Abstract
Recent reports have shown that upon expression of appropriate oncogenes, both stem cells and more differentiated progenitor populations can serve as leukemia-initiating cells. These studies suggest that oncogenic mutations subvert normal development and induce reacquisition of stem-like features. However, no study has described how specific mutations influence the ability of differentiating cell subsets to serve as leukemia-initiating cells and if varying such cellular origins confers a functional difference. We have examined the role of the tumor suppressor gene p19ARF in a murine model of acute lymphoblastic leukemia and found that loss of p19ARF changes the spectrum of cells capable of tumor initiation. With intact p19ARF, only hematopoietic stem cells (HSCs) can be directly transformed by BCR/ABL expression. In a p19ARF-null genetic background expression of the BCR/ABL fusion protein renders functionally defined HSCs, common lymphoid progenitors (CLP), and precursor B-lymphocytes competent to generate leukemia stem cells. Furthermore, we show that leukemias arising from p19ARF-null HSC versus pro-B cells differ biologically, including relative response to drug insult. Our observations elucidate a unique mechanism by which heterogeneity arises in tumor populations harboring identical genetic lesions and show that activity of p19ARF profoundly influences the nature of tumor-initiating cells during BCR/ABL-mediated leukemogenesis.
Introduction
Studies of myeloid leukemia have demonstrated the presence of malignant stem cells,1 which are thought to generate and perpetuate leukemic populations and have thus been termed “leukemia stem cells (LSCs)” or leukemia-initiating cells (L-ICs). Although the properties of L-ICs resemble normal stem cells in that self-renewal ability is retained, it is not clear that malignant stem cells necessarily arise from their normal counterparts. Indeed, recent data have demonstrated that not only multipotential hematopoietic stem cells (HSCs), but also committed progenitors can serve as L-ICs when engineered to express some but not all oncogenic mutations.2–5 These findings suggest that both the nature of specific mutations, as well as the developmental context (ie, stage of hematopoietic differentiation) can play a role in creating leukemia-initiating cells
While the origins of L-ICs in myeloid leukemia have been described in many reports, the biology of similar cells in lymphoid leukemia is less clear. Analyses of primary human specimens has shown that a small subpopulation of phenotypically distinct cells can drive the in vitro and in vivo growth of B-cell acute lymphoblastic leukemia (B-ALL) populations, thus suggesting that some form of L-IC may be relevant to lymphoid disease.6–9 However, it is not possible to retrospectively determine the origin of candidate L-IC in human specimens, and given the heterogeneity found between patient specimens, equally difficult to define the role of specific mutations in disease progression. Thus, it remains an open question whether human B-ALL arises from relatively primitive populations or more differentiated progenitor cells. In the present study we describe a murine model in which expression of human BCR/ABL together with the p19ARF-null manipulation differentially highlights the contribution of genetic and developmental factors in acute lymphoblastic leukemia.
Clinically, the BCR/ABL translocation lies at the heart of chronic myelogenous leukemia (CML) and is thought to be the first and only mutation required to induce chronic disease. Untreated, CML patients will inevitably acquire additional genetic alterations and progress to either a myeloid or lymphoid blast crisis stage.10 Among the most common secondary mutations found in lymphoid blast crisis is del(9p21), which encompasses the p16INK4A/p19ARF locus. Homozygous deletion of exon 2 at the p16INK4A/p19ARF locus is found in approximately 50% of BCR/ABL-mediated lymphoid blast crisis specimens,11–13 implicating dysfunction of this locus in lymphoid leukemogenesis. The overlapping p16INK4A/p19ARF locus encodes 2 tumor suppressors, p16INK4A and p14ARF (p19 in mouse), which are the negative regulators for Rb-mediated cell cycle progression and MDM2-regulated p53 degradation, respectively. Deletion or mutations within this gene locus have been frequently found in many human cancers, including both solid tumors and hematological malignancies.14–17 The 2 genes share exons 2 and 3 but have their own unique promoter and first exon (exon1α for Ink4a and exon 1β for p19ARF). Thus, deletion of exon 2 abolishes expression of both proteins. In vitro, transformation of precursor B cells by v-abl was shown to be greatly facilitated by null mutations at the p16INK4A/p19ARF locus; further studies associated this activity specifically with loss-of-function mutations involving the p19ARF gene.18–20 The aforementioned studies suggest that the gene products of this tumor suppressor locus, especially p19ARF, might be crucial for normal lymphocyte development. Indeed, the p16INK4A/p19ARF-null (exons 2 and 3) and p19ARF-null (exon 1β) mice develop aggressive lymphoproliferative diseases, among many other types of tumors seen in the adult mice.16,17 Based on the frequency of p16INK4A/p19ARF mutations in CML-associated lymphoid blast crisis, we reasoned that expression of the p210 BCR/ABL protein product of the 9;22 human translocation together with loss-of-function at the murine p16INK4A/p19ARF locus could be genetically sufficient to initiate B-ALL. This concept was recently validated in studies using murine precursor B-cell cultures as targets of transformation, wherein p19ARF loss enhances oncogenicity of BCR/ABL-induced B-ALL.21 In addition, studies have shown that expression of p185 BCR/ABL in a p19ARF-deficient background is genetically sufficient to induce the formation of fully transformed leukemia stem cells.22
In our experiments, expression of p210 BCR/ABL and loss of p16INK4A/p19ARF function was also shown to be sufficient to promote murine B-ALL. We further demonstrate that these specific mutations can induce malignant transformation at multiple stages of B-lineage differentiation. Remarkably, p210 B-ALLs originating from primitive versus committed progenitor populations have distinct biological phenotypes, including in vivo and in vitro growth characteristics, developmental outcomes, transcriptional profiles, and sensitivity to known leukemia drugs. Thus, we propose that both genetics and differentiation status can influence pathogenesis and in vivo biology of leukemias harboring specific mutations.
Methods
Mice
C57Bl/6 p16INK4A/p19ARF-null (Δ exon 2,3, strain number 01XB1) and p19ARF-null(Δ exon 1β, strain number 01XG7) mice were obtained from the National Cancer Institute (NCI; Frederick, MD) and bred at the University of Rochester under specific pathogen-free conditions. Female wild-type C57BL/6-Ly-5.1 mice (6-8 weeks of age) were purchased directly from the NCI or The Jackson Laboratory (Bar Harbor, ME). All animal studies were approved by the University of Rochester Committee on Animal Resources.
Plasmid and virus production
The MSCV-BCR/ABL-IRES-GFP vector was kindly provided by Dr Richard Van Etten. Viral plasmid DNA was isolated using the Maxi-Prep plasmid purification Kit (QIAGEN, Valencia, CA). At 24 to 48 hours prior to transfection, Phoenix-Eco packaging cells (ATCC, Manassas, VA) were seeded at 2 to 3 × 105 cells per 35-mm plate. When cell density reached 80% to approximately 90% confluence, 10 μg of retroviral plasmid DNA was tranfected using 5 μL lipofectamine 2000 (Invitrogen, Carlsbad, CA) per 35-mm plate. Five hours after transfection, the lipofectamine reagent was removed and replaced with 2 mL fresh culture medium (DMEM, 10% fetal bovine serum). The culture medium was harvested 24 hours later and centrifuged at 3000 rpm for 10 minutes to remove cells and debris. The supernatant was carefully collected without disturbing the cell pellet and stored in aliquots at −80 degrees centigrade.
