units/ml) and streptomycin (100 μg/ml) (Invitrogen-GIBCO, Carlsbad, CA). The next morning, differentiated ookinetes were counted and resuspended in 1× M199 media, pH 8.1, at a concentration of 500 ookinetes/50 nl of media.
Genetic screen injection protocol:
Fly stocks were maintained on standard yeasted dextrose food at 25°, 60% humidity. Only male flies were used in the genetic screen to avoid progeny growing in the vials. Male flies were aged 5–7 days at 25° before infection. Ten to 15 male flies from each stock were then injected with 500 P. gallinaceum ookinetes. Parasite suspension (50 nl) was injected into the fly's anterior abdomen on the ventrolateral surface. Injections were performed using pulled glass needles and a Picospritzer III injector (Parker Hannifin, Rohnert Park, CA). Following inoculation, flies were incubated at 25° for 9 days and then harvested to measure parasite load.
Measuring P. gallinaceum load by quantitative real-time PCR:
P. gallinaceum loads were determined in each line by quantitative real-time PCR (qRT–PCR). RNA was isolated from five flies from each line using a 96-well format RNeasy kit (Qiagen, Valencia, CA). The RNA was then treated with DNAse. qRT–PCR reactions were set up using the heterobifunctional rTth polymerase (Applied Biosystems, Foster City, CA) and run in an iCycler (Bio-Rad, Hercules, CA). Copies of P. gallinaceum small-unit ribosomal RNA (surRNA) were measured using the following oligomers: (5′ oligo pgrRNAbeg) 5′-agagttcgattccggagagg-3′, (3′ oligo pgrRNAbeg) 5′-tcattccaattgcaaaacca-3′, and (hybridization oligo pgrRNAbeg) 5′-FAM-accacatctaaggaaggcagcaggc-TAMRA-3′. surRNA values were normalized to the number of copies of D. melanogaster ribosomal protein 15a in each sample; expression of 15a does not change during infection and therefore this gene served as a control for the total amount of RNA in each sample (Schneider and Shahabuddin 2000).
Bacterial infections:
Flies were infected with either Salmonella typhimurium strain SL1344 (Hoiseth and Stocker 1981) or Listeria monocytogenes strain 10403s (Portnoy et al. 1988). SL1344 and 10403s were grown standing overnight at 37° in LB and brain heart infusion medium, respectively. One-week-old male flies were injected as described above with bacteria (for S. typhimurium, OD600 = 0.1, ∼10,000 CFU; for L. monocytogenes, OD600 = 0.01, ∼1000 CFU). Flies were maintained thereafter at 29°. All survival experiments were performed in triplicate with 10 flies/replicate. Experiments were repeated at least three times. Survival curves were plotted using the Kaplan–Meier method. Significance was determined by comparing survival curves of mutant flies to Oregon-R flies using the log-rank test.
Mosquito rearing and infections:
An. gambiae strain G3 mosquitoes were raised at 28°, 75% humidity, under a 12-hr light/dark cycle and maintained on a 10% sucrose solution during adult stages. Mosquito females were infected with P. berghei by feeding on anesthetized infected Balb/C mice 5–6 days post-emergence. Mice were infected with P. berghei by intraperitoneal injection of blood from an infected donor mouse. The infection was monitored by determining the parasitemia and by performing exflagellation assays as described previously (Billker et al. 1997). Mice with parasitemias between 5 and 15% and one to three exflagellations per 40× field were used to infect mosquitoes. Blood-fed mosquitoes were kept at 21° in a humidified environment and dissected 6 days post-infection.
Mosquito RNA isolation and qRT–PCR:
Poly(A+) RNA was extracted from 10 whole mosquitoes with an Oligotex direct mRNA kit (Qiagen, Valencia, CA). The first strand of cDNA synthesis was performed using a Quantitect reverse transcription kit (Qiagen). Gene-specific primers were designed using MacVector to amplify 100–200 bp PCR product (supplemental Table S1). The PCR reactions were assembled using DyNAmo SYBR Green qPCR master mix (Finnzymes) and run in the MJ Research detection system according to the manufacturer's instructions (Bio-Rad). All values were then normalized to the number of copies of ribosomal protein S7 in the sample as determined by qRT–PCR.
