Abstract
Two recombinant inbred (RI) populations having 194 and 222 lines each, derived, respectively, from a highly heterotic inter- (IJ) and intrasubspecific (II) hybrid, were backcrossed to their respective parents. The RI and two backcross populations along with F1 and its two parents of each hybrid were evaluated for nine important traits, including grain yield and eight other yield-related traits. A total of 76 quantitative trait loci (QTL) for the IJ hybrid and 41 QTL for the II hybrid were detected in the RI population, midparent heterosis of two backcross populations, and two independent sets of data by summation (L1 + L2) and by subtraction (L1 − L2) of two backcross populations (L1 and L2). The variance explained by each QTL ranged from 2.6 to 58.3%. In the IJ hybrid, 42% (32) of the QTL showed an additive effect, 32% (24) a partial-to-complete dominant effect, and 26% (20) an overdominant effect. In the II hybrid, 32% (13) of the QTL demonstrated an additive effect, 29% (12) a partial-to-complete dominant effect, and 39% (16) an overdominant effect. There were 195 digenic interactions detected in the IJ hybrid and 328 in the II hybrid. The variance explained by each digenic interaction ranged from 2.0 to 14.9%. These results suggest that the heterosis in these two hybrids is attributable to the orchestrated outcome of partial-to-complete dominance, overdominance, and epistasis.
HETEROSIS, a term to describe the superiority of heterozygous genotypes over their corresponding parental genotypes (Shull 1908), has been under investigation for ∼100 years, but no consensus exists about the genetic basis underlying this very important phenomenon. Two contending hypotheses, the dominance hypothesis and the overdominance hypothesis, were proposed to explain this phenomenon about one century ago. The dominance hypothesis attributes heterosis to canceling of deleterious or inferior recessive alleles contributed by one parent, by beneficial or superior dominant alleles contributed by the other parent in the heterozygous genotypes at different loci (Davenport 1908; Bruce 1910; Jones 1917). The overdominance hypothesis attributes heterosis to the superior fitness of heterozygous genotypes over homozygous genotypes at a single locus (East 1908; Shull 1908).
Molecular markers and their linkage maps have greatly facilitated the identification of individual loci conditioning heterosis and the estimation of gene action of underlying loci. Quantitative trait locus (QTL) mapping studies aiming at understanding the genetic basis of heterosis have been conducted in rice and other crops (Xiao et al. 1995; Li et al. 1997, 2001; Yu et al. 1997; Luo et al. 2001; Hua et al. 2002, 2003; Semel et al. 2006; Frascaroli et al. 2007; Melchinger et al. 2007a,b). Evidence from such studies suggests that heterosis may be attributable to dominance (Xiao et al. 1995; Cockerham and Zeng 1996), overdominance (Stuber et al. 1992; Li et al. 2001; Luo et al. 2001), pseudo-overdominance due to tightly linked loci with beneficial or superior dominant alleles in repulsion phase (Crow 2000; Lippman and Zamir 2007), or epistasis (Schnell and Cockerham 1992; Li et al. 2001; Luo et al. 2001).
Heterosis is the base of the great success in hybrid rice. Currently, hybrid rice accounts for ∼55% of the total planting acreage of paddy rice in China and the annual increased rice production resulting from planting hybrid rice amounts to ∼20 million metric tones, which can provide a main staple food for >70 million people (Lu et al. 2002). Hybrid rice varieties have a yield advantage of ∼10–20% over the best conventional inbred varieties using similar cultivation conditions (Lu et al. 2002). Besides the large planting in China, hybrid rice varieties are also widely planted in >20 countries around the world.
Previous studies indicated the genetic basis of heterosis in rice is very complicated and various, depending on study materials and analysis approaches (Xiao et al. 1995; Yu et al. 1997; Li et al. 2001; Hua et al. 2002, 2003). The objective of this study was to identify the main-effect QTL and digenic epistatic loci underlying heterosis of nine important agronomic and economic traits of rice and estimate the gene action of each QTL and interaction using a triple-testcross cross (TTC) design to shed light on the understanding of the genetic basis of heterosis in two diverse and highly heterotic rice hybrids.
MATERIALS AND METHODS
Populations:
Two highly heterotic rice hybrids, one intersubspecific between 9024 (indica) and LH422 (japonica) and one intrasubspecific between Zhenshan97 (indica) and Minghui63 (indica), were employed in this study. From the F1 of the intersubspecific hybrid (designated as IJ hybrid hereafter), 194 F7 lines were developed by single-seed descent. From the F1 of the intrasubspecific hybrid (designated as II hybrid hereafter), 222 F12 lines were developed through 11 consecutive selfing generations. Each of these F7 and F12 lines was derived from a different F2 plant. No positive or negative selection was performed during each of the selfing generations. A single plant from each of these 194 F7 lines and 222 F12 lines was chosen randomly and backcrossed to each of its two respective parents to produce backcross progeny and selfed to generate F8 or F13 lines.
Phenotypic variation:
For the IJ hybrid, two backcross populations having 194 lines each, 194 F8 recombinant inbred lines (RILs), along with the two parental lines and their F1, were arranged in a field in a randomized complete block design with two replications for phenotypic evaluation in the summer season of 1992 at the China National Hybrid Rice Research and Development Center, Changsha, Hunan, China. Twenty-seven plants (three rows × 9 plants per row) were planted at a density of 300,000 plants per hectare in each of 1170 plots. The middle 5 plants in the central row of each plot were used for phenotypic trait evaluation and data collection.
For the II hybrid, the two backcross populations with 222 lines each, the corresponding 222 F13 RILs, along with two parental lines and their F1, were laid out in a field in a randomized complete block design with two replications for phenotypic evaluation in the summer season of 2006 at the experimental farm of the Huazhong Agricultural University, Wuhan, Hubei, China. Twenty-one-day-old seedlings were transplanted into three-row plots with each plot consisting of a single row of a RIL and two rows of backcross (BC) hybrids. There were seven plants in each row, with 16.7 cm between plants within each row and 26.7 cm between rows. The middle five plants in each row were used for phenotypic trait evaluation and data collection.
Nine quantitative traits of agronomic and economic importance evaluated were heading date (HD) (in days), plant height (PH) (in centimeters), tillers per plant (TP), panicle length (PL) (in centimeters), filled grains per panicle (FGPP), percentage of seed set (SS), grain density (GD) (in grain numbers per centimeter of panicle length), 1000-grain weight (KGW) (in grams), and grain yield (YD) (in tons/hectare). Means over replications, for each trait, for the RIL and each of two backcross populations, were used for QTL and other analyses.
Analysis of field data and of heterosis:
For each hybrid, data of recombinant inbred (RI) and BC populations were analyzed separately. SAS PROC GLM (SAS Institute 1996) was used to test the differences among RILs and the corresponding BC hybrids. Heterosis was evaluated in BC populations by midparental heterosis (Hmp). Hmp = F1 − (RIL + recurrent parent)/2. F1's are mean trait values of individual BC hybrids while RIL is the corresponding RIL parent for each of the BC hybrids, and recurrent parent is 9024 or LH422 in the IJ hybrid and Zhenshan 97 or Minghui 63 in the II hybrid. To distinguish one from another, the RIL is designated as RILij in the IJ hybrid and as RILii in the II hybrid.
Following Kearsey et al. (2003) and Frascaroli et al. (2007), the crosses of the n RILs to the two recurrent parents are referred as “L1i” and “L2i” (i = 1 ∼ n), respectively. The two independent sets of data by summation (L1i + L2i) and by subtraction (L2i − L1i) of the two BC populations' values hereafter are referred to as the “SUM” data set and the “DIFF” data set, respectively. Variation within the SUM data set is due to additive effects and variation within the DIFF data set is due to dominance effects when combined over two BC populations.
In this study, for the IJ hybrid, L1i and L2i represent the n = 194 RILs to 9024 and LH422, respectively; while for the II hybrid, L1i and L2i represent the n = 222 RILs to Zhenshan97 and Minghui63, respectively. To distinguish one from another, the two data sets SUM and DIFF in the IJ hybrid are referred as SUMij and DIFFij and those in the II hybrid as SUMii and DIFFii.
NCIII and TTC analysis:
ANOVA was used to test for additive (L1i + L2i) and dominance (L2i − L1i) variation by following the standard North Carolina design III (NCIII) and for epistatic variation (L1i + L2i − P) by following the extended TTC design as described by Kearsey and Jinks (1968), with P indicated as the RI population in this study. Additive (VA) and dominance (VD) components of genetic variance were estimated and used to calculate the average degree of dominance [as √(2VD/VA)], which is a weighted mean of the level of dominance over all segregating loci (Kearsey and Pooni 1996).
Genetic linkage maps:
For the IJ hybrid, a subset of 141 polymorphic RFLP markers was selected from the rice high-density molecular map (Causse et al. 1994) to construct the linkage map of the RI population by Xiao et al. (1995). For the II hybrid, a linkage map was constructed by Xing et al. (2002), which consisted of 221 marker loci and covered a total of 1796 cM.
QTL mapping and detection of dominance degree of main-effect QTL and epistatic-effect QTL:
QTL mapping:
QTL analysis was performed separately for the RI, the midparental heterosis (Hmp) of two backcross populations, and two independent data sets SUM and DIFF in the IJ hybrid and the II hybrid. In the absence of epistasis, the analysis of RIL and SUM data sets identifies QTL with an additive effect (a), whereas the analysis of Hmp and DIFF data sets detects QTL with a dominance effect (d) (Frascaroli et al. 2007).
Analysis of main-effect QTL (M-QTL) was conducted in each mapping population by composite-interval mapping, using WinQTLcart (Zeng 1994). A LOD score of 2.0 was selected as the threshold for the presence of a main-effect QTL based on the total map distance and the average distance between markers. QTL detected in different populations or for different traits were considered as common if their estimated map position was within a 20-cM distance (Groh et al. 1998), which is a common approach in comparative mapping. Following Frascaroli et al. (2007), in the absence of epistasis, the expectation of genetic effects in RIL, SUM, Hmp, and DIFF data was a, a, d/2, and d.
Analysis of digenic interaction was conducted in each mapping population by the mixed linear approach and by the use of the computer software QTLMAPPER ver. 1.0 (Wang et al. 1999). The analysis was first conducted without considering epistasis to confirm the QTL detected with the method previously described and then with epistasis considered in the model. A threshold of LOD ≥ 3.0 (P < 0.001) was used for declaring the presence of a putative pair of epistatic QTL.
Genetic analysis methods for estimating QTL dominance degree:
North Carolina design III (NCIII) was put forward by Comstock and Robinson (1952). In a NCIII design, male progeny from generation 2 (F2, which were treated as a base population) of two inbred strains are backcrossed to their grandmothers (marked as L1 and L2), and their progeny are arranged in a completely randomized block design (Comstock and Robinson 1952). In 1968, an NCIII design was developed by Kearsey and Jinks. In their theory, the F3, F4, … , Fn, double haploid (DH), and RIL also can be treated as base populations. Following Kearsey, the base population was crossed to the two parents (P1 and P2) indicated as L1 and L2. With the data of L1 + L2 and L1 − L2, the genetic parameters of QTL such as additive effect, dominant effect, and the degree of dominance could be estimated.
On the basis of the correlation analysis of detected M-QTL and digenic interaction proposed by Hu et al. (1995, 2002), regression and variance analysis of two data L1 + L2 and L1 − L2 when the base population was the DH population could be deduced as follows (Tables 1 and 2).
TABLE 1.
Genetic expectation of regression coefficients of L1 + L2 and L1 − L2 when the base population was the DH population
| L1 + L2 | L1 − L2 | |(L1 − L2)/(L1 + L2)| | |
|---|---|---|---|
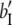 |
(1 − 2r1)a1 | −(1 − 2r1)d1 | d1/a1 |
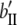 |
(1 − 2r1)(1 − 2r2)ia1a2 | (1 − 2r1)(1 − 2r2)ld1d2 |
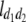 / /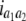
|
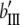 |
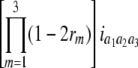 |
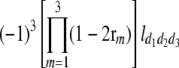 |
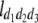 / /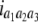
|
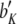 |
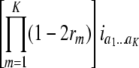 |
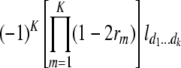 |
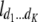 / /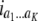
|
b′i (i = 1 ∼ K, where K is the total number of markers in linkage map) is indicated as a regression coefficient. ai (i = 1 ∼ K) and di (i = 1 ∼ K) are denoted as the additive effect and the dominant effect, respectively; i a1a2 is the additive × additive epistatic effect, i a1a2a3 is the additive × additive × additive epistatic effect, etc. l d1d2 is the dominance × dominance epistatic effect, l d1d2d3 is the dominance × dominance × dominance epistatic effect, etc. rm denotes the recombinant value. For the RI population, the expectations were similar to those in the DH population except for rm, which was replaced by 2r′m/(1 + 2r′m) and 4r″m/(1 + 6r″m), respectively. The r′m and r″m were recombinant values for two RI populations (selfing population and sib-mating population), respectively (Hu et al. 2002). d1/a1 is indicated as the dominant degree of main-effect QTL, l d1d2/i a1a2 as the epistasis dominance degree (EDD), and l d1d2d3/i a1a2a3 as the epistasis dominance degree among three markers, etc.