Generation of murine models
Six- to eight-week-old healthy donor mice (wild-type C57BL6/J, p16INK4A/p19ARF-null or p19ARF-null) were killed and marrow was flushed from femurs and tibiae. Single-cell suspension were incubated with NH4Cl solution for red blood cell lysis and depleted of lineage positive cells (Lin+) using the Becton Dickinson (BD, San Jose, CA.) IMag immunoaffinity system per manufacturer's instruction. For the experiments in Figures 1 and 2, Lin− cells were plated in dishes coated with retronectin (Takara, Shiga, Japan) and cultured overnight at 37°C and 5% CO2 (vol/vol) in IMDM containing 1% fetal bovine serum (FBS), 1% bovine serum albumin (BSA), 50% X-Vivo, stem cell factor (SCF; (25 ng/mL), Flt3 ligand (25 ng/mL), interleukin-3 (IL-3; 10 ng/mL), and IL-6 (10 ng/mL). The next day, cells were subjected to retroviral transduction and transplantation as described previously.5 Briefly, 50% of the culture media was replaced with viral supernatant twice a day for consecutive 2 days. The cells were then harvested and injected into the lateral tail vein of sublethally irradiated (6 Gy from a Rad Source 2000 X-ray irradiator [Alpharetta, GA]) female C57BL6/J recipient mice at a dose of 1 to 2 × 106 cells per mouse. For the experiments in Figures 3 to 7, specific bone marrow subsets were flow cytometrically sorted (FACSAria, BD) based on the expression of surface marker profiles: HSC (Lin−, c-Kit+, Sca1+), CLP (Lin−, c-Kit+, Sca1low, AA4.1+, CD127+), pro-B (B220low, CD43+, c-Kit+, CD2−, IgM−), and pre-B (B220low, CD43−, c-Kit−, CD2+, IgM−). The lymphoid progenitors (CLP, pro-B and pre-B) were incubated in IMDM containing 10% FBS, β-mercaptoethanol (55 μM), SCF (25 ng/mL) and IL-7 (10 ng/mL) while purified stem cells (HSCs) were cultured in the same medium as described for Lin− cells. Transplantations were performed using 104 to 105 cells per recipient. Recipients were evaluated daily for signs of morbidity, as indicated by hunched posture, roughened coat, abnormal weight loss, and inactivity after transplantation. Beginning at 10 to 14 days after transplantation, peripheral blood was sampled every 5 to 7 days. Total leukocyte count was obtained using the HESKA CBC-Diff System, and the blood smears were made and stained with the 3-Step Stain Set (Richard-Allan Scientific; Thermo-Fisher, Waltham, MA) to visualize the morphology of recipient leukocytes by light microscopy. To analyze the lineage of the engrafted cells (EGFP+), leukocytes from the recipients' blood were labeled with antibodies to lineage makers Gr-1, Mac1, B220, and CD19 (BD) for flow cytometric analysis (LSRII; BD). At advanced stages of disease, mice were killed and bone marrow, spleen, and blood were collected for phenotypic analyses and further applications. All the flow cytometric data were analyzed using FlowJo software (TreeStar, Ashland, OR).
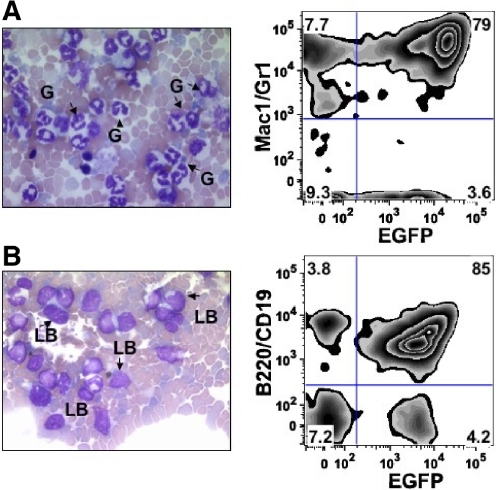
Figure 1.
Comparison of CML and lymphoid blast crisis models. Lin− cells from bone marrow of wild-type, p16INK4A/p19ARF-null, or p19ARF-null donors were isolated as described in “Generation of murine models.” The cells were transduced with vectors coexpressing p210BCR/ABL and EGFP, followed by injection into sublethally irradiated wild-type recipient mice. Leukocytes from peripheral blood of the recipients were collected for lineage and morphological analysis approximately 2 to 3 weeks after transplantation. (A) Expression of p210BCR/ABL in wild-type donors resulted in the development of CML-like disease in recipients. Histological staining of peripheral blood shows mature granulocytes (G), FACS analysis indicates that almost all EGFP+ cells are Mac1+/Gr1+. (B) Injection of p210BCR/ABL-expressing cells from p16INK4A/p19ARF or p19ARF-null donors showed high levels of immature lymphoblasts (LBs) in recipient mice. The immunophenotype of EGFP+ cells is almost entirely B220+/CD19+, indicating the disease type is B-lineage acute lymphoblastic leukemia (B-ALL).
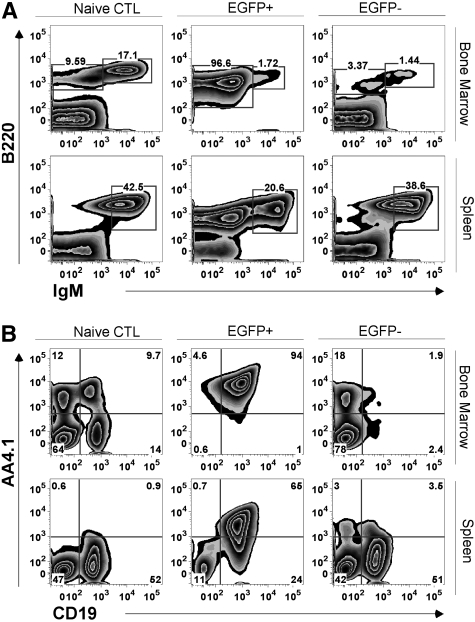
Figure 2.
Phenotypic analysis of BCR/ABL-mediated B-ALL. Representative analysis for recipients of p210BCR/ABL expressing Lin− cells from a p19ARF-null background. Animals were killed approximately 2 to 3 weeks after transplantation when there were significant signs of illness. Leukocytes from bone marrow and spleen of naive control (CTL) recipient mice were harvested for flow cytometric lineage analysis. FACS plots show comparison of (A) the expression of B220, IgM, and (B) the expression of AA4.1, CD19 between CTL mice, EGFP+ and EGFP− populations of recipient mice. The gates in panel A separate IgMhi and IgMlo∼neg expressing B cells.