Bioinformatics analysis:
D. melanogaster sequences were compared to the National Center for Biotechnology Information database by using the Basic Local Alignment Search Tool against the An. gambiae genome and translated database. Compiled EST sequences were aligned by using CLUSTAL, and gene-specific primers were designed in well-conserved regions to confirm expression in An. gambiae.
Double-stranded RNA production:
An An. gambiae cDNA for each gene was obtained from 10 naive 4-day-old females. This was used as a template to amplify a 500- to 600-bp amplicon for each gene with primers listed in supplemental Table S1. Flanking promoter sequences were added to the gene-specific primers M13 Fw (5′-GTAAAACGACGGCCAGT-3′) and Xho-T7-M13R (5′-CTCGAGTAATACGACTCACTATAGGGCAGGAAACAGCTATGAC-3′). The fragments were then cloned into the pCRII-TOPO vector (Invitrogen, Carlsbad, CA). Double-stranded RNA (dsRNA) was synthesized and purified with a Megasuperscript kit (Ambion, Austin, TX) and eluted in nuclease-free water.
Relative quantification of P. berghei load in An. gambiae by qRT–PCR:
Excised individual midguts (without blood) were placed into individual microcentrifuge tubes containing 10 μl of HotSHOT alkaline lysis reagent (25 mm NaOH, 0.2 mm EDTA, pH 12.0) (Truett et al. 2000). The tubes were boiled for 5 min and immediately placed on ice. Ten microliters of HotSHOT neutralizing reagent (40 mm Tris–HCl, pH 5.0) was added to each tube. The samples were centrifuged and stored at −20°.
Relative quantification of P. berghei by qRT–PCR:
qRT–PCR reactions were assembled using DyNAmo SYBR Green qPCR master mix (Finnzymes) and run in the MJ Research detection system according to the manufacturer's instructions (Bio-Rad). P. berghei-specific primers were designed and tested [Pb (28SrRNA) Fw, 5′-GTGGCCTATCGATCCTTTA-3′; Pb (28SrRNA) Rev, 5′-GCGTCCCAATGATAGGAAGA-3′] to detect the relative amount of P. berghei 28S gene copies. Template midgut genomic DNA (2 μl) was used to detect P. berghei 28S gene copies; 1 μl of template midgut genomic DNA was used to detect An. gambiae ribosomal protein S7 gene copies in a 20-μl PCR reaction. All P. berghei 28S values were then normalized to the number of copies of ribosomal protein S7 in the sample.
RESULTS AND DISCUSSION
To identify insect genes involved in regulating Plasmodium infection, we performed a forward genetic screen in the fruit fly primarily using publicly available mutant stocks. We chose these lines because the transposon insertion sites were already characterized, which permitted rapid identification of the affected genes. We injected mutant fly lines with P. gallinaceum ookinetes, harvested RNA at the peak of infection on day 9 post-injection, and measured parasite load by qRT–PCR. We tested 1045 homozygous viable P-element insertion lines of mixed background from the Bloomington Drosophila Stock Center, and 407 EMS-mutagenized fly lines.
Due to the heterogeneity of the mutant fly lines that we tested, we did not have a true parental strain to which we could compare parasite loads. Therefore, we identified interesting mutant lines by looking for large deviations from the group mean. We tested mutant lines in groups of ∼100 (no fewer than 50 and no more than 200) and a group average parasite load was determined. Lines with at least 10-fold more or fewer parasites than the group average were considered positive and were retested; this was a conservative selection criterion, as the 10-fold cutoff from the group mean was well outside the P < 0.05 significance by t-test for all screen runs (data not shown). During each round of retesting, positive lines were tested with a different random group of 50–200 mutant lines. The final group of mutant lines consisted of those that tested positive in three different rounds of screening.