TABLE 2.
Genetic expectation of variance components of L1 + L2 and L1 − L2 when the base population was the DH population
| ANOVA | L1 + L2 | L1 − L2 | 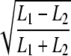 |
|---|---|---|---|
| One way |
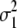 = (1 − 2r1)2 = (1 − 2r1)2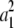
|
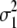 = (1 − 2r1)2 = (1 − 2r1)2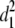
|
d1/a1 |
| Two way |
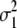 = (1 − 2r1)2(a1 + = (1 − 2r1)2(a1 + 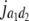 )2 )2
|
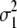 = (1 − 2r1)2(d1 + = (1 − 2r1)2(d1 + 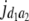 )2 )2
|
|
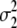 = (1 − 2r2)2(a2 + = (1 − 2r2)2(a2 + 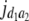 )2 )2
|
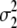 = (1 − 2r2)2(d2 + = (1 − 2r2)2(d2 + 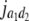 )2 )2
|
||
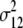 = (1 − 2r1)2(1 − 2r2)2 = (1 − 2r1)2(1 − 2r2)2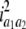
|
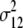 = (1 − 2r1)2(1 − 2r2)2 = (1 − 2r1)2(1 − 2r2)2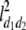
|
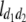 / /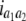
|
|
| Three way |
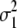 = (1 − 2r1)2(a1 + = (1 − 2r1)2(a1 + 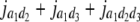 )2 )2
|
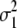 = (1 − 2r1)2(d1 + = (1 − 2r1)2(d1 + 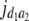 + + 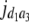 + + 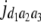 )2 )2
|
|
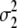 = (1 − 2r2)2(a2 + = (1 − 2r2)2(a2 + 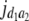 + + 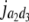 + + 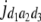 )2 )2
|
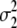 = (1 − 2r2)2(d2 + = (1 − 2r2)2(d2 + 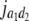 + + 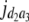 + + 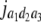 )2 )2
|
||
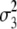 = (1 − 2r3)2(a3 + = (1 − 2r3)2(a3 + 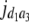 + + 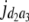 + + 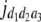 )2 )2
|
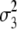 = (1 − 2r3)2(d3 + = (1 − 2r3)2(d3 + 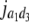 + + 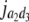 + + 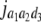 )2 )2
|
||
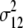 = (1 − 2r1)2(1 − 2r2)2( = (1 − 2r1)2(1 − 2r2)2(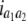 + + 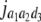 )2 )2
|
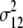 = (1 − 2r1)2(1 − 2r2)2( = (1 − 2r1)2(1 − 2r2)2(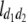 + + 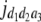 )2 )2
|
||
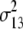 = (1 − 2r1)2(1 − 2r3)2( = (1 − 2r1)2(1 − 2r3)2(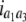 + + 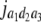 )2 )2
|
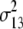 = (1 − 2r1)2(1 − 2r3)2( = (1 − 2r1)2(1 − 2r3)2(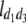 + + 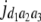 )2 )2
|
||
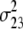 = (1 − 2r2)2(1 − 2r3)2( = (1 − 2r2)2(1 − 2r3)2(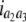 + + 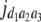 )2 )2
|
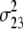 = (1 − 2r2)2(1 − 2r3)2( = (1 − 2r2)2(1 − 2r3)2(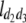 + + 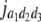 )2 )2
|
||
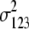 = (1 − 2r1)2(1 − 2r2)2(1 − 2r3)2 = (1 − 2r1)2(1 − 2r2)2(1 − 2r3)2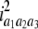
|
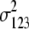 = (1 − 2r1)2(1 − 2r2)2(1 − 2r3)2 = (1 − 2r1)2(1 − 2r2)2(1 − 2r3)2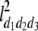
|
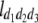 / /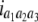
|
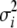 (i = 1 ∼ K),
(i = 1 ∼ K),  (i < j, i = 1 ∼ K, j = 2 ∼ K), and
(i < j, i = 1 ∼ K, j = 2 ∼ K), and  (i < j < l, i = 1 ∼ K, j = 2 ∼ K, l = 3 ∼ K) are denoted as variance components of a single marker, two markers, and three markers. The other parameters are the same as in Table 1.
(i < j < l, i = 1 ∼ K, j = 2 ∼ K, l = 3 ∼ K) are denoted as variance components of a single marker, two markers, and three markers. The other parameters are the same as in Table 1.
On the basis of the methodology proposed, we developed a software QTLIII (not published yet), which is suitable for analyzing the additive effect, dominant effect, and dominance degree of QTL (including one-factor, two-factor, and three-factor ANOVA, see Tables 1 and 2). In this study, it was used to estimate dominance degree of main-effect and epistatic-effect QTL.
The degree of dominance of a M-QTL was estimated as |d/a|. For this purpose, for all QTL declared as significant within any data set, dominant and additive effects were estimated in SUM and DIFF data sets by QTLIII with ANOVA analysis. These estimates were used to calculate |d/a| and classify the QTL as additive (A) (|d/a| < 0.2), partial dominance (PD) (0.2 ≤ |d/a| < 0.8), dominance (D) (0.8 ≤ |d/a| < 1.2), and overdominance (OD) (|d/a| ≥ 1.2) according to Stuber et al. (1987).
Genetic expectations of the parameters estimated in the epistatic models differ on the basis of genetic composition of data sets analyzed. For the SUM data set, the estimated interaction is expected to be predominantly additive × additive (aa), whereas for the DIFF data set it is expected to be predominantly dominance × dominance (dd). In this study, |dd/aa|, defined as epistasis dominance degree (EDD), was estimated by the software QTLIII with ANOVA analysis. These estimates were used to calculate |dd/aa| to classify the epistatic QTL as A (|dd/aa| < 0.2), PD (0.2 ≤ |dd/aa| < 0.8), D (0.8 ≤ |dd/aa| < 1.2), and OD (|dd/aa| ≥ 1.2).
Relationship between genomewide or chromosomewide molecular marker heterozygosity and phenotypic trait performance and heterosis:
GGT (Van 1999) was used to calculate genome ratios (percentage of total genome originated from one parental genome) for each line in the RI population, initially for the whole genome and then for each chromosome. Relationship between molecular marker heterozygosity and phenotypic performance was tested by regressing phenotypic performance on whole-genome heterozygosity in two backcross populations in both IJ and II hybrids. Meanwhile, to elucidate the relationship between observed heterosis and heterozygosity, (i) the Hmp and DIFF values were respectively regressed against heterozygosity across the whole genome using linear regression (when the DIFF data set was used as a dependent variable, genome heterozygosity of each backcross population was the independent variable), and (ii) the Hmp values were regressed against heterozygosity on individual chromosomes by multiple regression.
RESULTS
F1 heterosis:
In the IJ hybrid, LH422 showed significant higher mean trait values than 9024 (Table 3). All nine traits except heading date in F1 had a higher value than both parents. For midparental heterosis, yield showed the strongest significant heterosis (25.58%), followed by 1000-grain weight (15.82%), plant height (15.34%), panicle length (9.42%), tillers per plant (8.00%), seed set (4.06%), and heading date (1.74%). However, the F1 hybrid had a lower trait value for filled grains per panicle and grain density than the parental lines, with negative heterosis of 2.08 and 10.17%, respectively.
TABLE 3.
Mean values of nine important agronomic traits of P1, P2, F1, RIL, and their two backcross populations in two rice elite hybrids
| HD | PH | TP | PL | FGPP | SS | GD | KGW | YD | |
|---|---|---|---|---|---|---|---|---|---|
| IJ hybrid | |||||||||
| 9024 | 83.00 | 94.20 | 11.40 | 21.98 | 84.21 | 71.41 | 3.83 | 24.60 | 6.53 |
| LH422 | 86.00 | 104.00 | 8.60 | 23.88 | 105.88 | 70.03 | 4.43 | 22.18 | 6.02 |
| F1 | 86.00 | 114.30 | 10.80 | 25.09 | 93.07 | 73.59 | 3.71 | 27.09 | 7.88 |
| Heterosis (%) | 1.78 | 15.34 | 8.00 | 9.42 | −2.08 | 4.06 | −10.17 | 15.82 | 25.58 |
| RIL | 82.66 | 107.47 | 9.95 | 23.93 | 94.20 | 64.64 | 6.12 | 23.42 | 6.06 |
| 9024BC | 80.96 | 107.28 | 10.38 | 24.60 | 83.20 | 60.66 | 5.60 | 26.31 | 6.14 |
| LH422BC | 81.21 | 110.83 | 9.55 | 25.27 | 98.28 | 62.75 | 6.25 | 24.45 | 6.18 |
| II hybrid | |||||||||
| Zhenshan97 | 62.25 | 93.33 | 12.09 | 19.98 | 93.82 | 69.24 | 4.70 | 24.79 | 4.34 |
| Minghui63 | 91.00 | 112.55 | 12.04 | 24.95 | 118.51 | 64.89 | 4.75 | 29.81 | 6.05 |
| F1 | 90.00 | 125.52 | 13.40 | 25.48 | 137.09 | 78.25 | 5.38 | 29.54 | 9.52 |
| Heterosis (%) | 17.46 | 21.94 | 11.09 | 13.42 | 29.13 | 16.68 | 13.86 | 8.21 | 83.09 |
| RIL | 82.94 | 110.23 | 10.77 | 23.18 | 115.70 | 78.17 | 5.01 | 25.97 | 5.49 |
| Zhenshan97BC | 75.44 | 113.11 | 11.99 | 23.32 | 121.81 | 79.42 | 5.22 | 26.26 | 6.73 |
| Minghui63BC | 85.44 | 113.50 | 12.00 | 24.81 | 126.15 | 81.29 | 5.09 | 27.74 | 7.56 |
For a description of agronomic traits see materials and methods.
In the II hybrid, the parent Minghui63 had a significantly higher phenotypic value than Zhenshan97 for all nine traits investigated (Table 3). The F1 hybrid had 91 days to heading, similar to Minghui63, which took more days to heading than Zhenshan97. The values of the other traits were significantly higher in F1 than in both parents. The midparental heterosis of the F1 plants was strongest for yield (83.09%), followed by filled grains per panicle (29.13%), plant height (21.94%), heading date (17.46%), seed set (16.68%), grain density (13.86%), panicle length (13.42%), tillers per plant (11.09%), and 1000-grain weight (8.21%).
Heterosis in RI and BC populations:
RIL and parental inbred mean values (Table 3) were not significantly different for any trait in both IJ and II hybrids.
Significant heterosis for yield was observed in II hybrid BC populations, but not in IJ hybrid BC populations. Most of the other traits did not show significant heterosis in BC populations of both IJ and II hybrids.
For the IJ hybrid, the mean values of the 9024BC and LH422BC populations were 80.96 and 81.21 for heading date, 107.28 and 110.83 for plant height, 10.38 and 9.55 for tillers per plant, 24.60 and 25.27 for panicle length, 83.20 and 98.28 for filled grains per panicle, 60.66 and 62.75 for seed set, 5.60 and 6.25 for grain density, 26.31 and 24.45 for 1000-grain weight, and 6.14 and 6.18 for yield. The heterosis was 24.45 (29.5%) and 3.12 (7.0%) for heading date, 6.45 (6.4%) and 5.10 (4.6%) for plant height, −0.30 (−2.8%) and 0.28 (3.0%) for tillers per plant, 1.65 (7.2%) and 1.36 (5.5%) for panicle length, −5.90 (−6.6%) and −1.56 (−1.8%) for filled grains per panicle, −7.39 (−10.9%) and 4.62 (6.9%) for seed set, 0.62 (12.5%) and 0.97 (20.8%) for grain density, 2.19 (9.1%) and 1.58 (5.9%) for 1000-grain weight, and −0.16 (−2.5%) and 0.14 (2.3%) for yield, in the 9024BC and LH422BC populations, respectively.
For the II hybrid, the mean values of the Zhenshan97BC and Minghui63BC populations were 75.44 and 85.44 for heading date, 113.11 and 113.50 for plant height, 11.99 and 12.00 for tillers per plant, 23.32 and 24.81 for panicle length, 121.81 and 126.15 for filled grains per panicle, 79.42 and 81.29 for seed set, 5.22 and 5.09 for grain density, 26.26 and 26.74 for 1000-grain weight, and 6.73 and 7.56 for yield. The heterosis values were −11.53 (−15.9%) and −1.53 (−1.8%) for heading date, 1.72 (1.7%) and 2.11 (1.9%) for plant height, 0.59 (1.1%) and 0.59 (0.9%) for tillers per plant, −0.75 (−3.5%) and 0.74 (3.1%) for panicle length, 4.7 (4.5%) and 9.04 (7.7%) for filled grains per panicle, 4.89 (10.7%) and 8.75 (13.6%) for seed set, 0.34 (7.1%) and 0.21 (4.3%) for grain density, −1.64 (−0.64%) and −0.16 (−0.6%) for 1000-grain weight, and 1.82 (36.9%) and 1.04 (15.9%) for yield in the Zhenshan97BC and Minghui63BC populations, respectively.
NCIII and TTC analysis:
TTC analysis allows us to test nonallelic interactions. Significant additive × additive ([aa]) epistasis was detected for all traits in both IJ and II hybrids (Table 4). The epistasis due to additive × dominance or dominance × dominance ([ad] and [dd]) was significant for all traits in the IJ hybrid and all the traits except tillers per plant in the II hybrid.