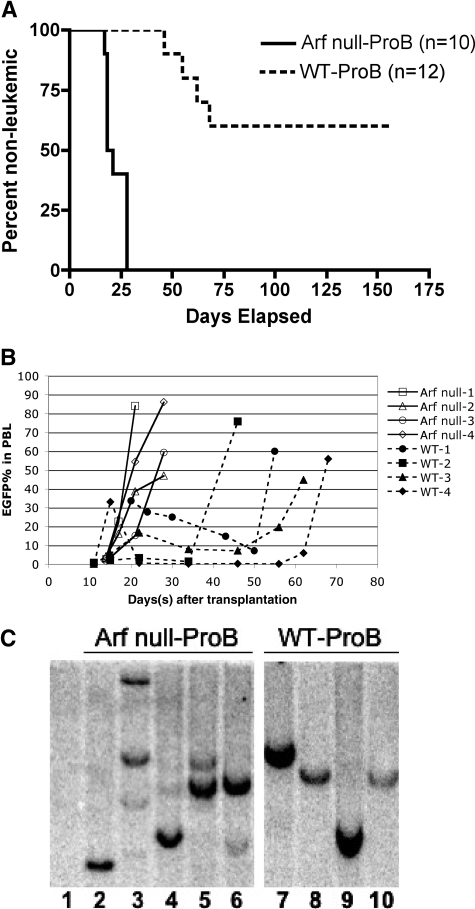
Figure 3.
Disease-onset kinetics and clonal patterns of B-ALL derived from wild-type versus p19ARF-null pro-B cells. Pro-B cells from wild-type and p19ARF-null bone marrow were flow sorted and transduced with vectors coexpressing p210BCR/ABL and EGFP, followed by transplantation into sublethally irradiated wild-type recipients. (A) Kaplan-Meier analysis of survival for the recipients of p19ARF-null (solid line) and wild-type (dashed line) proB cells. (B) Peripheral blood from recipients of p19ARF-null (solid lines, closed symbols) and wild-type (dashed lines, open symbols) proB cells were sampled starting on approximately 10 to 14 days after transplantation and continued approximately every 5 to 10 days until the mice succumbed to B-ALL. The percentage of EGFP+ leukocytes was measured by flow cytometric analysis. (C) Spleen DNA of naive control (lane 1), 5 recipients of p19ARF-null proB (lanes 2-6) and 4 recipients of wild-type pro-B (lanes 7-10) was digested with EcoRI and hybridized with an EGFP probe for Southern blot analysis to reveal the proviral integration pattern.
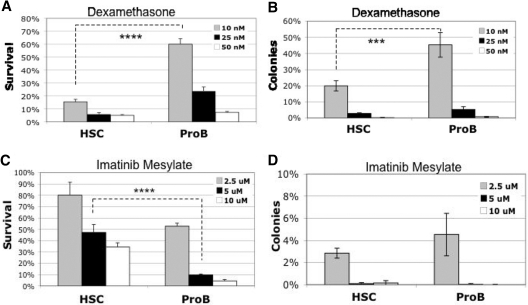
Figure 7.
In vitro drug response of B-ALL arising from p19ARF-null HSCs versus pro-B cells. Primary recipients injected with p210BCR/ABL-expressing p19ARF-null HSC or pro-B cells were killed approximately 2 to 3 weeks after transplantation. EGFP+ B-ALL (B220+, CD19+, IgM−) cells were isolated by FACS and cultured in vitro (IMDM + 10% FBS + 55 micromolar 2-ME, no cytokines) for 2 to 3 weeks. Cells were then treated with dexamethasone (A,B) or imatinib mesylate (C,D) at the concentrations indicated, 106 cells per mL. After 48 hours of treatment, half of each sample was stained with PE-conjugated anti–annexin V antibody and 7AAD to evaluate apoptotic response by flow cytometry. Results are presented as percent of survival relative to untreated control (A,C). A small portion of each culture was plated in methycellulose medium (with murine SCF, IL-3, IL-6, Flt3 ligand, and IL-7) to evaluate the clonal potential of residual cells. Results are presented as percent of colonies relative to untreated control (B,D). Significance was determined with Student paired, 1-tailed t test (****P < .001; ***P < .005).
In vitro colony-forming assays
The indicated cell populations were plated in methylcellulose medium (StemCell Technologies, Vancouver, BC; M3134) supplemented with 15% FBS, 20% BIT (BSA, insulin, transferrin; StemCell Technologies), β-mercaptoethanol (55 μM), SCF (50 ng/mL), Flt3L (50 ng/mL), IL-3 (10 ng/mL), IL-6 (10 ng/mL), and IL-7 (10 ng/mL). Cultures were incubated at 37°C and 5% CO2 (vol/vol) for 10 to 14 days before scoring colonies (60 or more cells/colony).
Southern blot analysis
Genomic DNA from spleen was digested with EcoRI, separated by electrophoresis on 0.8% agarose Tris-acetate-EDTA gels and transferred onto GeneScreen Plus membranes (Perkin Elmer, Waltham, MA). Blots were sequentially hybridized with [32P]-labeled probes for enhanced green fluorescent protein (EGFP), using the entire EGFP coding sequence from a pEGFPC2 plasmid (Clontech Laboratories, Mountain View, CA). The blots were washed at high stringency, and hybridization was detected after exposure on Molecular Dynamics Phosphor Screens using a Storm 860 Imaging System scanner and ImageQuant software (Molecular Dynamics, Piscataway, NJ).
Reverse-transcriptase polymerase chain reaction (RT-PCR)
EGFP+ B-ALL (B220+, CD19+, IgM−) cells (106) from recipients' bone marrow were sorted and lysed in 0.8 mL Trizol reagent (Invitrogen) for total RNA extraction per manufacturer's instructions. Total RNA was resuspended in RNase/DNase-free water followed by DNase I (Invitrogen) treatment. DNase I–treated RNA (0.5-1 μg) was used for first-strand cDNA synthesis by SuperScript II RT (Invitrogen) following the manufacturer's protocol.
Quantitative real-time PCR
The primer sets used for real-time PCR analysis were: mHPRT sense: GTTGGATACAGGCCAGACTTTGTTG; mHPRT antisense: GAGGGTAGGCTGGCCTATAGGCT; mRag2 sense: GAGATGTCCCTGAACCCAGA; mRag2 antisense: AACATGGGGTAGGCAGTCAG; mTdT sense: ATGCGAGCGTCCTCTGTACT; mTdT antisense: CAGCTGTCT TCAGTCCCACA; sterile kappa sense TCCACGCATGCTTGGAGAGGGGGTT; sterile kappa antisense: GTCCTGATCAGTCCAACTGTTCAG; mPax5 sense: GTCATCGGTGAGCACCGACTC; and mPax5 antisense: GAAGCCATGGCTGAATACTC.