Of the 1452 mutant fly lines tested, 5% (70 lines) tested positive in the first round (Table 1). Thirty-three percent (23 lines) of the first-round positive lines retested positive in the second round. Eighty percent (18 lines) of the second-round positive lines retested positive in the final round. The percentage of mutant lines testing positive in successive rounds increased with each round of screening, suggesting that our screening procedure effectively purified interesting mutants. After three rounds of screening, the final collection of mutant lines contained 18 mutants: 14 P-element lines and 4 EMS lines (Table 2). All of the mutant lines in this final collection were resistant to P. gallinaceum and contained fewer parasites than the average fly line. We did not identify mutants permitting more parasite growth than the average line in this screen.
TABLE 1.
Genetic screen statistics
| No. of mutant lines | % of previous round | |
|---|---|---|
| Total mutants tested | 1452 | |
| P-element lines | 1045 | |
| EMS lines | 407 | |
| First-round positives | 70 | 4.8 |
| P-element lines | 60 | 5.7 |
| EMS lines | 10 | 2.5 |
| Second-round positives | 23 | 33 |
| P-element lines | 18 | 30 |
| EMS lines | 5 | 50 |
| Final-round positives | 18 | 79 |
| P-element lines | 14 | 78 |
| EMS lines | 4 | 80 |
The selection of mutants through three rounds of screening is shown. It demonstrates a purification effect in which the percentage of mutants reisolated during each round rises from ∼5% in the first round to ∼80% in the final round.
TABLE 2.
Fly lines selected from genetic screen (fold-less from group average)
| Round
|
|||
|---|---|---|---|
| Line | 1 | 2 | 3 |
| EE2 | 47 | 14 | 14 |
| GC15 | 11 | 3890 | 25 |
| GE5 | 15 | 780 | 34 |
| iG1 | 150 | 16 | 54 |
| EP2425 | 70 | 3107 | 10 |
| 11603 | 100 | 33 | 100 |
| 12718 | 1403 | 11 | 100 |
| 12738 | 37 | 6 | 13 |
| 12516 | 16 | 46 | 22 |
| 13425 | 47 | 37 | 15 |
| 13462 | 136 | 9 | 7 |
| 13593 | 8 | 4 | 5 |
| 13596 | 1300 | 5 | 19 |
| 13739 | 100 | 2 | 100 |
| 13760 | 12 | 220 | 5 |
| 13918 | 37 | 7 | 100 |
| 13995 | 10 | 19 | 14 |
| 16497 | 10 | 5 | 8 |
The relative reduction in parasite numbers for each mutant during the three rounds of screening is shown. The numbers reported are the fold reduction with respect to the mean.
Past work on the fly's immune system revealed many mutations that damage the fly's immune response and result in increased growth of infecting microbes. These mutations typically block the antimicrobial immune response of the fly and thus the fly does not fight infections well. When we started this screen, we anticipated that we would find mutations that affected Plasmodium growth in a similar manner. We were therefore surprised that our screen revealed only mutants that reduced the ability of a parasite to grow. This could be due to a simple experimental reason, such as setting our threshold too high when looking for mutants that allow increased Plasmodium growth. A second explanation is that the logic of our screen differed from many that have been performed previously. Most fly immunity screens in the past have looked for a disruption in the transcription of antimicrobial peptide genes (Wu et al. 2001; Khush et al. 2002). These mutants are immunocompromised and permit higher growth rates of microbes. Few screens have been performed that measure microbial growth or survival or some other nonmechanistic output as a primary assay (Ayres et al. 2008). The results of this screen suggest that scientists studying Drosophila host–pathogen interactions may have been missing an important class of mutants because they concentrate on a single molecular mechanism controlling innate immunity, rather than assaying a broad endpoint.