TABLE 4.
NCIII and TTC analyses of the two rice hybrids
| Parameter | HD | PH | TP | PL | FGPP | SS | GD | KGW | YD |
|---|---|---|---|---|---|---|---|---|---|
| IJ hybrid | |||||||||
| VAa | 4.36 | 30.40 | 0.21 | 1.16 | 1069.24 | 75.67 | 0.36 | 50.70 | 3.12 |
| VDb | 3.61 | 24.24 | 0.23 | 1.25 | 71.22 | 17.00 | 0.30 | 1.06 | 0.16 |
| a.d.d. | 0.91 | 0.89 | 1.03 | 1.04 | 0.26 | 0.47 | 0.91 | 0.14 | 0.22 |
| [aa] | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| [ad], [dd] | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| II hybrid | |||||||||
| VAa | 33.49 | 36.24 | 0.79 | 1.06 | 128.57 | 29.88 | 0.44 | 2.96 | 0.32 |
| VDb | 23.11 | 67.18 | 0.48 | 0.69 | 84.86 | 35.34 | 0.08 | 0.68 | 0.70 |
| a.d.d. | 0.83 | 1.36 | 0.78 | 0.81 | 0.81 | 1.09 | 0.58 | 0.48 | 1.48 |
| [aa] | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| [ad], [dd] | *** | *** | NS | * | *** | *** | ** | *** | ** |
*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005.
Estimates of additive (VA) and dominance (VD) variance, average degree of dominance (a.d.d.), and tests for additive × additive ([aa]) and additive × dominance and dominance × dominance ([aa], [dd]) epistasis.
VA was highly significant (P ≤ 0.005) for all traits; VD was highly significant for all traits, except TP (significant at P ≤ 0.05) in the II hybrid.
In this study, NCIII analysis led to the estimates of VA (additive variance) and VD (dominance variance), which were always highly significant (P < 0.005) in both hybrids, except for the VD of tillers per plant in the II hybrid, which was significant at P < 0.05 (Table 4).
M-QTL:
QTL detected in RIL, SUM, two Hmp, and DIFF data sets in IJ and II hybrids are shown in Tables 5 and 6, respectively. In total, 76 and 41 QTL were revealed in five data sets of IJ and II hybrids, respectively. Most of these QTL explained <10% of variation individually. Five QTL (6.76%) in the IJ hybrid and 4 (9.76%) in the II hybrid accounted for >20% of phenotypic variation individually.
TABLE 5.
Main-effect QTL resolved in the IJ hybrid
| RILij
|
SUMij
|
9024Hmp
|
LH422Hmp
|
DIFFij
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Chr-Ina | Interval | Ab | R2 (%) | Ab | R2 (%) | Db | R2 (%) | Db | R2 (%) | Db | R2 (%) | d/ac |
| HD | 1-17 | RG811-RG173 | −0.99 | 2.60 | A | ||||||||
| 2-8 | RG544-RZ599 | −3.92 | 9.30 | A | |||||||||
| 3-6 | XNPB249-RZ16 | −4.06 | 9.40 | OD | |||||||||
| 3-8 | RZ993-CDO1081 | 1.71 | 7.90 | 1.10 | 6.80 | 1.20 | 6.50 | PD | |||||
| 5-1 | RZ390-RZ556 | −3.79 | 8.60 | A | |||||||||
| 6-16 | RZ682-RG653 | 3.92 | 8.70 | OD | |||||||||
| 6-17 | RG653-RZ828 | −1.05 | 4.80 | OD | |||||||||
| 7-6 | RG711-XNPB20 | −1.91 | 4.70 | PD | |||||||||
| 8-1 | RG333-RZ562 | −5.10 | 58.30 | −2.72 | 34.80 | −3.30 | 39.80 | −1.22 | 17.40 | PD | |||
| 11-2 | CDO534-XNPB179 | 1.07 | 4.90 | PD | |||||||||
| PH | 1-11 | RZ776-RG375 | −2.84 | 5.20 | A | ||||||||
| 1-17 | RG811-RG173 | −1.09 | 3.20 | A | |||||||||
| 2-8 | RG544-RZ599 | −2.82 | 6.30 | PD | |||||||||
| 3-3 | RG558-RG510 | 2.66 | 3.80 | OD | |||||||||
| 3-5 | XNPB232-XNPB249 | −2.14 | 8.80 | PD | |||||||||
| 3-8 | RZ993-CDO1081 | 1.89 | 9.70 | A | |||||||||
| 5-7 | RZ70-RG480 | 4.06 | 12.50 | 2.51 | 6.10 | A | |||||||
| 6-10 | RZ667-RG1028 | −3.20 | 10.10 | OD | |||||||||
| 6-16 | RZ682-RG653 | 3.45 | 8.70 | D | |||||||||
| 7-6 | RG711-XNPB20 | 1.63 | 7.00 | OD | |||||||||
| 7-9 | CDO533-RG528 | −3.40 | 9.50 | −3.79 | 29.00 | A | |||||||
| 8-1 | RG333-RZ562 | −5.10 | 58.30 | −4.81 | 15.80 | A | |||||||
| TP | 3-1 | RG1356-CDO87 | −0.27 | 5.00 | A | ||||||||
| 4-5 | RG214-CDO539 | 0.27 | 8.70 | A | |||||||||
| 5-11 | CDO1160-CDO202 | 0.20 | 5.10 | OD | |||||||||
| 9-9 | XNPB295-RZ404 | −0.38 | 5.10 | PD | |||||||||
| PL | 1-9 | RG233-XNPB302 | 0.63 | 5.80 | OD | ||||||||
| 2-4 | CDO1091-CDO395 | 0.46 | 4.10 | A | |||||||||
| 3-8 | RZ993-CDO1081 | 0.42 | 5.20 | A | |||||||||
| 4-13 | RZ602-CDO456 | −0.42 | 6.70 | OD | |||||||||
| 5-4 | RZ296-RG573 | −0.49 | 7.00 | PD | |||||||||
| 5-11 | CDO1160-CDO202 | −0.36 | 4.70 | −0.54 | 5.60 | D | |||||||
| 6-10 | RZ667-RG1028 | −0.81 | 9.90 | OD | |||||||||
| 7-9 | CDO533-RG528 | −1.07 | 34.60 | A | |||||||||
| 9-10 | RZ404-RG358 | 0.44 | 5.20 | A | |||||||||
| 10-4 | RZ400-RZ583 | 0.54 | 11.00 | OD | |||||||||
| 12-4 | RZ670-XNPB316 | −0.52 | 5.40 | A | |||||||||
| FGPP | 3-8 | RZ993-CDO1081 | −7.02 | 12.50 | −7.52 | 9.80 | A | ||||||
| 4-2 | RG143-RZ590 | −5.36 | 5.50 | A | |||||||||
| 4-5 | RG214-CDO539 | −7.80 | 15.70 | A | |||||||||
| 5-2 | RZ556-RG360 | −3.64 | 6.90 | 6.31 | 6.90 | PD | |||||||
| 5-3 | RG360-RZ296 | 5.96 | 9.00 | PD | |||||||||
| 8-1 | RG333-RZ562 | 5.52 | 5.80 | OD | |||||||||
| SS | 2-9 | RZ599-RG152 | −2.63 | 4.10 | A | ||||||||
| 3-8 | RZ993-CDO1081 | −2.44 | 5.10 | PD | |||||||||
| 4-9 | RZ565-XNPB271 | 2.38 | 4.80 | A | |||||||||
| 5-3 | RG360-RZ296 | 2.90 | 7.20 | A | |||||||||
| 6-17 | RG653-RZ828 | −1.44 | 7.50 | PD | |||||||||
| 7-9 | CDO533-RG528 | −4.41 | 12.30 | PD | |||||||||
| 7-10 | RG528-RG417 | 2.78 | 4.60 | PD | |||||||||
| 8-1 | RG333-RZ562 | 2.48 | 4.60 | 1.66 | 5.10 | OD | |||||||
| 11-3 | XNPB179-XNPB320 | 2.82 | 4.60 | A | |||||||||
| 12-8 | RZ816-XNPB189 | 1.57 | 4.90 | OD | |||||||||
| GD | 3-8 | RZ993-CDO1081 | −0.30 | 11.50 | −0.40 | 9.50 | A | ||||||
| 4-3 | RZ590-RZ262 | −0.38 | 17.80 | D | |||||||||
| 4-4 | RZ262-RG214 | −0.35 | 4.20 | PD | |||||||||
| 6-11 | RG1028-RG162 | 0.40 | 19.30 | OD | |||||||||
| 6-12 | RG162-CDO78 | −0.18 | 3.90 | OD | |||||||||
| 8-2 | RZ562-RZ66 | −0.34 | 14.00 | OD | |||||||||
| 10-3 | RZ561-RZ400 | 0.20 | 5.00 | A | |||||||||
| KGW | 1-3 | RG350-RZ739 | −1.85 | 45.80 | PD | ||||||||
| 1-4 | RZ739-XNPB370 | −0.59 | 4.80 | A | |||||||||
| 1-5 | XNPB370-RG541 | −0.40 | 7.10 | A | |||||||||
| 2-4 | CDO1091-CDO395 | 1.86 | 46.00 | A | |||||||||
| 3-7 | RG417-RG333 | −0.94 | 12.20 | PD | |||||||||
| 3-8 | RG333-RZ562 | −0.43 | 8.10 | PD | |||||||||
| 4-10 | XNPB271-RG449 | −0.65 | 5.50 | OD | |||||||||
| 6-9 | RZ144-RZ667 | −0.56 | 4.20 | A | |||||||||
| 7-9 | CDO533-RG528 | 1.53 | 4.70 | A | |||||||||
| 8-1 | RG333-RZ562 | 3.07 | 15.50 | OD | |||||||||
| YD | 3-8 | RZ993-CDO1081 | −0.49 | 7.20 | A | ||||||||
| 5-2 | RZ556-RG360 | 0.36 | 3.90 | A | |||||||||
| 6-4 | XNPB317-RZ247 | 0.33 | 8.00 | D | |||||||||
| 7-6 | RG711-XNPB20 | −0.23 | 11.90 | D | |||||||||
| 7-7 | XNPB20-RZ509 | −0.44 | 5.50 | D | |||||||||
| 8-1 | RG333-RZ562 | 0.66 | 11.30 | 0.17 | 5.90 | OD | |||||||
Effects estimated in 9024Hmp and LH422Hmp were multiplied by 2, and the values estimated in LH422Hmp and the DIFF were multiplied by (−1).
Chromosome number interval of the QTL detected in the study.
A and D represent additive effect and dominance effect of M-QTL.
The degree of dominance for all M-QTL declared as significant in any data set was determined after estimating their additive and dominance effects, respectively, in SUM and DIFF data sets. QTL were classified according to their |d/a| ratio as additive (A) (|d/a| < 0.2), partial dominance (PD) (0.2 ≤ |d/a| < 0.8), dominance (D) (0.8 ≤ |d/a| < 1.2), and overdominance (OD) (|d/a| ≥ 1.2) (Stuber et al. 1987).
TABLE 6.
Main-effect QTL resolved in the II hybrid
| RILii
|
SUMii
|
Zhenshan97Hmp
|
Minghui63Hmp
|
DIFFii
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Chr-Ina | Interval | Ab | R2 (%) | Ab | R2 (%) | Db | R2 (%) | Db | R2 (%) | Db | R2 (%) | d/ac |
| HD | 1-6 | RM243-RG173 | 1.02 | 4.10 | PD | ||||||||
| 2-7 | R712-RZ324 | 1.42 | 4.90 | 2.04 | 3.80 | PD | |||||||
| 2-16 | RM208-RM207 | 1.52 | 4.10 | A | |||||||||
| 6-24 | R2549-C962 | 1.28 | 5.20 | A | |||||||||
| 7-9 | RM234-R1789 | −1.60 | 6.60 | −2.28 | 7.60 | D | |||||||
| 8-2 | C1121-RG333 | −4.62 | 11.30 | PD | |||||||||
| 11-17 | RG118-C1237 | 1.50 | 5.70 | D | |||||||||
| 12-4 | C996-G1128a | −3.10 | 6.10 | A | |||||||||
| PH | 2-4 | R1738-RM53 | −2.86 | 6.70 | A | ||||||||
| 3-15 | RM227-R1925 | −2.86 | 4.60 | A | |||||||||
| 6-15 | RM204-R1014 | 2.06 | 4.20 | PD | |||||||||
| 10-10 | RG561-R2625 | 2.27 | 3.60 | A | |||||||||
| TP | 2-5 | RM53-RZ599 | 0.87 | 27.40 | OD | ||||||||
| 3-3 | C63-RM232 | −0.56 | 11.00 | OD | |||||||||
| 7-9 | RM234-R1789 | 1.60 | 6.60 | PD | |||||||||
| 10-5 | C148-RM239 | −0.58 | 4.60 | A | |||||||||
| 11-17 | RG118-C1237 | 1.50 | 5.70 | A | |||||||||
| PL | 5-2 | R3166-RG360 | 0.43 | 4.20 | A | ||||||||
| 6-27 | RG653-G342 | 0.62 | 5.20 | A | |||||||||
| 8-1 | R902-C1121 | 0.51 | 4.60 | A | |||||||||
| FGPP | 3-4 | RM232-G144 | −15.73 | 27.70 | PD | ||||||||
| 6-16 | R1014-RZ588 | 3.77 | 3.70 | OD | |||||||||
| SS | 12-6 | R887-G1314b | −2.66 | 3.40 | OD | ||||||||
| GD | 3-4 | RM232-G144 | −0.60 | 27.60 | PD | ||||||||
| 6-16 | R1014-RZ588 | 0.20 | 5.50 | A | |||||||||
| 9-7 | RG667-RM215 | 0.24 | 4.00 | A | |||||||||
| 10-12 | RM228-C371 | −0.27 | 4.90 | −0.27 | 4.90 | OD | |||||||
| KGW | 3-1 | C1176-C316 | 0.33 | 4.20 | 0.42 | 3.80 | D | ||||||
| 3-7 | C1087-RZ403 | 0.93 | 5.50 | PD | |||||||||
| 3-9 | R19-C746 | 0.47 | 8.90 | D | |||||||||
| 6-1 | C764-RM225 | −0.79 | 3.70 | A | |||||||||
| 6-15 | RM204-R1014 | −1.80 | 21.10 | A | |||||||||
| 8-13 | L363A-RZ66 | 0.50 | 5.50 | OD | |||||||||
| 11-14 | CDO127-R2918 | −0.63 | 8.30 | OD | |||||||||
| 11-15 | R2918-C794 | 0.35 | 4.10 | OD | |||||||||
| YD | 2-2 | RM211-RG634 | 0.72 | 3.30 | OD | ||||||||
| 7-8 | R1245-RM234 | −2.05 | 6.10 | OD | |||||||||
| 7-9 | RM234-R1789 | −1.99 | 5.20 | OD | |||||||||
| 11-11 | C104-RM20a | −1.47 | 4.00 | OD | |||||||||
| 11-23 | CDO534-RM21 | −2.05 | 11.30 | OD | |||||||||
| 11-30 | G389-G181 | 1.37 | 4.90 | A | |||||||||
Effects obtained in Zhenshan97Hmp and Minghui63Hmp were multiplied by 2, and the values obtained in LH422Hmp and the DIFF were also multiplied by (−1).