Each reaction mixture contained 1 μL cDNA template (diluted 1:10), 7.5 μL SYBR Green SuperMix (Bio-Rad), 5.75 μL water, and 250 nM of each primer. Real-time PCR was carried out using an iCycler IQ (Bio-Rad, Hercules, CA).
Drug treatment and apoptosis assays
EGFP-positive B-ALL (B220+, CD19+, IgM−) cells were flow sorted and expanded in vitro at 37°C and 5% CO2 (vol/vol) in IMDM containing 10% FBS, and β-mercaptoethanol (55 μM) for 2 to 3 weeks. For drug treatments, 106 cells per milliliter of B-ALL cells were incubated with various concentrations of dexamethasone or imatinib mesylate as indicated in Figure 7 for 48 hours (IMDM, 10% fetal bovine serum, 55 μM β-mercaptoethanol). For apoptosis analysis, cells were washed with cold phosphate-buffered saline (PBS) and resuspended in 200 μL annexin binding buffer (10 mM HEPES/NaOH, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2). Annexin V–PE (BD Pharmingen) and 0.25 μg/mL 7-aminoactinomycin D (7-AAD; Molecular Probes, Eugene, OR) were added, and the tubes were incubated at room temperature in the dark for 15 minutes. Cells were then diluted with 200 μL annexin binding buffer and analyzed immediately by flow cytometry.
Results
Loss of the p16INK4A/p19ARF locus and expression of p210BCR/ABL in hematopoietic cells induces B-ALL in mice
To recapitulate clinically observed features of human lymphoid blast crisis and to test whether loss of p16INK4A/p19ARF contributes to CML progressing from chronic phase to blast crisis stage, we coexpressed p210BCR/ABL and green fluorescent protein (EGFP) in adult bone marrow (BM) cells from wild-type C57BL/6, p16INK4A/p19ARF-null (exons 2 and 3), or p19ARF-null (exon1β) mice via retroviral mediated gene transfer. We then transplanted the cells into sublethally irradiated wild-type C57BL/6 mice as primary recipients to evaluate disease outcome. Transduced EGFP+ cells were detected in the peripheral circulation as early as 10 to 12 days after transplantation, and by 18 to 30 days after transplantation, recipient mice had elevated white blood cell counts and signs of advanced disease (hunched posture, roughened coat, anorexia, and inactivity). Wild-type cells transduced with the BCR/ABL retrovirus gave rise to a CML-like myeloproliferative disease (Figure 1A), as has frequently been reported by previous studies.23 In contrast, expression of BCR/ABL in the p16INK4A/p19ARF-null or p19ARF-null background resulted in predominantly B-cell engraftment (EGFP+, B220+, CD19+; Figure 1B). No discernable differences were evidenced between the phenotype or kinetics of disease arising from p16INK4A/p19ARF-null versus p19ARF-null donor cells. The recipient animals developed a classic leukemia, wherein there was moderate splenomegaly and occasional hepatomegaly, in the setting of striking bone marrow infiltration and high levels of circulating mononuclear cells. These EGFP+ mononuclear cells were found in bone marrow, spleen, and peripheral blood, with a prominent population of B lymphocytes expressing a phenotype consistent with precursor B cells (B220lo, IgMneg, AA4.1+, CD19+) (Figure 2A,B center column). In contrast, EGFP− cells in the same recipients (Figure 2A,B right-hand column) showed relatively normal lineage profiles, as compared with naive control mice (Figure 2A,B left-hand column). These data indicate that expression of BCR/ABL in bone marrow cells from p16INK4A/p19ARF-null mice induced lymphoid leukemia that clinically and phenotypically recapitulates typical features of lymphoid blast crisis in human. In addition, mutation of p19ARF alone, with intact p16INK4A, is also able to induce BCR/ABL-mediated blast crisis, a finding consistent with previous reports (data not shown).21,22
Distinct onset kinetics and clonal patterns of B-ALL derived from p19ARF-null and wild-type pro-B cells
Although expression of p210-BCR/ABL is most commonly associated with CML, some patients present with p210-BCR/ABL–positive acute B-lymphocytic leukemia (B-ALL).24 In murine models of CML, transduction with a p210-BCR/ABL retrovirus causes an increased incidence of B-ALL in mice where conditioning regimens likely to deplete donor lymphoid progenitors (such as 5-fluorouracil treatment) are omitted.25 These findings suggested that in some cases, BCR/ABL transformation of lymphoid progenitors, rather than an HSC or myeloid progenitor, might promote B-ALL, not the more common CML.
To better define the cellular origins of the disease, we employed flow cytometric sorting to purify pro-B cells (B220lo, cKit+, CD43+, sIgM−) from both wild-type and p19ARF-null marrow as targets for genetic manipulation using BCR/ABL-mediated transformation. During the course of 3 independent experiments, 4 of 12 recipient mice that received transplants of p210-BCR/ABL–expressing wild-type pro-B cells developed B-ALL between 40 to 70 days after transplantation (Figure 3A dashed line). Notably, of the 8 remaining recipients, 4 were killed between day 30 and day 55 after transplantation due to complications unrelated to BCR/ABL-induced illness (ie, anemia or infection arising as a consequence of the transplant conditioning regimen: 600 rads total body radiation). At day 17 and day 25, 2 more recipients died but did not appear to have leukemia, since the heterogeneous population of EGFP+ bone marrow cell failed to engraft disease in secondary recipients or establish in vitro cultures. Finally, 2 animals remained healthy and contained approximately 1% EGFP+ leukocytes in the periphery up to day 155 after transplantation (data not shown). In contrast, all recipients receiving p210-BCR/ABL–expressing pro-B cells from the p19ARF deficient background (n = 10, from 3 independent experiments) developed B-ALL within 28 days after transplantation (Figure 3A solid line) and readily established in vitro cultures of EGFP+ lymphoblasts (not shown).
From the peripheral blood screening, we also found that the B-ALL recipients from wild-type and p19ARF-null pro-B origin differed with respect to the percentage of EGFP+ leukocytes; between 10 to 14 days after transplantation, p19ARF-null pro-B recipients had a steady increase in EGFP+ cells until they succumbed to disease (Figure 3B solid lines, open symbols). In contrast, wild-type pro-B recipients displayed an initial elevation of EGFP+ cells between 10 to 20 days after injection, followed by a transient regression lasting 1 to 2 months. These animals then progressed to more aggressive disease, manifested by a rapid increase in EGFP+ cells, and a clinical decline necessitating euthanasia (Figure 3B dashed lines, closed symbols). In addition, in contrast to the very consistent surface antigen phenotype observed in leukemias arising in p19ARF-null pro-B recipients (IgM−, B220+, CD19+, AA4.1+), we observed immunophenotypic variations in expression of AA4.1 and B220 among the wild-type pro-B–derived B-ALL samples (data not shown). These results indicate that additional genetic alterations may be required for leukemogenesis in animals harboring p210-BCR/ABL–expressing wild-type pro-B cells. In contrast, the rapidly fatal disease in mice harboring p19ARF-null pro-B cells with p210-BCR/ABL suggests that these 2 mutations are genetically sufficient to induce B-cell malignancy.