The fly's immune response is best characterized with respect to bacterial and fungal challenges (Ferrandon et al. 2007); however, none of these well-characterized mutations are absolutely microbe specific and often broadly affect immunity against many microbes (Shirasu-Hiza and Schneider 2007). To determine how our collection of mutations might affect innate immunity in general, we challenged our final collection of mutants with two bacterial pathogens that cause lethal infections in wild-type flies. We challenged mutant flies with the gram-negative bacterium S. typhimurium and the gram-positive bacterium L. monocytogenes. We used these bacteria because they grow in wild-type flies and are fought using different mechanisms (Mansfield et al. 2003; Brandt et al. 2004). Mutants lacking Toll signaling are sensitive to L. monocytogenes while mutants lacking Imd signaling are sensitive to both S. typhimurium and L. monocytogenes. We selected microbes that cause disease in wild-type flies because immune pathways can be missed if they are tested only with nonpathogenic immune elicitors such as Escherichia coli or Micrococcus luteus. These experiments were not performed to test the contribution of Toll or Imd signaling to Plasmodium growth as we previously reported that these pathways play no role in fighting Plasmodium (Schneider and Shahabuddin 2000); rather, we were interested in challenging our mutants with a variety of pathogens that require the fly to use different diverse responses to fight the microbes. Survival of infected mutant flies was compared to survival of wild-type Oregon-R flies. These data are summarized in Table 3 and survival curves for fruit flies with altered immunity are shown in Figure 1. Two EMS lines, GC15 and iG1, were sensitive to S. typhimurium, and one P-element line, 16497, was sensitive to L. monocytogenes. The majority of the mutant fly lines behaved like Oregon-R flies when challenged with either S. typhimurium or L. monocytogenes.
TABLE 3.
Secondary screen for susceptibility of fly lines to bacterial infections
|
S. typhimurium (gram −)
|
L. monocytogenes (gram +)
|
|||||
|---|---|---|---|---|---|---|
| Line | Wild type | Sensitive | Resistant | Wild type | Sensitive | Resistant |
| EE2 | + | + | ||||
| GC15 | + | + | ||||
| GE5 | + | + | ||||
| iG1 | + | + | ||||
| EP2425 | + | + | ||||
| 11603 | + | + | ||||
| 12718 | + | + | ||||
| 12738 | + | + | ||||
| 12516 | + | + | ||||
| 13425 | + | + | ||||
| 13462 | + | + | ||||
| 13593 | + | + | ||||
| 13596 | + | + | ||||
| 13739 | + | + | ||||
| 13760 | + | + | ||||
| 13918 | + | + | ||||
| 13995 | + | + | ||||
| 16497 | + | + | ||||
All flies were ultimately killed by the infecting pathogen. Wild type, resistant, or sensitive refer to changes in survival relative to wild-type flies. Flies were injected with 50 nl S. typhimurium (OD600 = 0.1) or L. monocytogenes (OD600 = 0.01) and incubated at 29°. Significance was determined by log-rank test. n = 30 flies. See Figure 1 for survival curves of lines that were significantly sensitive or resistant to bacterial challenge.
Figure 1.—
Survival curves for P. gallinaceum-resistant lines that are susceptible to bacterial infection. Flies were injected with 50 nl S. typhimurium (OD600 = 0.1) or L. monocytogenes (OD600 = 0.01) and then incubated at 29°. Significance was determined by a log-rank test. n = 30 flies. (A) GC15 S. typhimurium challenge. (B) GC15 L. monocytogenes challenge. (C) iG1 S. typhimurium challenge. (D) iG1 L. monocytogenes challenge. (E) 11603 S. typhimurium challenge. (F) 11603 L. monocytogenes challenge. (G) 12516 S. typhimurium challenge. (H) 12516 L. monocytogenes challenge. (I) 13425 S. typhimurium challenge. (J) 13425 L. monocytogenes challenge. (K) 16497 S. typhimurium challenge. (L) 16497 L. monocytogenes challenge.