Chromosome number interval of the QTL detected in the study.
A and D represent additive effect and dominance effect of M-QTL.
The degree of dominance for all M-QTL declared as significant in any data set was determined after estimating their additive and dominance effects, respectively, in SUM and DIFF data sets. QTL were classified according to their |d/a| ratio as additive (A) (|d/a| < 0.2), partial dominance (PD) (0.2 ≤ |d/a| < 0.8), dominance (D) (0.8 ≤ |d/a| < 1.2), and overdominance (OD) (|d/a| ≥ 1.2) (Stuber et al. 1987).
HD:
In the IJ hybrid, 10 QTL were detected. Three showed an additive effect, 4 a partial-to-complete dominant effect, and 3 an overdominant effect. Six of the 9 QTL showing a dominant effect identified in Hmp and DIFFij were negative, with alleles from 9024 increasing the trait value. In the II hybrid, 8 QTL were found. Three exhibited an additive effect and 5 a partial-to-complete dominant effect. Four of the 5 QTL displaying a dominant effect revealed in Hmp and DIFFii were positive, with alleles from Minghui63 increasing the trait value.
PH:
In the IJ hybrid, 12 QTL were found. Six were classified as additive, 3 as partial-to-complete dominance, and 4 as overdominance. In the II hybrid, 4 QTL were detected. Three were found to be additive and 1 in Zhenshan97Hmp to be overdominant. No QTL was identified in SUMii.
TP:
In the IJ hybrid, four QTL were identified with two showing an additive effect, one an overdominant effect, and one a partial dominant effect. No QTL was found in the LH422Hmp and DIFFij data sets. In the II hybrid, five QTL were detected with two exhibiting an additive effect, one a dominant effect, and two an overdominant effect.
PL:
In the IJ hybrid, 11 QTL were found with 5 classified as an additive effect, 4 as an overdominant effect, and 2 as a partial-to-complete dominant effect. In the II hybrid, 2 QTL in RIL and 1 QTL in SUMii were detected, displaying an additive effect and with alleles from Minghui63 increasing the trait value.
FGPP:
In the IJ hybrid, six QTL were found with three behaving like an additive effect, two like a partial-dominant effect, and one like an overdominant effect. In the II hybrid, two QTL were detected with one appearing to be an overdominant effect and one a partial-dominant effect. No QTL was revealed in Hmp and DIFFii.
SS:
In the IJ hybrid, 10 QTL were found with 4 displaying an additive effect, 4 a partial-dominant effect, and 2 an overdominant effect. In the II hybrid, only 1 QTL was detected in DIFFii data, showing overdominant effect, and the alleles from Zhenshan97 increased the trait value.
GD:
In the IJ hybrid, seven QTL were identified with two exhibiting an additive effect, two a partial-to-complete dominant effect, and three an overdominant effect. No QTL was detected in 9024Hmp. In the II hybrid, four QTL were revealed with two showing an additive effect, one a partial-dominant effect, and one an overdominant effect. No QTL was found in Minghui63Hmp and DIFFii data sets.
KGW:
In the IJ hybrid, 10 QTL were revealed with 5 displaying an additive effect, 3 a partial-dominant effect, and 2 an overdominant effect. No QTL was found in 9024Hmp. In the II hybrid, 8 QTL were detected with 2 showing an additive effect, 3 a partial-to-complete dominant effect, and 3 an overdominant effect.
YD:
In the IJ hybrid, six QTL were identified with two exhibiting an additive effect, three a dominant effect, and one an overdominant effect. No QTL was found in SUMij and LH422Hmp. In the II hybrid, six QTL were detected with one showing an additive effect and five an overdominant effect. No QTL was found in Zhenshan97Hmp and SUMii data sets.
Digenic interaction:
Table 7 shows the digenic interactions detected in DIFFij data in the IJ hybrid. A total of 46 digenic interactions were found in DIFFij data. No significant interaction was found for yield. The variation explained by individual interaction ranges from 2.0 to 10.1%. The proportion of total variation explained by all digenic interaction was ∼30% in most traits. The highest value of total variation was observed for panicle length in the DIFFij data set (45.1%), which mainly reflected the dominance × dominance digenic interactions.
TABLE 7.
Digenic interactions in the DIFFij data set in the IJ hybrid
| Trait | Chria | Interval | Chrja | Interval | LOD | Aib | Ajb | AAijb | R2(AAij) (%)c | dd/aad |
|---|---|---|---|---|---|---|---|---|---|---|
| HD | 1 | RG233-XNPB302 | 2 | XNPB132-RG544 | 5.40 | 0.75*** | 6.20 | A | ||
| HD | 2 | CDO1091-CDO395 | 4 | RZ590-RZ262 | 3.25 | 0.51** | 2.90 | OD | ||
| HD | 2 | RG152-RG634 | 5 | RZ390-RZ556 | 5.50 | 0.59** | −0.61** | 4.16 | OD | |
| HD | 4 | CDO244-RG864 | 9 | RZ12-RG667 | 4.81 | 0.85*** | 7.96 | A | ||
| HD | 4 | XNPB271-RG449 | 5 | RZ70-RG480 | 2.87 | −0.53** | 3.08 | PD | ||
| HD | 5 | RZ495-RZ70 | 6 | RG653-RZ828 | 3.15 | −0.49** | 2.65 | D | ||
| HD | 6 | XNPB317-RZ247 | 11 | CDO127-RZ597 | 3.07 | −0.54** | 3.21 | OD | ||
| HD | 10 | RZ561-RZ400 | 12 | RG9-RZ670 | 2.89 | −0.63** | 4.47 | OD | ||
| 34.63 | ||||||||||
| PH | 1 | RG406-RG462 | 5 | RG697-CDO1083 | 3.20 | −1.58** | 6.24 | PD | ||
| PH | 1 | MK16-RG811 | 11 | RZ536-CDO534 | 4.41 | −1.83*** | 8.33 | OD | ||
| PH | 6 | XNPB317-RZ247 | 9 | XNPB103-XNPB317 | 4.16 | 1.68*** | 7.03 | OD | ||
| PH | 11 | RZ536-CDO534 | 11 | RZ638-CDO127 | 3.70 | −1.69*** | 7.09 | OD | ||
| 28.69 | ||||||||||
| TP | 3 | XNPB249-RZ16 | 11 | XNPB320-RG1022 | 4.06 | 0.23*** | 6.88 | PD | ||
| TP | 5 | RG480-RG697 | 6 | RG1028-RG162 | 3.75 | −0.24*** | 7.39 | A | ||
| TP | 6 | RG213-RZ144 | 12 | RG901-RZ76 | 3.77 | −0.22*** | 6.18 | PD | ||
| TP | 7 | CDO405-RG146 | 9 | RZ927-RZ12 | 3.89 | 0.25** | 8.10 | A | ||
| TP | 9 | RZ927-RZ12 | 12 | RG901-RZ76 | 3.27 | 0.24** | 7.27 | PD | ||
| 35.82 | ||||||||||
| PL | 2 | RG634-RZ825 | 9 | XNPB385-RZ422 | 4.53 | −0.58*** | 6.36 | PD | ||
| PL | 4 | RZ569-RG143 | 5 | RG480-RG697 | 3.04 | −0.51** | 5.02 | A | ||
| PL | 5 | RG360-RZ296 | 11 | CDO534-XNPB179 | 4.21 | 0.66*** | 8.18 | PD | ||
| PL | 5 | RZ70-RG480 | 12 | RG9-RZ670 | 3.29 | −0.56*** | 5.89 | PD | ||
| PL | 6 | RG653-RZ828 | 7 | XNPB20-RZ509 | 3.48 | 0.53*** | 5.25 | PD | ||
| PL | 7 | CDO497-RZ626 | 11 | CDO127-RZ597 | 5.04 | −0.28* | 0.48** | 4.28 | A | |
| PL | 9 | RZ404-RG358 | 10 | RZ892-RZ561 | 4.90 | −0.73*** | 10.14 | PD | ||
| 45.12 | ||||||||||
| FGPP | 1 | RZ776-RG375 | 12 | RZ76-RG9 | 3.99 | 5.76*** | 5.61 | A | ||
| FGPP | 2 | RZ987-XNPB132 | 3 | CDO87-RG558 | 3.83 | 5.76** | 5.59 | PD | ||
| FGPP | 2 | CDO395-RZ987 | 4 | RZ569-RG143 | 4.00 | −3.22* | −5.29** | 4.73 | A | |
| FGPP | 3 | XNPB249-RZ16 | 11 | RZ536-CDO534 | 3.17 | 5.51** | 5.13 | D | ||
| FGPP | 6 | XNPB317-RZ247 | 9 | XNPB103-XNPB317 | 3.93 | −3.00* | 5.33** | 4.80 | PD | |
| 25.86 | ||||||||||
| SS | 1 | XNPB370-RG541 | 9 | RZ404-RG358 | 3.41 | 3.86** | 8.80 | PD | ||
| SS | 1 | RG406-RG462 | 5 | RZ390-RZ556 | 3.62 | 1.86* | 2.04 | PD | ||
| SS | 1 | RG233-XNPB302 | 3 | XNPB232-XNPB249 | 7.30 | −3.32*** | 6.50 | A | ||
| SS | 1 | RG462-RG233 | 6 | RZ247-RZ450 | 2.86 | 2.13** | 2.68 | A | ||
| SS | 1 | CDO962-RG811 | 12 | RG121-RG98 | 4.25 | −2.25** | 2.98 | OD | ||
| SS | 5 | CDO1083-CDO1160 | 12 | RG901-RZ76 | 2.76 | 3.16** | 5.90 | A | ||
| 28.90 | ||||||||||
| GD | 1 | RG233-XNPB302 | 5 | RZ296-RG573 | 3.88 | −0.34*** | 5.92 | D | ||
| GD | 1 | XNPB302-RZ776 | 7 | XNPB20-RZ509 | 4.66 | −0.34*** | 5.78 | A | ||
| GD | 1 | RG233-XNPB302 | 9 | RG662-XNPB295 | 3.80 | −0.36*** | 6.43 | A | ||
| GD | 1 | RG375-CDO348 | 10 | RZ583-RZ811 | 4.30 | 0.17* | 0.30*** | 4.62 | A | |
| GD | 2 | RG555-TW500 | 8 | RZ562-RZ66 | 4.94 | 0.20* | −0.36*** | 6.50 | A | |
| GD | 5 | RZ495-RZ70 | 10 | RG257-RZ892 | 4.19 | −0.38*** | 7.30 | A | ||
| GD | 5 | RG480-RG697 | 6 | RG213-RZ144 | 3.14 | −0.32** | 5.19 | A | ||
| 41.74 | ||||||||||
| KGW | 2 | RZ123-RZ913 | 4 | RZ569-RG143 | 3.76 | 0.37*** | 5.92 | PD | ||
| KGW | 3 | RG1356-CDO87 | 4 | XNPB271-RG449 | 4.16 | −0.44*** | 8.50 | PD | ||
| KGW | 4 | RZ569-RG143 | 10 | RZ561-RZ400 | 2.75 | 0.33** | 4.71 | PD | ||
| KGW | 7 | CDO497-RZ626 | 9 | RZ927-RZ12 | 4.05 | 0.45*** | 9.14 | A | ||
| 28.27 |
*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Chri and Chrj represent the chromosomes that loci i and loci j are located on, respectively.