To further investigate this hypothesis, we looked at the clonality of EGFP+ splenocytes by Southern blot analysis of genomic DNA isolated from mice with wild-type or p19ARF-null derived B-ALL. Using an EGFP probe to assay for proviral integration, we found that B-ALL specimens derived from wild-type pro-B cells consistently displayed a single-band integration pattern (Figure 3C lanes 7-10), while specimens from p19ARF-null B-ALL samples had multiple bands consistent with oligo- or polyclonal integration patterns (Figure 3C lanes 1-6, and SF1, lanes 1-10). Together with the observed differences in disease kinetics and phenotype of disease in the recipient animals, these data support the notion that wild-type pro-B cells transduced with BCR/ABL accumulate secondary mutation(s) before acquiring competence to mediate acute B-ALL, whereas lesions combining BCR/ABL expression and p19ARF deficiency are sufficient for leukemic transformation of relatively differentiated B-lymphoid targets.
Defining the developmental stages of B-ALL–initiating events
Studies of p210-BCR/ABL in myeloid leukemia models have demonstrated that the mutation is oncogenic only when expressed in the HSC fraction (linneg/c-kit+/Sca1+), but not in highly purified later stage progenitors.3,5 However, if BCR/ABL is coexpressed with a second facilitating oncogenic event, such as Nup98/HoxA9, committed myeloid progenitors can then be successfully transformed. Thus, BCR/ABL-mediated transformation in myeloid leukemia is a function of both the cellular context and additional cooperating mutations. To investigate the potential origins/targets of B-ALL, we performed p210-BCR/ABL retrovirus transduction experiments on enriched populations of stem cells and different lymphoid progenitor populations. Common lymphoid progenitors (CLPs) were prospectively identified as c-kit+/AA4.1+/CD127+/linneg populations, pro-B cells as IgMneg/CD43+/CD2neg/B220lo subsets, distinct from B220lo/IgMneg/CD43neg/CD2+ pre-B-cell subsets in wild-type versus p19ARF deficient murine bone marrow. Within 30 days, expression of p210 BCR/ABL in CLP, pro-B and pre-B cells from mutant mice reproducibly generated B-ALL, unlike wild-type cells, which, at all stages only showed delayed and incomplete transformation. These experiments demonstrated that, during B-lymphoid differentiation, several functionally and phenotypically distinct stages of progenitor populations are rendered competent to serve as L-IC in the absence of p19ARF function (Figure 4A,B). Notably, mutant HSCs also gave rise to B-ALL, but, in addition, showed a prevalent myeloid hyperproliferation, similar to the CML-like phenotype observed in the wild-type background (Figure 4A).
Figure 4.
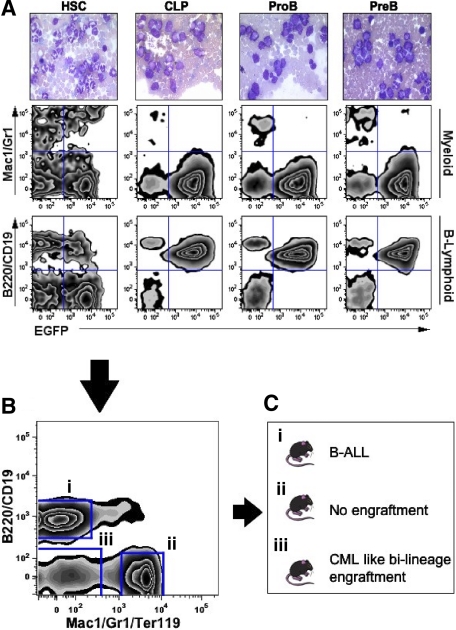
Defining the developmental stages of B-ALL–initiating events. HSCs, CLP, proB, and preB cells from p19ARF-null bone marrow were flow sorted and transduced with vectors coexpressing p210BCR/ABL and EGFP, followed by transplantation into sublethally irradiated wild-type recipients. (A) Peripheral blood of recipient mice was sampled at day 17 after transplantation for histological (top row) and flow cytometric analyses to assess the GFP expression and lineage distribution of leukocytes (center and bottom rows). (B) At day 23 after transplantation, bone marrow cells from the p19ARF-null HSC primary recipients were flow sorted into 3 distinct cell groups, (1) B-lymphoid marker positive, (2) myeloid marker positive, and (3) lineage negative, followed by transplantation into sublethally irradiated secondary wild-type recipients. The plot shown was gated on EGFP+ and T-cell negative (CD3−) cell population. (C) Outcome of the secondary recipients that received different sorted cells from p19ARF-null HSC primary recipients indicated in panel B.
Since HSC-transplanted mice showed expansion of cells representing distinct hematopoietic lineages (Figure 4B, CML-like myeloid cells as well as 5%-15% lymphoblasts), we further characterized the bi-lineage leukemia in the experiment outlined in Figure 4B,C. Three distinct transduced subpopulations, B220+/CD19+ precursor-B, Linneg primitive cells, and Mac-1+/Gr-1+/Ter119+ myeloid fractions were sorted from the bone marrow of p19ARF-null BCR/ABL+ primary recipients and transplanted into secondary recipients. Despite the absence of p19ARF and in accord with findings from BCR/ABL transduction of wild-type cells, mature myeloid cells (fraction II) did not induce disease in recipient mice. In contrast, lymphoid lineage cells (fraction I) transmitted B-ALL to the recipients in a characteristically short period of time (2 to 6 weeks, depending on inoculum), confirming the presence of B-ALL leukemia-initiating activity in this population. Intriguingly, mice receiving the more primitive EGFP+ fraction (fraction III) reprised bi-lineage engraftment, with a more chronic myeloid component, suggesting that a small number of transformed HSCs retained multilineage potential (data not shown). This final cohort of mice, recipients of fraction III, developed CML-like disease similar to that observed in the p210-BCR/ABL–transduced wild-type HSCs. Taken together, these data demonstrate that p19ARF loss-of-function mutations allow committed, relatively differentiated precursor B-cell populations, but not myeloid populations from the same animals, to function as leukemia-initiating cells.