We focused our attention on the P-element lines because their insertion sites were known. We obtained the location of the inserted P elements from FlyBase (http://flybase.net) or the P screen database (http://flypush.imgen.bcm.tmc.edu/pscreen) and we confirmed a subset of five P-element insertion sites by inverse PCR (Sullivan et al. 2000). We assumed the putative gene affected in each P-element line to be the gene closest to the location of the inserted P element (Table 4). Half of the P elements sat within genes; two P elements sat within 5′ untranslated regions and five P elements within introns. Three other P elements sat within 500 bp upstream of the gene. Ten of 14 of these genes had strong homologs in An. gambiae (Table 4). The genes identified in the screen had diverse functions. None of the genes had been previously implicated in innate immune responses or the mosquito response to Plasmodium; likewise, none of the genes had been reported to genetically or biochemically interact with each other or with immunity genes.
TABLE 4.
Insertion sites and putative identities of resistant Drosophila lines
| Line | Insertion site | Closest gene | Function | Anopheles homolog |
|---|---|---|---|---|
| EP2425 | 2R: 2551842a | tsp42ej | Signaling | |
| 11603 | 3L: 13424707a | CG10133 | Phospholipase A2 | ENSANGG00000007839 |
| 12718 | 3R: 10902878 | CG12601 | Unknown | ENSANGG00000016829 |
| 12738 | X: 10660983 | CG1691 | mRNA binding | ENSANGG00000014401 |
| 12516 | X: 6478974 | CG3168 | Transporter | ENSANGG00000010074 |
| 13425 | 3R: 3645219a | CG31473 | Pyridoxamine-phosphate oxidase | |
| 13462 | X: 11636313 | Hsc70-3 | Heat-shock protein | ENSANGG00000010404 |
| 13593 | X: 2439599 | trol | Unknown | ENSANGG00000017722 |
| 13596 | X: 14436517a | CG9413 | Amino acid permease | ENSANGG00000017734 |
| 13739 | X: 2760775 | CG11086 | Unknown | ENSANGG00000019046 |
| 13760 | X: 1129267 | l(3)82Fd | lysM domain: peptidoglycan-binding hydrolase | ENSANGG00000006384 |
| 13918 | 2L: 6252967 | CG9486 | N-acetyltransferase | |
| 13995 | 2L: 7782431 | CG7164 | Unknown | |
| 16497 | 3L: 9031537a | ArgK | Arginine kinase | ENSANGG00000019341 |
The insertion sites for the P. gallinaceum-resistant mutants as recorded by FlyBase or the P screen database are shown. The identity of the strongest mosquito homolog for each putative mutant is listed in the last column.
The insertion site for these lines were confirmed by inverse PCR.
Although none of the genes that we identified in this screen are obviously involved in immunity, it is possible to generate plausible hypotheses about some of them. The phospholipase A2, for example, is easy to implicate in immunity because arachadonic acid release plays an important role in the regulation of prostaglandin synthesis in vertebrates and prostaglandins also play some roles in insect immunity. Tetraspanins are important for the activity of immune cells in humans. At this point, we are not concentrating on the mechanism behind these genes but on the phenotypes, for which we have much more data.
It is an important point that none of the genes that we identified in this Drosophila screen have been followed previously in mosquitoes. These genes are not induced during immune responses in the fly and either lack an exciting identity or have no predictable function; these genes would be missed by data mining yet they have strong phenotypes in Drosophila. This screen demonstrates the importance of performing random forward genetics because experiments have the power to reveal previously unexplored physiologies.
At this point, as the typical next step in a Drosophila genetic screen, we would revert the mutations by hopping out the P elements and rescuing the mutations using P-element-mediated transgenic flies. The end goal of this experiment, however, was not to identify genes that affected Plasmodium growth in Drosophila as that is a very artificial system; rather, we wanted to identify genes for further study in the mosquito. Therefore, we directly tested whether mosquito orthologs of the genes that we identified in the fly are also required to support Plasmodium development.
