Abstract
Most of the hypodermis of a rhabditid nematode such as Caenorhabditis elegans is a single syncytium. The size of this syncytium (as measured by body size) has evolved repeatedly in the rhabditid nematodes. Two cellular mechanisms are important in the evolution of body size: changes in the numbers of cells that fuse with the syncytium, and the extent of its acellular growth. Thus nematodes differ from mammals and other invertebrates in which body size evolution is caused by changes in cell number alone. The evolution of acellular syncytial growth in nematodes is also associated with changes in the ploidy of hypodermal nuclei. These nuclei are polyploid as a consequence of iterative rounds of endoreduplication, and this endocycle has evolved repeatedly. The association between acellular growth and endoreduplication is also seen in C. elegans mutations that interrupt transforming growth factor-β signaling and that result in dwarfism and deficiencies in hypodermal ploidy. The transforming growth factor-β pathway is a candidate for being involved in nematode body size evolution.
Do large animals have more cells than small animals, or do they have a similar number of larger cells? Studies done a century ago showed that the cells of most mammalian tissues were much the same size whether taken from an elephant or a mouse (1, 2). In 1912, Conklin (3) showed that snails of the genus Crepidula had identical cell sizes, regardless of adult body size. By 1925, E. B. Wilson, summarizing these and other studies in The Cell (4), was able to conclude that variation in body size is explained by cell number alone; his successors have generally agreed (5–7).
They must surely be largely correct. It is difficult to see how increases in cell size could account for much of the vast gulf that separates the mymarid wasps from the balaenopteran whales five orders of magnitude longer (6, 8). Even so, it remains possible that cell size does influence body size in smaller taxa such as nematodes and insects. Recent studies have shown that cell size accounts for much body size evolution among populations and species of Drosophila (9, 10).
In this paper we study the cellular basis of body size evolution in the nematode order Rhabditida. Of the 12 species we study, 8 belong to the family Rhabditidae, among them Caenorhabditis elegans; they vary between 0.5 and 3.0 mm in length or 100-fold in volume. We focus on the hypodermis, which covers the entire worm, secretes the exoskeletal cuticle, and is likely the single most important organ in the developmental control of body size. In all these species, most of the hypodermis is a single large syncytium (hyp7 in C. elegans) which grows as a succession of cells fuse with it. These are daughters of the lateral seam cells (H, V, and T cells in C. elegans), which divide only during larval growth (11–14). Body size in nematodes might evolve by changes in the complexity of the H, V, and T lineages so that, by maturity, large species have more hypodermal nuclei than smaller species. Indeed, the nematodes that we study here vary 4-fold in hypodermal nuclear number (15).
But growth in nematodes probably does not depend upon cell proliferation alone. During larval growth the nuclei embedded within the hypodermis endoreduplicate so that by late L4 they are tetraploid (16). The DNA content of the adult C. elegans hypodermis is a function of both the number of nuclei that fuse with it and their ultimate ploidy. Later, the mature C. elegans continues to grow in the absence of cell divisions, albeit at gradually diminishing rate, to give the S-shaped growth curve of a typical nematode (17, 18). Perhaps body size evolution depends on the extent of endoreduplication of hypodermal nuclei, so that large worms have highly polyploid nuclei, whereas small worms do not. Alternatively, the DNA content of hypodermal nuclei might not evolve by endoreduplication (which is confined to somatic tissues), but by changes in haploid genome size (as seen in germ cells). Genome size is known to be positively correlated with cell size in many taxa (19–24), and a syncytium is arguably just a special kind of cell. Here we investigate the relative roles of cell proliferation, endoreduplication, and haploid genome size in the evolution of nematode body size.
Materials and Methods
Strains and Culture Conditions.
We studied free-living terrestrial nematodes belonging to three families: Rhabditidae, Cephalobidae, and Panagrolaimidae in the order Rhabditida. The wild-type C. elegans N2 strain used in this study was derived from the Caenorhabditis Genetics Center (Minneapolis) in 1996 and has been kept in continuous culture in the Leroi laboratory since; it is designated RX008 to distinguish it from other N2s. Other species were Acrobeloides nanus (DF5047), Acrobeloides maximus (DF5048), Panagrellus redivivus (MT8872), Panagrolaimus rigidus (AF36), Rhabditoides regina (DF5012), Pellioditis sp. (EM434), Pellioditis typica (DF5025), Rhabditella octopleura (DF5044), Oscheius myriophila (BW290), Oscheius dolichuroides (DF5018), and Oscheius sp. (DF5000). The following mutant C. elegans strains were used: LGIII: daf-4 (m63), sma-2 (e502), dpy-2 (e8); all are loss-of-function mutations (25–27). All strains were cultured on agar plates seeded with Escherichia coli (OP50) and incubated at 20°C. (28).