Distinct phenotype of B-ALL arising from p19ARF-null HSC and pro-B cells
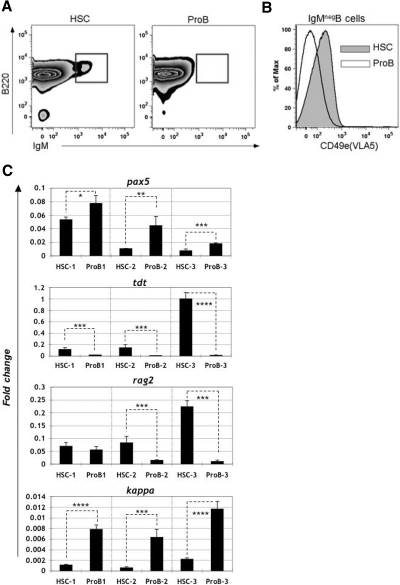
Based on the data above, we hypothesized that B-ALL arising from different p210 BCR/ABL p19ARF-null developmental stages (ie, HSC vs CLP vs pro-B, etc) may possess distinct biological properties. We reproducibly observed that a small fraction (1%-5%) of HSC-derived BCR/ABL+ p19ARF-null lymphocytes could mature to the IgM+ stage; consistently 3- to 4-fold more than that observed in mice harboring sorted pro-B populations with the same genotype (Figure 5A boxed), suggesting differences may exist in the overall differentiation potential of these 2 leukemias. Indeed, analysis of precursor B-cell fractions expressing a B220low CD43+ phenotype shows marked heterogeneity of BP-1 and CD24 surface antigen density on the pro-B cell populations that is quite distinct when compared with HSC-derived cell populations (Figure S2, available on the Blood website; see the Supplemental Materials link at the top of the online article). Finally, analysis of circulating or bone marrow–derived blast populations (EGFP+, IgMneg, B220+, CD19+) showed that B-ALL cells arising from HSC had reproducibly higher expression of the integrin VLA-5/CD49e (Figure 5B) than cells derived from the purified pro-B stage. The interaction of CD49e and its ligand has been implicated in bone marrow microenvironmental interactions that modulate leukemia cell response to extrinsic signals.26,27
Figure 5.
Phenotypic analyses of B-ALL arising from p19ARF-null HSC versus pro-B cells. Primary recipients injected with p210BCR/ABL-expressing p19ARF-null HSC or pro-B cells were killed approximately 2 to 3 weeks after transplantation. Leukocytes from the recipients' bone marrow were harvested for flow cytometric phenotypic analysis. (A) Cells derived from HSC (left) contained approximately 3-fold more (2.1% vs 0.7%) mature B cells (IgM+) as cells derived from pro-B stage (right). The plots shown were gated on EGFP+ lymphocytes. (B) EGFP+ B-ALL cells (B220+, CD19+, IgM−) derived from HSCs (shaded in gray) showed significantly higher CD49e (VLA5) expression than the cells arising from pro-B (white area). (C) 106 flow-sorted EGFP+ B-ALL cells from bone marrow of both HSC and pro-B recipients were reverse-transcribed to cDNA and analyzed by real-time quantitative PCR. Results are shown for 3 independent experiments, with each bar representing the average of triplicates. Error bars are standard deviation. Each gene expression value is shown relative to expression of hypoxanthine-guanine phosphoribosyl transferase (HPRT). Significance was determined with Student paired, 1-tailed t test (*P < .05; **P < .01; ***P < .005; ****P < .001).
Bone marrow B-lymphoid differentiation is directed by well-characterized patterns of gene transcription linked to lineage commitment, cellular function, and the hierarchical assembly of B-lymphoid receptor components.28 To further characterize the BCR/ABL+ p19ARF-null leukemias derived from the HSC versus the more differentiated pro-B cell fractions, Q-PCR was used to compare expression profiles of genes that are regulated during B-cell differentiation. Notably, the expression profiles of terminal deoxy transferase (TdT), sterile kappa, and pax-5/BSAP were consistently different between the 2 forms of B-ALL (Figure 5C, 3 independent experiments). In addition, for 2 of 3 experiments and recombinase activating gene (rag)–2 also differed (Figure 5C). Since precursor B-cell differentiation is characterized by distinct and interacting transcriptional programs, the differences in transcription profiles between HSC- and pro-B–derived ALL raised the possibility of functional differences between these cell populations.
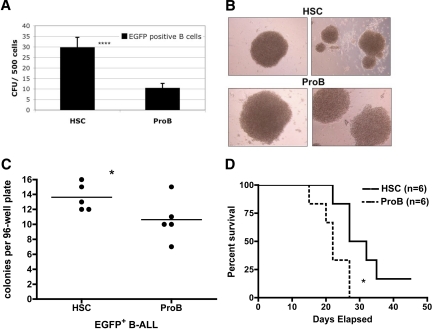
To probe for such biological differences, we purified EGFP+, IgMneg, B220+, CD19+ cells from primary recipients with HSC- versus pro-B–derived p19ARF deficient B-ALL and performed in vitro colony forming assays. Leukemic specimens derived from BCR/ABL-transduced p19ARF-null HSCs showed an approximately 3-fold higher frequency of colony-forming cells (CFUs) than equivalent populations derived from pro-B cells (Figure 6A). We also observed that the colonies derived from HSC-derived B-ALL were generally smaller than those from pro-B cells (Figure 6B), suggesting that, while CFU potential was greater in the HSC fraction, colony expansion is more vigorous in the pro-B–derived fraction. Further analysis of clonal potential was performed by isolating single cells from HSC versus pro-B–derived leukemias and analyzing colony outgrowth in the absence of cytokine supplementation. Under these conditions, the frequency of clones was again higher for HSC- than pro-B–derived B-ALL single-sorted into U-bottom 96-well plates (Figure 6C) and pro-B–derived clones were again readily distinguishable from the smaller BCR/ABL-transduced HSC colonies (data not shown). These observations are consistent with the notion that the developmental progression of lymphoblastic leukemia progenitor cells may differ as a function of the originating cell type.
Figure 6.
Assessments of clonal and growth potential of B-ALL arising from p19ARF-null HSC versus pro-B cells. Primary recipients injected with p210BCR/ABL-expressing p19ARF-null HSC or pro-B cells were killed approximately 2 to 3 weeks after transplantation. EGFP+ B-ALL (B220+, CD19+, IgM−) cells from bone marrow were isolated by FACS for analyses. (A) B-ALL cells were directly sorted into semi-solid methycellulose medium (with murine SCF, IL-3, IL-6, Flt3 ligand, and IL-7), 500 cells per well, and colonies were scored 2 weeks later. Significance was determined with Student paired, one-tailed t test (****P < .001). (B) Photographs of colonies (100× magnified) grown from cultures in panel A. (C) B-ALL cells were directly sorted into U-bottom 96-well culture plate, 1 cell per well, 5 plates per group, with 100 μL culture medium (IMDM + 10% FBS + 55 μM 2-ME). Cell clones that arose from each well were scored 2 weeks later. Significance was determined with Student paired, one-tailed t test (*P < .05). (D) Fresh isolated B-ALL cells from both groups were transplanted into sublethally irradiated wild-type secondary recipients (6 per group) at a dose of 5000 cells per mouse. Results are presented as Kaplan-Meier analysis to compare the survival of recipients that engrafted with B-ALL arising from p19ARF-null HSC and pro-B cells. Significance was determined with survival analysis method of Prism software (*P < .05).