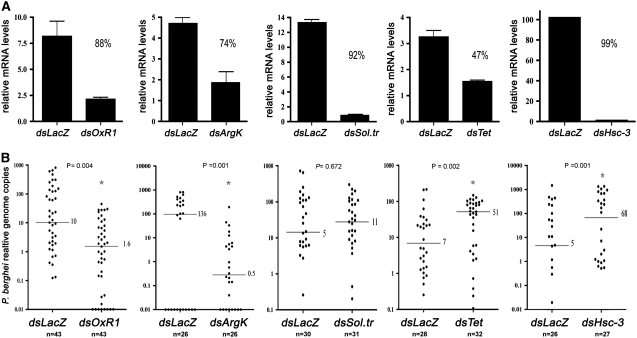
Five genes identified in the screen were carefully analyzed by comparing all homologous genes present in the An. gambiae, D. melanogaster, and Aedes aegypti genomes. It was possible to predict clear putative one-to-one orthologs in An. gambiae for four genes: l(3)82Fd (similar to the Saccharomyces cerevisiae gene, oxidation resistance 1), CG3168 (major facilitator transporter), argk (arginine kinase), and Hsc70-3 (heat-shock protein) (supplemental Table S3). The tetraspanin family, however, has undergone extensive gene expansion in these three species, making it impossible to identify a putative An. gambiae ortholog of Drosophila tsp42ej. To explore this gene family, we decided to silence one member of a group of genes that may have resulted from an An. gambiae-specific gene expansion. One-day-old G3 An. gambiae female mosquitoes were injected with dsRNA to a target host gene of interest or lacZ as a control; RNAi knockdown of target genes was confirmed by qRT–PCR (Figure 2A). Four days later, mosquitoes were fed on anesthetized P. berghei-infected mice. Six days after the feeding, P. berghei load was measured by qRT–PCR (Figure 2B). RNAi knockdown of the An. gambiae putative orthologs of oxr1 and argk resulted in a significant decrease in the P. berghei parasite load, P < 0.0001 by Kolmogorov–Smirnov test, which resembles the phenotype that we found in Drosophila. We found no significant difference in parasite load in mosquitoes treated with CG3168 dsRNA (putative solute transporter) in comparison to control mosquitoes. Mosquitoes treated with AgTetraspanin or Hsc70-3 dsRNA contained significantly more P. berghei than control mosquitoes.
Figure 2.—
Gene knockdown in An. gambiae. (A) Validation of gene silencing. One-day-old female mosquitoes were injected with double-stranded LacZ (dsLacZ) or dsRNA of the target gene and whole-body mRNA was extracted 5 days later. Gene expression was determined by qRT–PCR. Each sample was normalized using ribosomal protein S7 expression as an internal control. Samples were analyzed in triplicate. Columns indicate averages and standard deviations. The silencing efficiency is expressed as a percentage reduction in mRNA levels relative to the dsLacZ control. (B) Effect of gene silencing on P. berghei infection. One-day-old female mosquitoes were injected with either dsLacZ or dsRNA of the target gene. Four days later, mosquitoes fed on anesthetized P. berghei-infected mice. Midguts were dissected and genomic DNA extracted 6 days post-infection. The intensity of P. berghei infection was established on the basis of the abundance of the parasite 28S RNA gene relative to the mosquito ribosomal protein S7 gene in genomic DNA extracted from individual infected midguts determined by qRT–PCR. Medians were compared using the Kolmogorov–Smirnov test and the P-values <0.05 were considered significantly different.
Our success rate of finding genes that have RNAi phenotypes is in line with that found using data mining or testing genes induced by Plasmodium infections. Two of the genes that we tested that had strong homologs between the fly and mosquito had the same loss-of-function phenotype in both the fruit fly and the mosquito. The tetraspanin gene silenced in mosquitoes is not the predicted ortholog of Drosophila. This could explain the increase in infectivity observed when the mosquito tetraspanin gene was silenced. Even when there was not a clear ortholog in An. gambiae, the Drosophila screen was useful in pointing out a gene family in mosquitoes that has an interesting phenotype and warrants more detailed analysis.