Ai and Aj are the main effects of loci i and loci j, and AAij is the epistatic effect between loci i and j.
Percentage of the total variation explained by AAij.
The epistasis dominance degree (EDD) of digenic interaction. Digenic interactions were classified according to their |dd/aa| ratio as additive (A) (|dd/aa| < 0.2), partial dominance (PD) (0.2 ≤ |dd/aa| < 0.8), dominance (D) (0.8 ≤ |dd/aa| < 1.2), and overdominance (OD) (|dd/aa| ≥ 1.2).
Table 8 shows the digenic interaction identified in DIFFii data in the II hybrid. In total, 81 digenic interactions were revealed. Each interaction generally showed modest R2 < 10% for all significant interactions except one interaction with 18.1%. However, in the IJ hybrid, the total variation explained by all digenic interactions was >40% for most of the traits. The highest value of total R2 was observed for SS in the DIFFii data set (52.7%).
TABLE 8.
Digenic interactions in the DIFFii data set in the II hybrid
| Trait | Chria | Interval | Chrja | Interval | LOD | Aib | Ajb | AAijb | R2(AAij) (%)c | dd/aad |
|---|---|---|---|---|---|---|---|---|---|---|
| HD | 1 | G359-RG532 | 1 | RM243-RG173 | 4.69 | 2.50*** | 8.83 | A | ||
| HD | 1 | C904-R2632 | 10 | RM258-RG561 | 4.91 | 2.11*** | 6.27 | OD | ||
| HD | 1 | R2201-RM212 | 10 | RM222-R2174 | 3.38 | −1.73** | 4.22 | A | ||
| HD | 2 | R1738-RM53 | 11 | C104-RM20a | 3.27 | 1.56** | 3.45 | OD | ||
| HD | 2 | RZ386-G1314a | 11 | C794-RG118 | 3.09 | −1.87** | 4.91 | D | ||
| HD | 4 | RZ467-C2807 | 6 | P-R2147 | 4.78 | −1.68*** | 3.97 | OD | ||
| HD | 4 | C107-RG620 | 8 | C1121-RG333 | 2.73 | −1.20** | 2.02 | OD | ||
| HD | 6 | C688-R1952a | 6 | R2147-G200 | 3.65 | 0.97* | −1.38** | 2.70 | A | |
| HD | 8 | C483-C347 | 10 | R2625-RM228 | 4.12 | −1.76*** | 4.35 | D | ||
| HD | 9 | RM215-R1952b | 12 | C996-G1128a | 3.06 | 1.47*** | 3.05 | A | ||
| 43.77 | ||||||||||
| PH | 1 | RM259-RM243 | 11 | G4001-RM254 | 6.86 | −1.93*** | 1.74** | 3.48 | PD | |
| PH | 1 | RM243-RG173 | 7 | R1440-RG678 | 5.88 | −1.89*** | 4.08 | D | ||
| PH | 2 | R1738-RM53 | 4 | RM255-G235 | 2.78 | −1.57** | 2.83 | A | ||
| PH | 3 | C316-C63 | 6 | C226-RZ398 | 5.44 | −2.01*** | 4.64 | OD | ||
| PH | 3 | RM200-RM227 | 4 | R78-C1016 | 4.13 | −1.91*** | 4.19 | A | ||
| PH | 4 | RZ467-C2807 | 9 | C472-R2638 | 3.85 | −1.70*** | 3.30 | PD | ||
| PH | 6 | C226-RZ398 | 12 | G1314b-R643 | 4.35 | 0.87* | 1.53** | 2.68 | PD | |
| PH | 7 | R1245-RM234 | 8 | R902-C1121 | 7.71 | −1.71*** | −1.87*** | 3.99 | PD | |
| PH | 8 | R1394-G2132 | 11 | CDO127-R2918 | 4.47 | 1.73*** | 3.45 | PD | ||
| PH | 8 | G1149-R2272 | 9 | C153B-C2 | 3.00 | 1.27** | 1.84 | OD | ||
| PH | 10 | C1633-C677 | 4 | C56-C820 | 7.26 | −2.55*** | 7.46 | D | ||
| 41.94 | ||||||||||
| TP | 1 | RM259-RM243 | 1 | C2340-C86 | 3.25 | 0.48** | 3.39 | OD | ||
| TP | 2 | RM53-RZ599 | 2 | RZ386-G1314a | 5.31 | 0.33* | −0.52** | 4.11 | OD | |
| TP | 5 | C734b-RZ649 | 11 | RG118-C1237 | 4.29 | 0.63*** | 5.92 | D | ||
| TP | 5 | RM26-C1447 | 10 | RM222-R2174 | 7.22 | −0.82*** | 10.00 | OD | ||
| TP | 6 | RM204-R1014 | 9 | C472-R2638 | 3.10 | 0.50** | 3.78 | OD | ||
| TP | 6 | RZ667-RG424 | 7 | RM70-R1245 | 2.70 | −0.44** | 2.88 | OD | ||
| TP | 7 | RG528-RG128 | 11 | CDO127-R2918 | 3.66 | 0.41** | 2.46 | OD | ||
| TP | 7 | RG528-RG128 | 12 | G1128a-R887 | 5.98 | −0.49** | 0.40** | 2.42 | OD | |
| TP | 8 | RZ66-G1149 | 10 | C1633-C677 | 4.95 | 0.62*** | 5.80 | OD | ||
| TP | 10 | C909A-C148 | 11 | RM20a-R3203 | 3.86 | −0.28* | −0.43** | 2.78 | OD | |
| TP | 10 | C405a-C223 | 12 | R496-C909B | 2.61 | 0.41** | 2.48 | OD | ||
| TP | 11 | G257-RM229 | 11 | R543a-RZ536 | 4.14 | −0.61*** | 5.62 | OD | ||
| 51.64 | ||||||||||
| PL | 1 | C161-R753 | 11 | C104-RM20a | 6.10 | 0.66*** | 6.15 | OD | ||
| PL | 1 | C567-C2340 | 12 | C732-R2672 | 2.52 | −0.42** | 2.48 | D | ||
| PL | 5 | RM26-C1447 | 6 | R1962-C764 | 4.88 | 0.64*** | 5.91 | D | ||
| PL | 6 | R2549-C962 | 10 | C153A-RM222 | 3.67 | −0.46*** | 3.05 | OD | ||
| PL | 7 | RZ471-RM70 | 4 | C56-C820 | 5.03 | 0.73*** | 7.48 | OD | ||
| PL | 11 | RM224-MP12 | 11 | RZ536-TEL3 | 3.92 | 0.61* | 1.13** | 18.09 | A | |
| 43.16 | ||||||||||
| FGPP | 1 | G359-RG532 | 5 | RM26-C1447 | 3.86 | 4.28*** | 4.02 | D | ||
| FGPP | 1 | G1128b-C904 | 2 | RM53-RZ599 | 3.84 | 4.75** | 4.94 | A | ||
| FGPP | 1 | G393-R2201 | 5 | C624-C246 | 3.77 | 4.80*** | 5.05 | A | ||
| FGPP | 3 | C1176-C316 | 6 | RZ667-RG424 | 4.04 | −4.80*** | 5.05 | OD | ||
| FGPP | 4 | C107-RG620 | 6 | RG424-R2549 | 4.24 | 4.48*** | 4.40 | OD | ||
| FGPP | 5 | C734b-RZ649 | 9 | R1952b-RZ404 | 2.95 | 4.46** | 4.36 | OD | ||
| FGPP | 6 | R1962-C764 | 6 | R2549-C962 | 3.60 | −4.92*** | 5.31 | A | ||
| FGPP | 6 | R2549-C962 | 10 | R2174-C909A | 5.00 | −4.83*** | 5.12 | D | ||
| FGPP | 7 | RZ471-RM70 | 4 | C56-C820 | 3.68 | 4.91** | 5.29 | OD | ||
| FGPP | 8 | RM223-L363A | 9 | RM242-RG570 | 2.65 | −3.75** | 3.08 | OD | ||
| FGPP | 9 | RM257-RM242 | 12 | C732-R2672 | 3.49 | −4.63*** | 4.69 | D | ||
| 51.31 | ||||||||||
| SS | 2 | RZ386-G1314a | 6 | RG653-G342 | 2.69 | −2.14* | 3.44 | OD | ||
| SS | 3 | C1087-RZ403 | 10 | C153A-RM222 | 6.21 | 3.34*** | 8.34 | OD | ||
| SS | 4 | G102-RM255 | 5 | RZ649-C624 | 3.58 | −2.61*** | 5.09 | OD | ||
| SS | 4 | C1016-C107 | 7 | C1023-R1440 | 3.80 | −2.44*** | 4.46 | OD | ||
| SS | 6 | RM204-R1014 | 4 | C56-C820 | 2.59 | 2.14** | 3.42 | OD | ||
| SS | 6 | RG424-R2549 | 4 | C820-C933 | 2.99 | 2.18** | 3.56 | OD | ||
| SS | 7 | RG128-C1023 | 12 | G1314b-R643 | 3.35 | −2.28** | 3.89 | D | ||
| SS | 8 | C347-RG978 | 9 | C1232-R265 | 3.39 | −2.19*** | 3.59 | OD | ||
| SS | 8 | RZ66-G1149 | 12 | C87-R496 | 4.23 | −2.52*** | 4.74 | OD | ||
| SS | 9 | RM201-C472 | 9 | RG667-RM215 | 4.32 | 3.48*** | 9.08 | OD | ||
| SS | 10 | R2174-C909A | 12 | R887-G1314b | 2.74 | −2.04** | 3.12 | OD | ||
| 52.73 | ||||||||||
| GD | 1 | G393-R2201 | 5 | C624-C246 | 4.66 | 0.17*** | 6.64 | OD | ||
| GD | 1 | G393-R2201 | 11 | C794-RG118 | 3.16 | 0.13** | 3.80 | PD | ||
| GD | 1 | RG236-C112 | 9 | C472-R2638 | 4.31 | 0.17*** | 6.26 | OD | ||
| GD | 2 | RZ599-R712 | 5 | RM26-C1447 | 3.60 | 0.15** | 4.86 | D | ||
| GD | 2 | R712-RZ324 | 3 | C1087-RZ403 | 4.24 | −0.15*** | 5.41 | OD | ||
| GD | 3 | RM232-G144 | 11 | C794-RG118 | 2.49 | −0.13** | 4.08 | PD | ||
| GD | 6 | P-R2147 | 9 | C153B-C2 | 3.32 | −0.14*** | 4.45 | PD | ||
| GD | 6 | R2549-C962 | 9 | C153B-C2 | 2.91 | −0.13** | 3.84 | OD | ||
| GD | 7 | RG528-RG128 | 10 | C405a-C223 | 3.00 | 0.07* | 0.10** | 2.31 | OD | |
| 41.65 | ||||||||||
| KGW | 2 | RZ324-RM29 | 5 | R830-R3166 | 4.86 | −0.58*** | 7.14 | A | ||
| KGW | 2 | RM213-RM208 | 5 | R830-R3166 | 2.90 | −0.46** | 4.64 | D | ||
| KGW | 5 | C1447-RM31 | 6 | C952-Waxy | 4.36 | −0.51*** | 5.55 | PD | ||
| KGW | 8 | R727-RM223 | 11 | R3203-CDO127 | 5.79 | 0.64*** | 8.71 | OD | ||
| 26.04 | ||||||||||
| YD | 1 | RG236-C112 | 11 | RZ536-TEL3 | 3.28 | 1.84** | 3.62 | OD | ||
| YD | 2 | RM48-RG520 | 6 | C474-R3139 | 3.14 | 1.95** | 4.07 | OD | ||
| YD | 3 | C63-RM232 | 4 | G102-RM255 | 8.83 | −3.47*** | 12.96 | OD | ||
| YD | 4 | C2807-RM241 | 6 | R1962-C764 | 3.38 | −2.01** | 4.35 | PD | ||
| YD | 4 | C2807-RM241 | 9 | C477-C1232 | 5.70 | −2.77*** | 8.24 | OD | ||
| YD | 7 | RG128-C1023 | 11 | R3203-CDO127 | 5.30 | 2.48*** | 6.61 | OD | ||
| YD | 9 | RG570-RG667 | 11 | R543a-RZ536 | 3.43 | −1.84** | 3.65 | OD | ||
| 43.50 |
*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Chri and Chrj represent the chromosomes that loci i and loci j are located on, respectively.
Ai and Aj are the main effects of loci i and loci j, and AAij is the epistatic effect between loci i and j.
Percentage of the total variation explained by AAij.
The epistasis dominance degree (EDD) of digenic interaction. Digenic interactions were classified according to their |dd/aa| ratio as additive (A) (|dd/aa| < 0.2), partial dominance (PD) (0.2 ≤ |dd/aa| < 0.8), dominance (D) (0.8 ≤ |dd/aa| < 1.2), and overdominance (OD) (|dd/aa| ≥ 1.2).