Morphometrics.
Growth curves were determined for each strain from worms grown individually on 5-cm Petri dishes. Worms were measured at 8-h intervals after hatching until maturity and then every 24 h until death (sample size n = 25). Images were captured at ×50 (Wild dissecting microscope) by using a video camera (JVC KY-F50E) attached to a Power Macintosh running nih image 1.62. Length and area of each image were determined by using object image 1.62; volume was estimated assuming cylindrical body shape. Maximum volume was estimated from a logistic function fitted to each growth curve by least-squares nonlinear regression. Age at maturity was taken as age at final molt as scored by vulval eversion. Maximum body size of mutant strains was determined 96 h after hatching (n = 20).
Cellular Analysis.
For hypodermal cell counts, young adult worms were anesthetized with 1% sodium azide, mounted as for cell lineage determination (11), and viewed at ×1000 under differential interference contrast optics with a Nikon Eclipse E600 microscope. Images were captured with a CV-M300 camera attached to a Power Macintosh running nih image 1.62 and reconstructed by using Adobe PhotoShop 4.0. For comparisons among species, all nuclei, except neuronal nuclei, between the posterior pharyngeal bulb and anus were counted; for comparisons among mutants, all nuclei between the mouth and tail-tip, except neuronal nuclei, were counted. Bilateral symmetry was assumed and counts are on one side of each animal only. n = 10–13 for each species and mutant strain.
DNA content was determined by microdensitometry. Upon completing growth, worms were fixed in Carnoy's solution for a minimum of 24 h, stained in a 0.007 mg/ml solution of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (28), and viewed with a Leitz epifluorescence microscope. Images of nuclei were collected by using a CV-M300 video camera, and a Scion LG3 framegrabber mounted in a Power Macintosh running scion image 1.62a. On average, 17 nuclei per worm were measured; n = 6–10 for each species; n = 16–22 for each mutant strain.
Haploid genome sizes were estimated, using DAPI-based densitometry, from sperm taken from freeze-fractured nematodes. For the two Acrobeloides species, which are parthenogenetic and so have no sperm, we inferred a haploid value from a variety of neuronal cells associated with the pharynx, the tail, and the ventral cord. These are the smallest nuclei within the organism and were therefore assumed to be diploid, as they are in C. elegans (16). DNA contents were converted from pixels to megabases (Mb) by taking the haploid genome size of C. elegans, 97 Mb, as a standard (29). We confirmed the known linear relationship between DNA content and DAPI fluorescence (30) by using haploid and diploid cells from C. elegans N2 and from 4N C. elegans, SP346 (31). Estimates of DNA contents for the sperm of different species and the hypodermal nuclei of different C. elegans mutants were done twice; the correlation between blocks was 95% and 96%, showing the high repeatability of the technique and justifying the pooling of blocks.
Confocal images were taken with an MRC 600 and reconstructed by using VoxelView on a Silicon Graphics workstation.
Statistical Analysis.
The phylogeny is based on 18S rDNA data (15, 32, 33). Phylogenetic contrasts on log-transformed data were obtained from caic 2.0, assuming equal branch lengths (34, 35), and relationships between variables were tested by regression through the origin. Basic statistics were calculated by using jmp 3.2.2 (SAS Institute) or Excel 98 (Microsoft); unless stated otherwise, all analyses are on log-transformed data.
Results
Body Size, Cell Number, Haploid Genome Size, and Somatic Ploidy Have All Evolved Repeatedly in Rhabditid Nematodes.
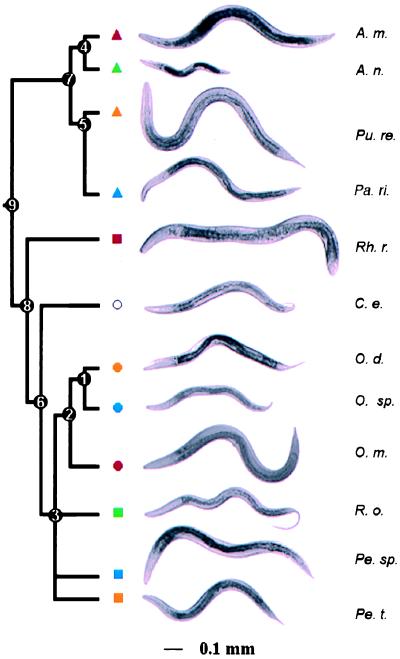
Fig. 1 shows the phylogenetic relationships of these species and demonstrates that body size has evolved repeatedly within several genera. Table 1 shows that haploid genome size, hypodermal nuclear number, and their degree of somatic polyploidization have also evolved repeatedly. We have not studied enough species, nor is the phylogeny of the Rhabditida sufficiently well known, for us to be able to determine the ancestral states for any of these traits or clades.