Our forward screen assessing Plasmodium growth gives a different range of phenotypes to different classes of microbes than has been seen previously for immunity mutants in Drosophila. None of the genes that we identified have been previously implicated in immunity in either the fruit fly or the mosquito. This work shows the importance of performing unbiased forward genetics to explore new physiologies and demonstrates that genes identified in Drosophila that affect Plasmodium growth can also affect Plasmodium growth in the mosquito.
Acknowledgments
This work was supported by the Ellison Medical Foundation (D.S.S.) and the National Science Foundation (S.M.B.).
References
- Alavi, Y., M. Arai, J. Mendoza, M. Tufet-Bayona, R. Sinha et al., 2003. The dynamics of interactions between Plasmodium and the mosquito: a study of the infectivity of Plasmodium berghei and Plasmodium gallinaceum, and their transmission by Anopheles stephensi, Anopheles gambiae and Aedes aegypti. Int. J. Parasitol. 33 933–943. [DOI] [PubMed] [Google Scholar]
- Ayres, J. S., N. Freitag and D. S. Schneider, 2008. Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics 178 1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billker, O., M. K. Shaw, G. Margos and R. E. Sinden, 1997. The roles of temperature, pH and mosquito factors as triggers of male and female gametogenesis of Plasmodium berghei in vitro. Parasitology 115(Pt. 1): 1–7. [DOI] [PubMed] [Google Scholar]
- Brandt, S. M., M. S. Dionne, R. S. Khush, L. N. Pham, T. J. Vigdal et al., 2004. Secreted bacterial effectors and host-produced eiger/TNF drive death in a Salmonella-infected fruit fly. PLoS Biol. 2 e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y., R. Aguilar, Z. Xi, E. Warr, E. Mongin et al., 2006. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2 e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon, D., J. L. Imler, C. Hetru and J. A. Hoffmann, 2007. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7 862–874. [DOI] [PubMed] [Google Scholar]
- Hoiseth, S. K., and B. A. Stocker, 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291 238–239. [DOI] [PubMed] [Google Scholar]
- Khush, R. S., W. D. Cornwell, J. N. Uram and B. Lemaitre, 2002. A ubiquitin-proteasome pathway represses the Drosophila immune deficiency signaling cascade. Curr. Biol. 12 1728–1737. [DOI] [PubMed] [Google Scholar]
- Kumar, S., G. K. Christophides, R. Cantera, B. Charles, Y. S. Han et al., 2003. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 100 14139–14144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levashina, E. A., L. F. Moita, S. Blandin, G. Vriend, M. Lagueux et al., 2001. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell 104 709–718. [DOI] [PubMed] [Google Scholar]
- Mansfield, B. E., M. S. Dionne, D. S. Schneider and N. E. Freitag, 2003. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell. Microbiol. 5 901–911. [DOI] [PubMed] [Google Scholar]
- Meister, S., S. M. Kanzok, X. L. Zheng, C. Luna, T. R. Li et al., 2005. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA 102 11420–11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy, D. A., P. S. Jacks and D. J. Hinrichs, 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167 1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, D., and M. Shahabuddin, 2000. Malaria parasite development in a Drosophila model. Science 288 2376–2379. [DOI] [PubMed] [Google Scholar]
- Shirasu-Hiza, M. M., and D. S. Schneider, 2007. Confronting physiology: How do infected flies die? Cell. Microbiol. 9 2775–2783. [DOI] [PubMed] [Google Scholar]
- Sullivan, W., M. Ashburner and R. S. Hawley (Editors), 2000. Drosophila Protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Truett, G. E., P. Heeger, R. L. Mynatt, A. A. Truett, J. A. Walker et al., 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29(52): 54. [DOI] [PubMed] [Google Scholar]
- Wu, L. P., K. M. Choe, Y. Lu and K. V. Anderson, 2001. Drosophila immunity: genes on the third chromosome required for the response to bacterial infection. Genetics 159 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov, E. M., C. von Mering, I. Letunic, D. Torrents, M. Suyama et al., 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298 149–159. [DOI] [PubMed] [Google Scholar]