Table 9 summarizes the digenic interaction detected in RIL, SUM, Hmp, and DIFF data sets of IJ and II hybrids. Most of the detected interactions involved QTL without a significant main effect and each interaction showed a modest R2 < 10% for all traits. However, it should be noted that an interaction occurred between two significant M-QTL in Minghui63Hmp for 1000-grain weight, which explained 43.4% of phenotypic variation (data not shown here).
TABLE 9.
Summaries of digenic interaction in five data sets of the two hybrids
| Hmp (1)a
|
Hmp (2)a
|
DIFF
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RIL
|
SUM
|
Additive
|
Partial-to-complete dominance
|
Overdominance
|
Additive
|
Partial-to-complete dominance
|
Overdominance
|
Additive
|
Partial-to-complete dominance
|
Overdominance
|
||||||||||||
| Trait | No. | R2b | No. | R2b | No. | R2b | No. | R2b | No. | R2b | No. | R2b | No. | R2b | No. | R2b | No. | R2b | No. | R2b | No. | R2b |
| IJ hybrid | ||||||||||||||||||||||
| HD | 10 | 16.8 | 4 | 10.9 | 1 | 5.2 | 2 | 14.6 | — | — | — | — | — | — | 1 | 4.7 | 2 | 14.2 | 2 | 5.7 | 4 | 14.7 |
| PH | 6 | 29.3 | 2 | 13.9 | 1 | 6.0 | 3 | 23.5 | 2 | 8.5 | 5 | 18.8 | 2 | 13.8 | 1 | 11.0 | — | — | 1 | 6.2 | 3 | 22.5 |
| TP | 8 | 39.1 | 4 | 34.0 | 1 | 5.4 | 2 | 13.4 | 1 | 4.6 | — | — | 1 | 11.8 | 1 | 6.8 | 2 | 15.5 | 3 | 20.3 | — | — |
| PL | — | — | 6 | 24.9 | — | — | 1 | 3.5 | 1 | 10.0 | — | — | 2 | 17.3 | 1 | 5.9 | 2 | 9.3 | 5 | 35.8 | — | — |
| FGPP | — | — | 7 | 44.7 | 2 | 13.2 | 1 | 6.0 | 3 | 20.2 | 1 | 4.1 | 4 | 22.8 | 2 | 14.2 | 2 | 10.3 | 3 | 15.5 | — | — |
| SS | 3 | 28.3 | 6 | 36.4 | 2 | 10.9 | 3 | 14.4 | 4 | 22.3 | — | — | 6 | 32.1 | 2 | 8.7 | 3 | 15.1 | 2 | 10.8 | 1 | 3.0 |
| GD | 4 | 17.8 | 4 | 23.7 | 3 | 13.4 | 4 | 27.9 | 3 | 14.4 | — | — | 6 | 31.7 | 2 | 10.0 | 6 | 35.8 | 1 | 5.9 | — | — |
| KGW | 8 | 35.6 | 6 | 25.2 | — | — | 1 | 9.5 | — | — | 2 | 18.5 | 1 | 7.3 | 1 | 5.8 | 1 | 9.1 | 3 | 19.1 | — | — |
| YD | 6 | 26.5 | 4 | 32.3 | — | — | 2 | 19.3 | 2 | 15.5 | 2 | 16.6 | — | — | 1 | 9.0 | — | — | — | — | — | — |
| Mean | 3.22 | 16.4 | 3.44 | 21.8 | 1.11 | 6.0 | 2.11 | 14.7 | 1.78 | 10.6 | 1.11 | 6.4 | 2.44 | 15.2 | 1.33 | 8.4 | 2.00 | 12.2 | 2.22 | 13.3 | 0.89 | 4.5 |
| II hybrid | ||||||||||||||||||||||
| HD | 11 | 56 | 14 | 59.4 | 1 | 5.5 | 2 | 9.1 | 1 | 7.6 | — | — | — | — | — | — | 4 | 18.8 | 2 | 9.3 | 4 | 15.7 |
| PH | 7 | 39 | 9 | 54.8 | 1 | 6.3 | 9 | 43.6 | — | — | — | — | — | — | — | — | 2 | 7 | 7 | 28.4 | 2 | 6.5 |
| TP | 8 | 42.8 | 9 | 53.5 | 3 | 14.7 | 3 | 18.6 | 5 | 19.8 | — | — | 2 | 19.1 | 1 | 3.9 | — | — | 1 | 5.9 | 11 | 45.7 |
| PL | 7 | 54.4 | 3 | 23.3 | 1 | 3.5 | 7 | 41.6 | — | — | — | — | 1 | 9 | — | — | 1 | 18.1 | 2 | 8.4 | 3 | 16.7 |
| FGPP | 8 | 50.3 | — | — | 2 | 8.2 | 5 | 31.5 | 3 | 14.7 | — | — | 1 | 5.9 | 1 | 5.3 | 3 | 15.3 | 2 | 13.8 | 5 | 22.2 |
| SS | — | — | 3 | 25.8 | 2 | 10.6 | 3 | 21.5 | 2 | 16.8 | 1 | 6.1 | 3 | 15.4 | 3 | 21.2 | — | — | 1 | 3.9 | 11 | 48.8 |
| GD | 12 | 53.9 | 10 | 56.9 | 1 | 4.9 | 12 | 46.1 | 3 | 11.6 | — | — | 1 | 9.2 | — | — | — | — | 4 | 17.2 | 5 | 24.5 |
| KGW | 9 | 47.5 | 10 | 49.2 | 1 | 5.1 | 5 | 21.4 | 4 | 20.7 | — | — | 3 | 47.5 | 1 | 3.7 | 1 | 7.1 | 2 | 14.5 | 6 | 39.2 |
| YD | 11 | 87 | 9 | 41.1 | 1 | 4 | 3 | 15.9 | 4 | 18.5 | — | — | 3 | 17.7 | 2 | 13.2 | — | — | 1 | 4.4 | 1 | 8.7 |
| Mean | 8.11 | 47.9 | 7.44 | 40.4 | 1.44 | 6.98 | 5.44 | 27.7 | 2.44 | 12.2 | 0.11 | 0.68 | 1.56 | 13.8 | 0.89 | 5.3 | 1.22 | 7.37 | 2.44 | 11.8 | 5.33 | 25.3 |
In the IJ hybrid, Hmp (1) and Hmp (2) represent 9024Hmp and LH422Hmp, respectively; while in the II hybrid, Hmp (1) and Hmp (2) represent Zhenshan97Hmp and Minghui63Hmp, respectively.
The total variation explained by each trait (%).
In the IJ hybrid, the number of digenic interactions detected for each trait varies from none to 10 in the RILij population with an average of 3.22, and the variance explained (R2) by each pair was up to 39.1% with an average of 16.4%. The number of digenic interactions detected in the SUMij data set varies from two to 7 with an average 3.44, and the R2 of each pair varies from 10.9 to 44.7% with an average of 21.8%. For digenic interaction of dominance × dominance, on average, 1.11, 1.11, and 2.00 QTL pairs with an additive effect were detected in 9024Hmp, LH422Hmp, and DIFFij and had a contribution rate of 6.0, 6.4, and 12.2%, respectively; 2.11, 2.44, and 2.22 QTL pairs with partial-to-complete dominance were detected in 9024Hmp, LH422Hmp, and DIFFij and had a contribution rate of 14.7, 15.2, and 13.3%, respectively; and 1.67, 1.33, and 0.89 QTL pairs with overdominance were detected in 9024Hmp, LH422Hmp, and DIFFij and had a contribution rate of 10.6, 8.4, and 4.5%, respectively.
For the II hybrid, the number of digenic interactions identified for each trait varies from none to 12 in the RILii population with an average of 8.11 and had a contribution rate (R2) up to 87.0%, with an average of 47.9%. The number of digenic interactions detected in the SUMii data set varies from none to 14 with an average of 7.44, and each pair had an R2 up to 59.4% with an average of 40.4%. For digenic interaction of dominance × dominance, on average, 1.44, 0.11, and 1.22 QTL pairs with additive effect were detected in Zhenshan97Hmp, Minghui63Hmp, and DIFFii and had a contribution rate of 7.0, 0.7, and 7.4%, respectively; 5.44, 1.56, and 2.44 QTL pairs with partial-to-complete dominance were detected in Zhensha97Hmp, Minghui63Hmp, and DIFFii and had a contribution rate of 27.7, 13.8, and 11.8%, respectively; and 2.44, 0.89, and 5.33 QTL pairs with overdominance were detected in Zhenshan97Hmp, Minghui63Hmp, and DIFFii and had a contribution rate of 12.2, 5.3, and 25.3%, respectively.
Relationship between trait performance and genomewide or chromosomewide marker heterozygosity:
The correlation coefficients (Table 10) between level of genomewide heterozygosity and performance per se of the two backcross populations were not significant for most of the traits in both IJ and II hybrids (except plant height in 9024BC and 1000-grain weight in Minghui63BC). The analysis of the relationship between level of heterozygosity and level of heterosis (as evaluated in Hmp and DIFF) showed that correlation coefficients, for several traits, were slightly higher than those previously shown, but still not significant for most traits. The significant correlation coefficients were found for plant height, heading date, and 1000-grain weight in the IJ hybrid and for tillers per plant in the II hybrid.
TABLE 10.
Correlation coefficients between genomewide molecular marker heterozygosity and phenotypic values
|
IJ hybrid
|
II hybrid
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Performance per se
|
Heterosis
|
Performance per se
|
Heterosis
|
|||||||||
| Trait | 9024BC | LH422BC | 9024Hmp | LH422Hmp | DIFF (1)a | DIFF (2)a | Minghui63BC | Zhenshan97BC | Minghui63Hmp | Zhenshan97Hmp | DIFF (3)a | DIFF (4)a |
| HD | 0.010 | 0.008 | 0.201** | 0.044 | 0.034 | −0.011 | 0.080 | 0.008 | 0.079 | 0.033 | −0.101 | 0.098 |
| PH | 0.209** | 0.062 | 0.097 | 0.243** | 0.228** | −0.289** | 0.003 | −0.002 | −0.012 | 0.017 | −0.020 | 0.000 |
| TP | −0.079 | 0.002 | 0.006 | −0.099 | −0.022 | 0.002 | 0.093 | −0.012 | 0.150* | −0.062 | −0.064 | 0.072 |
| PL | 0.107 | −0.044 | 0.054 | 0.037 | 0.104 | −0.085 | 0.001 | 0.029 | 0.033 | 0.005 | −0.021 | 0.016 |
| FGPP | 0.046 | −0.063 | −0.023 | 0.035 | −0.092 | 0.071 | −0.044 | −0.014 | −0.005 | −0.077 | 0.066 | −0.064 |
| SS | 0.063 | −0.034 | 0.104 | −0.071 | −0.010 | 0.015 | −0.039 | 0.049 | −0.110 | 0.099 | 0.019 | −0.006 |
| GD | −0.028 | −0.020 | −0.109 | 0.072 | 0.006 | −0.014 | −0.048 | −0.031 | −0.014 | −0.094 | 0.088 | −0.083 |
| KGW | 0.115 | 0.132 | 0.193** | 0.069 | 0.232** | −0.177* | 0.166* | −0.110 | 0.046 | 0.029 | −0.090 | 0.071 |
| YD | 0.091 | 0.025 | 0.119 | 0.000 | 0.090 | −0.101 | 0.099 | −0.062 | 0.090 | −0.065 | −0.029 | 0.040 |
*P ≤ 0.05, **P ≤ 0.01.
DIFF (1) and DIFF (2) represent that, in the IJ hybrid, when the DIFF data set was used as a dependent variable, genome heterozygosity of the 9024BC and LH422BC hybrids was the independent variable, respectively. While in the IJ hybrid, DIFF (3) and DIFF (4) represent that when the DIFF data set was used as a dependent variable, genome heterozygosity of the Zhenshan97BC and Minghui63BC hybrids was the independent variable, respectively.
In this study, the Hmp value was regressed against heterozygosity on individual chromosomes using multiple linear regression (Table 11). The hybrid performance was also poorly associated with marker heterozygosity in most chromosomes. There were 8, 6, 8, and 5 significant regressions between trait value and marker heterozygosity in individual chromosomes resolved in 9024Hmp, LH422Hmp, Zhenshan97Hmp, and Minghui63Hmp, respectively. Nineteen of these 27 (70.3%) significant regressions were associated with one or two M-QTL and/or digenic interaction. In the IJ hybrid, the F-test value was significant for panicle length and grain density in 9024Hmp and for plant height and heading date in LH422Hmp. While in the II hybrid, the F-test value was significant for plant height in Zhenshan97Hmp and for yield in Minghui63Hmp. The coefficients (r2) for most traits were <0.10 in both IJ and II hybrids.
TABLE 11.