EGFP+ bulk cultures expand aggressively in cultures plated from the peripheral blood, spleen, or bone marrow populations derived from p210 BCR/ABL B-ALL p19ARF-null animals; these cells do not require stromal cell or cytokine support (data not shown). We tested the in vivo growth potential of both HSC- and pro–B cell–derived B-ALL cells by injecting samples of each population into secondary recipient mice. Whether inoculated directly from euthanized primary recipients, or from established in vitro cultures, all such experiments led to clinically evident disease in recipient mice (data not shown). Reproducibly, however, mice that received cells from pro-B–derived B-ALL succumbed earlier than those with B-ALL from HSCs (Figure 6D), indicating that leukemias derived from the pro-B–cell fraction behave more aggressively in vivo. Limiting dilution analysis of pro-B versus HSC-derived BCR/ABL+ p19ARF-null leukemia-initiating activity in vivo supported this observation, in that roughly 1 in 350 cells transferred fatal disease, whereas more than twice that number were necessary for a similar mortality in HSC-derived B-ALL.
Finally, we compared the responses of HSC- versus pro-B–derived B-ALL to clinically relevant drugs by analyzing the in vitro survival and progenitor expansion (CFU) potential after challenge with either the corticosteroid dexamethasone (Dex) or the Abl kinase inhibitor imatinib mesylate (IM). As shown in Figure 7 (panels A and C), suspension cultures of the respective B-ALL types display differential sensitivity to the 2 drugs. HSC-derived B-ALL is relatively sensitive to Dex but somewhat more resistant to IM; whereas pro–B-derived B-ALL shows the opposite behavior. After drug treatment, cells from each suspension culture were plated in methylcellulose media to assay for CFU potential (Figure 7B,D). Consistent with the data observed for suspension cultures, the 2 forms of B-ALL show differential responses to Dex, with HSC-derived disease displaying greater sensitivity. CFU potential after IM challenge was similar for HSC- versus pro–B-derived B-ALL, with a high degree of drug sensitivity observed for both tumor types. Taken together, these results confirm and extend the finding that p19ARF deficient HSC-derived versus pro–B-derived disease arising in the same genetic background possesses multiple distinct biological features.
Discussion
The findings reported here establish several important parameters with regard to the pathogenesis of p210 BCR/ABL-mediated B-ALL. First, using the experimental system described herein, we find that p210 BCR/ABL expression and loss of p19ARF function are sufficient to mediate full transformation to B-ALL in mice. These experimental results are entirely consistent with the work of Williams et al, wherein expression of p185 BCR/ABL in p19ARF-null B-lymphoid cells was shown to be genetically sufficient to induce B-ALL in vivo.21,22 The rapidly fatal lymphoblastic leukemia evolving in these experimental animals is particularly informative, given the prevalence of this genotype in the poor prognosis precursor B-cell leukemias known as human CML lymphoid blast crisis.10 A harbinger of the aggressive clinical behavior and short latency of p19ARF-null BCR/ABL lymphoblastic leukemias can be found in observations published a decade ago, wherein cells harboring the p19ARF-null mutation expand almost immediately, bypassing the typical period of culture “crisis” expected in v-abl–transformed wild-type precursor B-lymphoid cell lines.18
Our second observation of importance is that a second, highly specific mutagenic event is closely linked to the evolution of acute lymphoblastic leukemia not only from hematopoietic stem cells, but also from B-cell precursors undergoing the highly ordered and strictly regulated B-lymphoid differentiation program. As predicted by a substantial body of literature, we observed that leukemia arising as a consequence of p210 BCR/ABL expression alone in p19ARF+/+ (ie, wild-type) control animals was almost exclusively myeloid, and disease evolved from cells representing only the earliest stage of hematopoiesis.29 Coexisting null mutations of p19ARF did not change the oncogenic spectrum of BCR/ABL in committed myeloid progenitor populations. In contrast, in mice harboring an p19ARF-null mutation, BCR/ABL-mediated lymphoblastic leukemia arose not only from HSCs, but also evolved readily from common lymphoid progenitor (CLP) as well as the relatively differentiated pro-B– and pre-B–cell populations. Within 2 to 4 weeks, highly purified populations representing distinct developmental stages (HSCs, CLP, pro-B and pre-B cells) but harboring identical leukemogenic mutations gave rise to fully transformed primary populations and readily transferred aggressive leukemia to secondary recipients, confirming the pathogenic nature of the p19ARF-null, BCR/ABL genotype.
Lymphoid precursor populations are highly proliferative; normal B-precursor populations undergo iterative bursts of transcriptional and cell cycle activity during assembly of immunoglobulin receptor components and in response to microenvironmental signals. Despite these proliferative and genetic events, lymphoblastic leukemia represents only a small subset of the life-threatening transformation to acute leukemia observed in patients with CML. Our data show that the tumor-suppressor properties encoded by the p19ARF gene locus, a key regulator of p53-dependent cellular response, is required for the prevention of lymphoblastic evolution at multiple developmental stages during p210 BCR/ABL B-cell differentiation. In this light, our data are extremely informative regarding B-lymphopoiesis in CML patients, by elucidating the remarkable spectrum of vulnerability in this pre-leukemic compartment. Given these observations, and the similarity of developmental programs shared by T lymphocytes, it is perhaps not surprising that emerging data implicate p14ARF function in T leukemogenesis as well.30,31
Finally, our experimental model and initial observations allowed us to ask important questions regarding the contribution of developmental origin to disease pathogenesis in vivo. Clearly, the stage at which the 2 mutations (BCR/ABL translocation and p19ARF deficiency) occur during differentiation could have a significant influence on the biological properties of the resulting tumor. Although B-lymphoid differentiation is a dynamic process involving multiple developmental transitions, populations representative of specific B-lymphoid differentiation stages and functional responses in vitro are readily distinguishable. Using some of these canonical profiles and functional assays, we probed for differences between leukemic populations of p19ARF-null, BCR/ABL ALL derived from transformation of HSC populations versus lineage-committed pro-B–cell populations. Notably, HSC- and pro-B–derived p19ARF-null, BCR/ABL leukemias could be distinguished by immunophenotype, the transcriptional activity of critical genes, and in vitro assay responses. Most importantly, we demonstrated that p19ARF-null BCR/ABL leukemias from developmentally distinct populations have differential drug sensitivity.
If B-ALL arising from double-mutant HSC versus pro-B cells demonstrates multiple phenotypic and functional differences, including differential drug responses, these observations support the notion that independent human B-ALLs arising from identical genetic changes could display variable clinical features (ie, drug responsiveness) as a function of the cell of origin. The underlying reason for the differences we detected in HSC versus pro-B–derived B-ALL are as yet unknown, but it is attractive to speculate that changes in epigenetic status could modulate the changes observed in tumor biology. Indeed, upon differentiation from the HSCs to pro-B stage, a complex set of gene regulatory changes must occur, wherein signals associated with levels of transcriptional activity, concurrence of transcription programs, and competition for critical factors all influence B-lymphoid differentiation. If such changes carry through to the malignant state induced by BCR/ABL expression and loss of p19ARF function, it is possible they could represent a fundamental difference among tumor cells arising from these distinct developmental stages. The appreciation of such considerations is highly important as the use of epigenetic modifying agents to influence therapeutic responses in cancer therapy becomes an increasingly common strategy.