Figure 1.
Body size has evolved repeatedly within the Rhabditida. Species: Acrobeloides maximus (A. m.), Acrobeloides nanus (A. n), Panagrellus redivivus (Pu. re.), Panagrolaimus rigidus (Pa. ri.), Rhabditoides regina (Rh. r.), Caenorhabditis elegans (C. e.), Oscheius dolichuroides (O. d.), Oscheius sp. (DF5000) (O. sp.), Oscheius myriophila (O. m.), Rhabditella octopleura (R. o.), Pellioditis sp. (EM434) (Pe. sp.), and Pellioditis typica (Pe. t.). All species are depicted at young adulthood. Numbers correspond to nodes used in comparative contrasts analysis.
Table 1.
Body size and cellular nature of rhabditid nematode species
| Species | Body size, mm3 | Nucleus number | Genome size, Mb | Hypodermal DNA content, Mb | Hypodermal ploidy | % adult growth |
|---|---|---|---|---|---|---|
| A. m. | 0.0080 (±0.0003) | 216.1 (±4.6) | 237.9 (±52.7) | 1188.2 (±195.2) | 5.0 (±0.8) | 41.4 |
| A. n. | 0.0004 (±0.0001) | 69.2 (±1.4) | 125.9 (±10.0) | 686.0 (±138.9) | 5.4 (±1.0) | 36.6 |
| C. e. | 0.0050 (±0.0003) | 64.6 (±0.7) | 97.0 (±5.4) | 1037.6 (±71.7) | 10.7 (±0.7) | 57.2 |
| O. d. | 0.0037 (±0.0003) | 108.0 (±2.5) | 92.8 (±5.4) | 724.4 (±61.9) | 7.8 (±0.7) | 51.0 |
| O. sp. | 0.0016 (±0.0002) | 81.9 (±2.2) | 72.4 (±3.9) | 666.2 (±90.6) | 9.2 (±1.3) | 51.4 |
| O. m. | 0.0250 (±0.003) | 81.9 (±0.6) | 137.7 (±5.0) | 2054.4 (±648.5) | 16.6 (±3.4) | 87.5 |
| Pu. re. | 0.0097 (±0.001) | 245.9 (±18.2) | 147.3 (±12.8) | 822.8 (±103.5) | 5.5 (±0.7) | 81.4 |
| Pa. ri. | 0.0023 (±0.0004) | 202.4 (±2.1) | 200.9 (±17.3) | 906.7 (±145.9) | 4.5 (±0.2) | 49.7 |
| Pe. sp. | 0.0318 (±0.005) | 140.2 (±7.4) | 258.9 (±14.9) | 2202.4 (±230.6) | 8.5 (±0.9) | 89.4 |
| Pe. t. | 0.0029 (±0.001) | 67.3 (±1.1) | 240.3 (±9.9) | 1089.2 (±193.7) | 4.5 (±0.8) | 74.5 |
| R. o. | 0.0046 (±0.001) | 142.1 (±1.8) | 139.0 (±8.9) | 541.8 (±51.5) | 3.9 (±0.3) | 78.8 |
| Rh. r. | 0.0406 (±0.008) | 203.6 (±8.8) | 151.3 (±10.7) | 1651.7 (±160.2) | 10.9 (±1.1) | 79.1 |
Species are abbreviated as in Fig. 1. Body size and hypodermal DNA content are taken at age of maximal growth. Results are presented as mean with 95% confidence interval in parentheses.
The number of nuclei visible in the adult hypodermal syncytia ranges from 65 in C. elegans to 246 in Panagrellus redivivus; lineaging of several species has confirmed that all these cells, except for a few visible in the hatchling, are the product of the lateral seam cells, H, V, and T (12, 14). Haploid genome size, as measured by DNA densitometry, also varies 3-fold, with Oscheius sp. (DF5000) having the smallest genome, 72.4 (±3.9) Mb, and the two Pellioditis species having the largest at 258.9 (±14.9) Mb and 240.3 (±9.9) Mb (means and 95% confidence interval).