Significant regression coefficients of midparent values of backcross populations on individual chromosome marker heterozygosity for the indicated traits
| Population | Trait | Chr1 | Chr2 | Chr3 | Chr4 | Chr5 | Chr6 | Chr7 | Chr8 | Chr9 | Chr10 | Chr11 | Chr12 | Fa | r2b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IJ hybrid | |||||||||||||||
| 9024Hmp | HD | — | — | 0.044* | — | — | — | — | — | — | — | — | — | 1.720 | 0.10 |
| 9024Hmp | PH | — | — | — | — | — | 0.032* | — | — | — | — | — | — | 1.311 | 0.08 |
| 9024Hmp | TP | 0.004* | — | — | — | — | — | — | — | — | — | — | — | 1.350 | 0.08 |
| 9024Hmp | PL | — | — | — | — | — | — | — | — | — | −0.005* | — | — | 1.821* | 0.11 |
| 9024Hmp | FGPP | — | — | — | — | — | — | — | — | — | — | — | — | 1.333 | 0.08 |
| 9024Hmp | SS | — | — | — | — | — | — | — | — | — | — | — | — | 0.583 | 0.04 |
| 9024Hmp | GD | — | — | −0.004* | — | — | −0.004* | — | — | — | — | — | — | 1.821* | 0.11 |
| 9024Hmp | KGW | — | — | — | 0.007* | — | — | — | — | — | — | — | — | 1.334 | 0.09 |
| 9024Hmp | YD | — | — | — | 3.039* | — | — | — | — | — | — | — | — | 1.649 | 0.11 |
| LH422Hmp | HD | — | — | — | — | — | — | — | 0.023*** | — | — | −0.012* | — | 4.000*** | 0.21 |
| LH422Hmp | PH | — | — | — | — | — | — | 0.045** | — | — | — | — | — | 2.117** | 0.12 |
| LH422Hmp | TP | — | — | — | — | — | — | — | — | — | — | — | — | 1.115 | 0.07 |
| LH422Hmp | PL | −0.008* | — | — | — | — | 0.007* | — | — | — | — | — | — | 1.733 | 0.10 |
| LH422Hmp | FGPP | — | — | — | — | — | — | — | — | — | — | — | — | 0.904 | 0.06 |
| LH422Hmp | SS | — | — | — | — | — | — | — | — | — | — | — | — | 1.292 | 0.08 |
| LH422Hmp | GD | — | — | — | — | — | — | — | — | −0.005* | — | — | — | 1.256 | 0.08 |
| LH422Hmp | KGW | — | — | — | — | — | — | — | — | — | — | — | — | 0.990 | 0.07 |
| LH422Hmp | YD | — | — | — | — | — | — | — | — | — | — | — | — | 0.907 | 0.06 |
| II hybrid | |||||||||||||||
| Zhenshan97Hmp | HD | — | — | — | — | — | — | −0.055* | — | — | — | — | — | 0.989 | 0.05 |
| Zhenshan97Hmp | PH | — | — | 0.211** | — | — | — | — | — | — | — | — | — | 2.292** | 0.12 |
| Zhenshan97Hmp | TP | — | — | — | — | 0.017* | — | — | — | — | — | — | — | 1.135 | 0.06 |
| Zhenshan97Hmp | PL | — | — | — | — | — | — | — | — | — | — | — | — | 0.581 | 0.03 |
| Zhenshan97Hmp | FGPP | — | — | — | — | — | — | — | — | — | — | — | — | 0.936 | 0.05 |
| Zhenshan97Hmp | SS | — | — | — | — | — | — | — | 0.107* | — | — | — | — | 1.063 | 0.06 |
| Zhenshan97Hmp | GD | 0.006* | — | — | — | — | — | — | — | — | −0.005** | — | — | 1.445 | 0.08 |
| Zhenshan97Hmp | KGW | — | — | — | — | — | — | — | — | −0.017* | 0.016** | — | — | 1.565 | 0.08 |
| Zhenshan97Hmp | YD | — | — | — | — | — | — | — | — | — | — | — | — | 0.631 | 0.04 |
| Minghui63Hmp | HD | — | — | 0.062* | — | — | — | — | — | — | — | — | — | 1.372 | 0.07 |
| Minghui63Hmp | PH | — | — | — | — | — | — | — | — | — | — | — | — | 0.619 | 0.03 |
| Minghui63Hmp | TP | — | — | — | — | — | — | — | — | — | — | — | 0.017* | 1.294 | 0.07 |
| Minghui63Hmp | PL | — | — | — | — | — | — | — | — | −0.019* | — | — | — | 0.865 | 0.05 |
| Minghui63Hmp | FGPP | — | — | — | — | — | — | — | — | — | — | — | — | 0.428 | 0.02 |
| Minghui63Hmp | SS | — | — | — | — | — | — | — | — | — | — | — | — | 0.961 | 0.05 |
| Minghui63Hmp | GD | — | — | — | — | — | — | — | — | — | — | — | — | 0.665 | 0.04 |
| Minghui63Hmp | KGW | — | — | — | — | — | — | — | — | — | — | — | — | 0.676 | 0.04 |
| Minghui63Hmp | YD | — | — | — | — | — | — | — | — | — | — | 0.095** | 0.080** | 1.954* | 0.10 |
*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Italics indicate the presence of a QTL on a particular chromosome.
F-test value.
Determination coefficients.
DISCUSSION
Choice of the experimental design and statistical methods:
NCIII and TTC designs are most suitable for studies of heterosis in the presence of epistasis because they provide estimates of augmented dominance effects (Kusterer et al. 2007a,b). Meanwhile, compared with the F2 or the F3 population, RILs as parents for producing testcross progenies offer few advantages. First, the effects of linkage are reduced because linkage disequilibrium between tightly linked loci is almost half of that in the F2 population. Second, use of homozygous parents (RIL) maximizes the genetic variance among testcross progenies and leads to an increased power in F-tests and reduced standard errors of variance component and dominance effect estimates since RILs are homozygous at almost all of the genetic loci while F2 plants have 50% heterozygous loci. Third, RILs are immoral and testcross progeny can be repeatedly generated and tested as needed.
Up to now, several studies have been conducted to try to understand the genetic basis of heterosis in rice (Xiao et al. 1995; Li et al. 2001; Luo et al. 2001; Hua et al. 2002, 2003). However, the causes underlying this important phenomenon have remained unclear and none of these studies quantified the gene action of QTL. In this study, with two derived data sets (SUM and DIFF) and the software developed by us, we resolved the dominance degree for all of the M-QTL and digenic interactions. The statistical method employed in this study is much more precise and informative to understand the causes of heterosis in rice since it classifies underlying QTL into A, PD, D, and OD on the basis of degree of dominance.
It should be noted that the A, PD, D, and OD referred to in this study are different from the additive effect, dominant effect, and overdominant effect in a traditional dominant–additive model. In fact, as well as in hybrid F1, since each locus is in heterozygosis, only gene action of dominance, dominance × dominance, dominance × dominance × dominance, etc., existed in Hmp and DIFF. Therefore, in this study, A, PD, D, and OD were treated only as a scale for quantifying the degree of dominance (d) or dominance × dominance (dd) effect.
Heterosis for the traits studied:
In the two hybrids investigated here, grain yield showed the strongest heterosis among the nine traits studied (25.58% in the IJ hybrid and 83.09% in the II hybrid), consistent with the findings of previous studies conducted on rice (Li et al. 2001; Luo et al. 2001) as well as other cereal crops (Tollenaar et al. 2004; Hoecker et al. 2006). Heterosis for the other traits was <20% in the IJ hybrid and <30% in the II hybrid. Negative heterosis for filled grains per panicle and grain density was observed in the IJ hybrid. These results confirm that heterosis of yield components was much less than grain yield itself (Li et al. 2001).
For the IJ hybrid, the Hmp of some backcross lines was stronger than that of F1, while some other backcross lines expressed an Hmp in the opposite direction. This result is in harmony with the study conducted by Mei et al. (2005) in which an indica/japonica hybrid was also used. It can be concluded that heterosis was generally related to the average level of heterozygosity in a hybrid population but poorly correlated with heterozygosity at the individual level (Zhang et al. 1995; Yu et al. 1997). This conclusion also can be confirmed by the fact that the correlation between marker heterozygosity and trait expression is negligible.
For the II hybrid, the heterosis in BC populations was much lower than that in F1. This may be due to the fact that the two intraspecific parents are more genetically similar than the two interspecific parents of IJ hybrids. The reduction in the proportion of heterozygous loci in the BCF1 population probably caused the reduced average level of heterosis in the BCF1 compared to the hybrid between two parents.
NCIII and TTC analysis:
For the traits showing highly significant epistasis, VA and VD estimates are to some extent biased (Kearsey and Pooni 1996) and so are the average degree of dominance estimates. In the IJ hybrid, highly significant [aa], [ad], and [dd] epistasis was observed for all the traits studied. In the II hybrid, the average degree of dominance for most traits was <1.00, except for plant height (1.18) and grain yield (1.20), suggesting an important contribution of overdominance to the heterosis of these two traits. For epistasis conducted by TTC analysis, [aa] was highly significant (P ≤ 0.005) for all traits, and [ad] and [dd] for most of traits, except for yield and grain density (significant at P ≤ 0.01), panicle length (significant at P ≤ 0.05), and tillers per plant (not significant). Therefore, epistasis appeared to be of more importance than intralocus interaction in affecting heterosis in these two elite hybrids. A similar conclusion was drawn in Arabidopsis by Kusterer et al. (2007a) in which a TTC family derived from the Arabidopsis C24 × Col-0 was analyzed, and it was found that epistasis across environments was more important for most traits. However, in the TTC design with recombinant inbred lines of the maize B73 × H99 (Frascaroli et al. 2007), the epistasis was found not significant for most traits.
Genetic basis of heterosis in two highly heterotic hybrids of rice:
Our analyses allowed the identification of several QTL for each of the traits investigated. Most individual QTL explained modest variation (<10%), and only four QTL in the IJ hybrid and five QTL in the II hybrid contributed >20% variation individually (Tables 5 and 6), confirming that the heterosis is a polygenic phenomenon (Hallauer and Miranda 1981; Kusterer et al. 2007a).
The proportion of QTL with an additive or a dominant effect is different between the two hybrids. Among the 74 main-effect QTL detected in the IJ hybrid, 24 (32%) showed a gene action of partial-to-complete dominance, 20 (26%) showed overdominance, and 32 (42%) showed an additive effect; while among the 41 main-effect QTL identified in the II hybrid, 12 (29%) exhibited partial-to-complete dominance, 16 (39%) showed overdominance, and 13 (32%) showed an additive effect. These results indicate that dominance and overdominance played an important role in conditioning the heterosis in these two hybrids. Also, the results from the dominance degree (|d/a|) of main-effect QTL estimated by QTLIII with regression analysis and by WinQTLcart (Zeng 1994) show that, although the dominance degrees were not exactly consistent with each other by the three approaches (ANOVA, regression analysis, WinQTLcart), the proportions of QTL detected with dominance and with overdominance were >25% each.
The importance of dominance and overdominance conditioning the heterosis of these two hybrids seems different. In the IJ hybrid, the proportion of QTL showing a gene action of overdominance is less than that with partial-to-complete dominance. This result was also found in the study conducted by Xiao et al. (1995) using the same materials, but a different analysis method. However, in the II hybrid, the proportion of QTL exhibiting a gene action of overdominance is more than the proportion of those having a gene action of partial-to-complete dominance. This result is in harmony with other studies, especially the work conducted on the F2:3 families derived from the cross between Zhenshan97 and Minghui63 by Yu et al. (1997). However, although a relatively higher portion of QTL demonstrated overdominance in the II hybrid, QTL exhibiting high levels of overdominant effects are not necessarily indicative of true overdominance, but rather can be the result of dominant alleles linked in repulsion (pseudo-overdominance).
Compared to M-QTL detected in these two hybrids, only two QTL for heading date were found in a similar genomic region bordered by the same molecular markers. This may be due to the fact that very few markers were common across these two linkage maps. On chromosome 1, one QTL was detected between RG811 and RG173 in the IJ hybrid, showing an additive effect. One QTL between RM243 and RG173 was detected in the II hybrid, displaying a partial-dominant effect. On chromosome 8, one QTL between RG333 and RZ562 in the IJ hybrid and one between C1121 and RG333 in the II hybrid exhibited an additive effect, thus suggesting that, even in the same or a similar genomic region bordered by the same molecular markers in different hybrids, the gene action of QTL could be different due to interaction of different alleles at the QTL. It should be noted that, for the two hybrids that were planted in different environments, the type of gene action may be influenced by environmental effect.
Various levels of negative dominance were observed at some QTL for each trait, indicating that heterozygosity was not necessarily always favorable for the expression of the trait even in highly heterotic hybrids. For both hybrids studied here, dominant effects of the detected QTL were always bidirectional, resulting in the cancellation of positive and negative dominant effects contributed by different QTL controlling the trait, which explains the poor relationship observed between marker heterozygosity and trait expression. A good consistency was also found in other studies of rice (Yu et al. 1997; Mei et al. 2005), but in contrast with the study (Frascaroli et al. 2007) in maize.
There were a large number of digenic interactions found to have effects on the traits of the two hybrids studied here. Two pronounced features were notably found for the epistasis in this study. First, although individual interaction had a modest R2 (phenotypic variation), <10% in most cases (data not shown) for each trait of the two hybrids, the total variation explained by all the significant digenic interactions for the trait was much greater than that by all the M-QTL affecting the same trait for most traits.
Similar to a large number of empirical studies in other selfing and outcrossing plant species (Allard 1988; Li et al. 2001; Mei et al. 2005), most epistasis occurred between complementary loci with no detectable main effects. In many fewer cases, epistasis occurred between a M-QTL and a complementary locus and in only seven cases in the IJ hybrid and two in the II hybrid between M-QTL. By using the same population of IJ hybrids reported here, Xiao et al. (1995) was unable to detect epistasis due to the unavailability of appropriate mapping methodology (Li et al. 2001).