These observations provide a clear illustration of the critical importance of underlying genetic and developmental programs on the manifestation of oncogenic mutations and their impact on the malignant transformation process. These reflect both the epigenetic status of the target cell and epistatic effects between the transforming oncogene(s) and other differentiation-specific cellular factors. While a thorough understanding of these complex interactions is beyond our current capabilities, it is evident that their elucidation at the genomic level, both in terms of epigenomics and of global gene expression changes, may be critical for strategies aimed at preventing oncogenic progression/transformation and for developing optimal targeted therapies.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of John Ashton and Min Yee.
This work was supported by grants from the American Cancer Society (RSG-03-096-01-LIB); United States Department of Defense (CM050044), and National Institutes of Health (R01-CA122206, C.T.J. and R21-CA107355 to A.B.). C.T.J. is a Scholar of the Leukemia & Lymphoma Society.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.Y.W. and F.Y. designed research, performed research, analyzed data, and helped write the paper. C.Y.C., B.M.S., S.J.N., R.M.R., T.B. I.K., and D.H. performed research. A.B. designed research and analyzed data. C.T.J. designed research, analyzed data, and helped write the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Craig T. Jordan, University of Rochester Medical Center, 601 Elmwood Avenue, Box 703, Rochester, NY 14642; e-mail: craig_jordan@urmc.rochester.edu.
References
- 1.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 2.Cozzio A PE, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huntly BJ, Shigematsu H, Deguchi K, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 5.Neering SJ, Bushnell T, Sozer S, et al. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood. 2007;110:2578–2585. doi: 10.1182/blood-2007-02-073031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castor A, Nilsson L, Astrand-Grundstrom I, et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat Med. 2005;11:630–637. doi: 10.1038/nm1253. [DOI] [PubMed] [Google Scholar]
- 7.Cobaleda C, Gutierrez-Cianca N, Perez-Losada J, et al. A primitive hematopoietic cell is the target for the leukemic transformation in human philadelphia-positive acute lymphoblastic leukemia. Blood. 2000;95:1007–1013. [PubMed] [Google Scholar]
- 8.Cox CV, Evely RS, Oakhill A, Pamphilon DH, Goulden NJ, Blair A. Characterization of acute lymphoblastic leukemia progenitor cell. Blood. 2004;104:2919–2925. doi: 10.1182/blood-2004-03-0901. [DOI] [PubMed] [Google Scholar]
- 9.George AA, Franklin J, Kerkof K, et al. Detection of leukemic cells in the CD34(+)CD38(-) bone marrow progenitor population in children with acute lymphoblastic leukemia. Blood. 2001;97:3925–3930. doi: 10.1182/blood.v97.12.3925. [DOI] [PubMed] [Google Scholar]
- 10.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 11.Sill H, Goldman JM, Cross NC. Homozygous deletions of the p16 tumor-suppressor gene are associated with lymphoid transformation of chronic myeloid leukemia. Blood. 1995;85:2013–2016. [PubMed] [Google Scholar]
- 12.Hernandez-Boluda JC, Cervantes F, Colomer D, et al. Genomic p16 abnormalities in the progression of chronic myeloid leukemia into blast crisis: a sequential study in 42 patients. Exp Hematol. 2003;31:204–210. doi: 10.1016/s0301-472x(02)01075-5. [DOI] [PubMed] [Google Scholar]
- 13.Mullighan CG, Williams RT, Downing JR, Sherr CJ. Failure of CDKN2A/B (INK4A/B-ARF)-mediated tumor suppression and resistance to targeted therapy in acute lymphoblastic leukemia induced by BCR-ABL. Genes Dev. 2008;22:1411–1415. doi: 10.1101/gad.1673908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharpless NE, Bardeesy N, Lee KH, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 15.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 16.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 17.Kamijo T, Zindy F, Roussel MF, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 18.Radfar A, Unnikrishnan I, Lee HW, DePinho RA, Rosenberg N. p19(Arf) induces p53-dependent apoptosis during abelson virus-mediated pre-B cell transformation. Proc Natl Acad Sci U S A. 1998;95:13194–13199. doi: 10.1073/pnas.95.22.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mostecki J, Halgren A, Radfar A, et al. Loss of heterozygosity at the Ink4a/Arf locus facilitates Abelson virus transformation of pre-B cells. J Virol. 2000;74:9479–9487. doi: 10.1128/jvi.74.20.9479-9487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randle DH, Zindy F, Sherr CJ, Roussel MF. Differential effects of p19(Arf) and p16(Ink4a) loss on senescence of murine bone marrow-derived preB cells and macrophages. Proc Natl Acad Sci U S A. 2001;98:9654–9659. doi: 10.1073/pnas.171217498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103:6688–6693. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams RT, den Besten W, Sherr CJ. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes Dev. 2007;21:2283–2287. doi: 10.1101/gad.1588607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Etten RA. Retroviral transduction models of Ph+ leukemia: advantages and limitations for modeling human hematological malignancies in mice. Blood Cells Mol Dis. 2001;27:201–205. doi: 10.1006/bcmd.2000.0370. [DOI] [PubMed] [Google Scholar]
- 24.Ponzetto C, Guerrasio A, Rosso C, et al. ABL proteins in Philadelphia-positive acute leukaemias and chronic myelogenous leukaemia blast crises. Br J Haematol. 1990;76:39–44. doi: 10.1111/j.1365-2141.1990.tb07834.x. [DOI] [PubMed] [Google Scholar]
- 25.Li S, Ilaria RL, Jr., Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 Forms of the BCR/ABL Oncogene Induce a Similar Chronic Myeloid Leukemia-like Syndrome in Mice but Have Different Lymphoid Leukemogenic Activity. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whetton AD, Graham GJ. Homing and mobilization in the stem cell niche. Trends Cell Biol. 1999;9:233–238. doi: 10.1016/s0962-8924(99)01559-7. [DOI] [PubMed] [Google Scholar]
- 27.Suda T, Arai F, Hirao A. Hematopoietic stem cells and their niche. Trends Immunol. 2005;26:426–433. doi: 10.1016/j.it.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 29.Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5:311–321. doi: 10.1038/nrc1592. [DOI] [PubMed] [Google Scholar]
- 30.Shank-Calvo JA, Draheim K, Bhasin M, Kelliher MA. p16Ink4a or p19Arf loss contributes to Tal1-induced leukemogenesis in mice. Oncogene. 2006;25:3023–3031. doi: 10.1038/sj.onc.1209326. [DOI] [PubMed] [Google Scholar]
- 31.Kohno T, Yamada Y, Tawara M, et al. Inactivation of p14ARF as a key event for the progression of adult T cell leukemia/lymphoma. Leuk Res. 2007;31:1625–1632. doi: 10.1016/j.leukres.2006.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.