It should be noted that the two digenic interactions in the II hybrid occurred between M-QTL accounting for a large variation for 1000-grain weight detected in Minghui63Hmp and for panicle length detected in Zhenshan97BC, explaining 43.4 and 23.8% of the variation, respectively (data not shown). When a M-QTL is involved in the epistatic interaction, the effect of the single-locus QTL is mostly dependent on the genotypes of the other locus and can sometimes be negated by the genotypes of a second locus. Thus an attempt to utilize the QTL in the breeding programs needs to consider such epistatic effects, especially the interaction occurring between two significant M-QTL and having a high phenotypic variation.
Another feature of digenic interaction in this study is that both partial-to-complete dominance and overdominance played an important role in conditioning heterosis. Shown in Table 9 is the relative importance of additive and nonadditive gene action of digenic interaction summarized by comparing the genetic effects detected in the SUM and DIFF data sets by QTL with ANOVA analysis.
For the additive × additive digenic interactions, there were an average of 3.22 and 3.44 pairs detected in the RILij and SUMij data sets for each trait in the IJ hybrid, contributing 16.4 and 21.8% phenotypic variation, respectively; while in the II hybrid, an average of 8.11 and 7.44 pairs were detected in the RILii and SUMii data sets for each trait, explaining 47.9 and 40.4% of the phenotypic variation, respectively.
There were a total of 135 and 188 dominance × dominance digenic interactions detected in Hmp and DIFF in the IJ and II hybrids, respectively. The proportion of digenic interactions displaying partial-to-complete dominance was a little more than that showing overdominance in both hybrids. There were 62 (45.2%) and 85 (45.2%) digenic interactions that behaved like partial-to-complete dominance, 36 (26.7%) and 78 (41.5%) digenic interactions that exhibited overdominance, and 37 (28.1%) and 25 (13.3%) digenic interactions that displayed an additive effect, in IJ and II hybrids, respectively.
The poor relationship between total genomewide molecular marker heterozygosity and phenotypic trait performance was observed for almost all the traits in this study (Table 10). This result is different from the study of maize performed by Frascaroli et al. (2007) in which they found that there was a high relationship between marker heterozygosity level and performance per se and heterosis (as evaluated in Hmp and DIFF) for most traits. To further investigate the relationship between observed heterosis and heterozygosity, Hmp value was regressed against heterozygosity on individual chromosomes, using multiple linear regression. As shown in Table 11, the hybrid performance was also poorly associated with marker heterozygosity in most chromosomes, although it was relatively more significant than that with whole-genome heterozygosity. Nineteen of the 27 (70.4%) significant regressions by individual chromosomes were associated with one or two M-QTL and/or digenic interaction, indicating that marker heterozygosity in individual chromosomes in QTL regions was important for phenotypic variation. This finding is consistent with Syed and Chen's (2005) result of the relationship between heterozygosity and heterosis in Arabidopsis. Therefore, the hybrid vigor is poorly related to heterozygosity of the whole genome and on individual chromosomes in rice, which further confirms that the genetic basis or mechanism of heterosis of rice is different from that of maize.
Our results indicate that heterosis in rice is very complex, reflected by the large number of loci involved, their wide genomic distribution, and complex epistatic relationships, and that the nonallelic interactions (epistasis) play a relatively more important role than allelic interactions (M-QTL) in conditioning the heterosis of these two highly heterotic hybrids, implicating that marker-assisted selection in heterosis breeding to significantly enhance the heterosis of desirable traits may be very challenging.
So far almost all of the documented studies on revealing the genetic basis of heterosis are limited to classical quantitative genetics and QTL mapping using molecular markers. The advancements in functional genomics have created a novel avenue to study the genetic basis of heterosis at the gene-expression level. DNA microarrays can quantify expression of tens of thousands of genes on a single DNA chip (Schena et al. 1998). The timing, level, and relationship of the transcription of two different alleles of the same gene in the hybrids can be compared with that of their corresponding parental lines by using microarrays (Stupar and Springer 2006; Swanson-Wagner et al. 2006). Functional genomics approaches to elucidating the genetic basis of heterosis would turn the study of this very important and still controversial issue into a new chapter in its history. Evidence from functional expression studies of genes underlying heterosis would elevate our understanding of the genetic basis of heterosis to a new level.
Acknowledgments
We gratefully acknowledge the expert technical assistance of Qifa Zhang on trial design and analysis and the skillful assistance of Yingguo Zhu in field trials. We thank Jinhua Xiao and Yunchun Song for valuable suggestions for improving the manuscript. This work was financially supported by the 973 Program (no. 2006CB101707), the 863 Program (no. 2003AA207160), the National Natural Science Foundation of China (no. 30270760), and the Key Grant Project of the Chinese Ministry of Education (no. 307018).
References
- Allard, R. W., 1988. Genetic changes associated with the evolution of adaptedness in cultivated plants and their wild progenitors. J. Hered. 79 225–238. [DOI] [PubMed] [Google Scholar]
- Bruce, A. B., 1910. The Mendelian theory of heredity and the augmentation of vigor. Science 32 627–628. [DOI] [PubMed] [Google Scholar]
- Causse, M., T. M. Fultony, G. Cho, S. N. Ahn, K. Wu et al., 1994. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138 1251–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham, C. C., and Z. B. Zeng, 1996. Design III with marker loci. Genetics 143 1437–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock, R. E., and H. F. Robinson, 1952. Estimation of average dominance of genes, pp. 494–516 in Heterosis, edited by J. W. Growen. Iowa State College Press, Ames, IA.
- Crow, J. F., 2000. The rise and fall of overdominance. Plant Breed. Rev. 17 225–257. [Google Scholar]
- Davenport, C. B., 1908. Degeneration, albinism and inbreeding. Science 28 454–455. [DOI] [PubMed] [Google Scholar]
- East, E. M., 1908. Inbreeding in corn, pp. 419–428 in Reports of the Connecticut Agricultural Experiment Station for Years 1907–1908.
- Frascaroli, E., M. A. Cane, P. Landi, G. G. Pea, M. Villa et al., 2007. Classical genetic and quantitative trait loci analyses of heterosis in a maize hybrid between two elite inbred lines. Genetics 176 625–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh, S., D. Gonzàles-de-Leòn, M. M. Khairallah, C. Jiang, D. Bergvinson et al., 1998. QTL mapping in tropical maize: III. Genomic regions for resistance to Diatraea spp. and associated traits in two RIL populations. Crop Sci. 38 1062–1072. [Google Scholar]
- Hallauer, A. R., and B. Miranda, 1981. Quantitative Genetics in Maize Breeding. Iowa State University Press, Ames, IA.
- Hoecker, N., B. Keller, H. P. Piepho and F. Hochholdinger, 2006. Manifestation of heterosis during early maize (Zea mays L.) root development. Theor. Appl. Genet. 112 421–429. [DOI] [PubMed] [Google Scholar]
- Hu, Z. L., X. F. Zhang, C. Xie, G. R. McDaniel and D. L. Kuhlers, 1995. A correlation method for detecting and estimating linkage between a marker locus and a quantitative trait locus using inbred lines. Theor. Appl. Genet. 90 1074–1078. [DOI] [PubMed] [Google Scholar]
- Hu, Z. L., Q. X. Sun, X. F. Zhang, Y. C. Song and Q. F. Zhang, 2002. A correlation method for detecting epistatic QTLs using inbred lines. Chin. Sci. Bull. 1 122–126. [Google Scholar]
- Hua, J. P., Y. Z. Xing, C. G. Xu, X. L. Sun, S. B. Yu et al., 2002. Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics 162 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J., Y. Xing, W. Wu, C. Xu, X. Sun et al., 2003. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 100 2574–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D. F., 1917. Dominance of linked factors as a means of accounting for heterosis. Proc. Natl. Acad. Sci. USA 3 310–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey, M. J., and J. L. Jinks, 1968. A general method of detecting additive, dominance and epistatic variation for metrical traits. I. Theory. Heredity 23 403–409. [DOI] [PubMed] [Google Scholar]
- Kearsey, M. J., and H. S. Pooni, 1996. The Genetical Analysis of Quantitative Traits. Chapman & Hall, London.
- Kearsey, M. J., H. S. Pooni and N. H. Syed, 2003. Genetics of quantitative traits in Arabidopsis thaliana. Heredity 91 456–464. [DOI] [PubMed] [Google Scholar]
- Kusterer, B., J. Muminovic, H. F. Utz, H. P. Piepho, S. Barth et al., 2007. a Analysis of a triple testcross design with recombinant inbred lines reveals a significant role of epistasis in heterosis for biomass-related traits in Arabidopsis. Genetics 175 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusterer, B., H. P. Piepho, H. F. Utz, C. C. Schön, J. Muminovic et al., 2007. b Heterosis for biomass-related traits in Arabidopsis investigated by quantitative trait analysis of the triple-testcross design with recombinant inbred lines. Genetics 177 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. K., S. R. M. Pinson, A. H. Paterson, W. D. Park and J. W. Stansel, 1997. Epistasis for three grain yield components in rice (Oryza sativa L.). Genetics 145 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. K., L. J. Luo, H. W. Mei, D. L. Wang, Q. Y. Shu et al., 2001. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics 158 1737–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman, Z. B., and D. Zamir, 2007. Heterosis: revisiting the magic. Trends Genet. 23 60–66. [DOI] [PubMed] [Google Scholar]
- Lu, Q. S., Y. Sun and Z. T. Hua, 2002. Heterosis of Cereal Crops, p. 240. Science and Technology of China Agricultural Press, Beijing (in Chinese).
- Luo, L. J., Z. K. Li, H. W. Mei, Q. Y. Shu, R. Tabien et al., 2001. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. II. Grain yield components. Genetics 158 1755–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, H. W., Z. K. Li, Q. Y. Shu, L. B. Guo, Y. P. Wang et al., 2005. Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two backcross populations. Theor. Appl. Genet. 110 649–659. [DOI] [PubMed] [Google Scholar]
- Melchinger, A. E., H. P. Piepho, H. Friedrich Utz, J. Muminović, T. Wegenast et al., 2007. a Genetic basis of heterosis for growth-related traits in Arabidopsis investigated by testcross progenies of near-isogenic lines reveals a significant role of epistasis. Genetics 177 1827–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchinger, A. E., H. F. Utz, H. P. Piepho, Z. B. Zeng and C. C. Schön, 2007. b The role of epistasis in the manifestation of heterosis: a systems-oriented approach. Genetics 177 1815–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute, 1996. SAS Users Guide: Statistic. SAS Institute, Cary, NC.
- Schena, M., R. A. Heller, T. P. Theriault, K. Konrad, E. Lachenmeier et al., 1998. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol. 16 301–306. [DOI] [PubMed] [Google Scholar]
- Schnell, F. W., and C. C. Cockerham, 1992. Multiplicative vs. arbitrary gene action in heterosis. Genetics 131 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semel, Y., J. Nissenbaum, N. Menda, M. Zinder, U. Krieger et al., 2006. Overdominant quantitative trait loci for yield and fitness in tomato. Proc. Natl. Acad. Sci. USA 103 12981–12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull, G. H., 1908. The composition of a field of maize. Ann. Breed. Assoc. Rep. 4 296–301. [Google Scholar]
- Stuber, C. W., M. D. Edwards and J. F. Wendel, 1987. Molecular marker-facilitated investigation of quantitative trait loci in maize. II. Factors influencing yields and its component traits. Crop Sci. 27 639–648. [Google Scholar]
- Stuber, C. W., S. E. Lincoln, D. W. Wolff, T. Helentjaris and E. S. Lander, 1992. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics 132 823–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar, R. M., and N. M. Springer, 2006. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner, R. A., Y. Jia, R. DeCook, L. A. Borsuk, D. Nettleton et al., 2006. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. USA. 103 6805–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed, N. H., and Z. J. Chen, 2005. Molecular marker genotypes, heterozygosity and genetic interactions explain heterosis in Arabidopsis thaliana. Heredity 94 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollenaar, M., A. Ahmadzadeh and E. A. Lee, 2004. Physiological basis of heterosis for grain yield in maize. Crop Sci. 44 2086–2094. [Google Scholar]
- Van, B. R., 1999. GGT: software for the display of graphical genotypes. J. Hered. 90 328–329. [Google Scholar]
- Wang, D. L., J. Zhu, L. Li and A. H. Paterson, 1999. Mapping QTLs with epistatic effects and QTL × environment interactions by mixed linear model approaches. Theor. Appl. Genet. 99 1255–1264. [Google Scholar]
- Xiao, J., J. Li, L. Yuan and S. D. Tanksley, 1995. Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y. Z., Y. F. Tan, J. P. Hua, J. P. Sun, C. G. Xu et al., 2002. Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor. Appl. Genet 105 248–257. [DOI] [PubMed] [Google Scholar]
- Yu, S. B., J. X. Li, C. G. Xu, Y. F. Tan, Y. J. Gao et al., 1997. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA. 94 9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Y. J. Gao, M. A. Saghai, S. H. Maroof and J. X. Li Yang, 1995. Molecular divergence and hybrid performance in rice. Mol. Breed. 1 133–142. [Google Scholar]