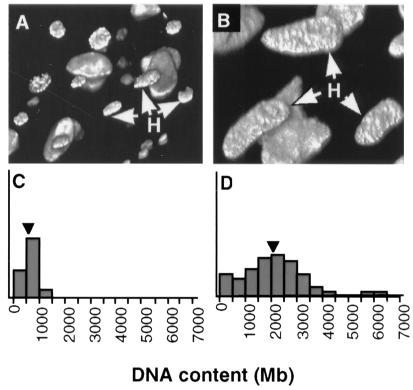
Independent of haploid genome size, the ploidy of hypodermal nuclei is also evolutionarily variable, and is most likely the result of varying numbers of rounds of endoreduplication. As discussed above, Hedgecock and White (16) showed that in C. elegans lateral seam cell descendants endoreduplicate once shortly after fusion with the hypodermal syncytium, giving 4C nuclei (C being the haploid DNA content). They did not follow endoreduplication beyond the late L4. We have found that by early adulthood (45 h) hypodermal nuclei have a mean ploidy of 8.0C (±0.8) and, by late adulthood (211 h) when the worm is fully grown, a mean ploidy of 10.7C (±0.7). This result implies that some, but not all, nuclei undergo two postlarval rounds of endoreduplication; however, some nuclei of even higher ploidy were observed. All species show at least some degree of polyploidization of hypodermal nuclei, but the extent varies from 3.9C (±0.3) in Rhabditella octopleura to 16.6C (±3.4) in Oscheius myriophila. In all species somatic polyploidization was taken to be due to endoreduplication, as no obvious chromosomal condensation was seen. The adult ploidy of hypodermal nuclei can differ dramatically even among congeners; Oscheius sp. (DF5000), for example, has a ploidy of 9.2C (±1.3), nearly half that of O. myriophila. Fig. 2 shows a confocal reconstruction of the nuclei of O. myriophila and O. sp. (DF5000). The difference in the final DNA content of the hypodermal nuclei is a consequence of difference between these species in both haploid genome size and the number of rounds of endoreduplication that the nuclei of these species undergo (Table 1). A characteristic of our data is that by late adulthood the endoreduplicate nuclei do not fall into clear polyploid series (2C, 4C, 8C, etc.). This could be because of measurement error, partial endoreduplication by the selective loss or amplification of some genome regions, or partial endoreduplication of the entire genome. The repeatability of our technique argues against measurement error (see above). As regards the second and third possibilities, we can exclude only chromosomal diminution of the sort found in ascarids because diploid larval nuclei have been detected in all species studied here (data not shown); chromosomal diminution has, in any case, not been found in C. elegans or in any other free-living nematode so far (36).
Figure 2.
Evolution of nuclear DNA content. Oscheius sp. (DF5000) (A and C) and O. myriophila (B and D) differ in adult hypodermal DNA content at late adulthood. (A and B) Reconstructions of nematode nuclei based on confocal images of whole-mounted nematodes; white arrows indicate hypodermal nuclei (surrounded by larger intestinal and smaller neuronal nuclei). (C and D) Distribution of densitometrically estimated DNA contents of hypodermal nuclei; black arrowheads are means.
Given this evolutionary variation in cell number, genome size, and ploidy, we now ask which of these variables explains the evolution of body size.
Cell Number and Somatic Ploidy Account for Body Size Evolution in Nematodes.
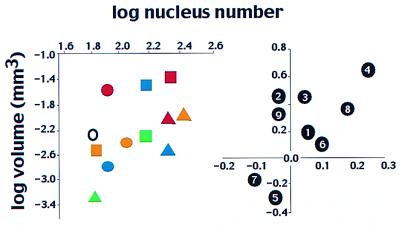
Fig. 3 shows the relationship across species (or independent contrasts) between body size and the number of hypodermal nuclei. Across species, hypodermal volume is not significantly correlated with nucleus number (r = 0.44; P = 0.15) but is across independent contrasts (P = 0.02). Examination of the data shows why this relationship is weak. Pellioditis sp. (EM434) with 140 nuclei is 13 times larger than Panagrolaimus rigidus with 202. Similar discrepancies between nucleus number and body size are found at lower taxonomic levels as well. O. dolichuroides with 108 nuclei is larger than O. sp. (DF5000) with 82, but smaller than O. myriophila, also with 82. We conclude that body size is partly independent of the number of hypodermal nuclei.
Figure 3.
The relationship between nucleus number and body size. (Left) Comparison of species means (log transformed). Hypodermal nuclear number at adulthood is from Cunha et al. (15); maximum body volume is at adulthood. (Right) Phylogenetic contrasts of the same data. See text for regression statistics. Species and contrasts as in Fig. 1.
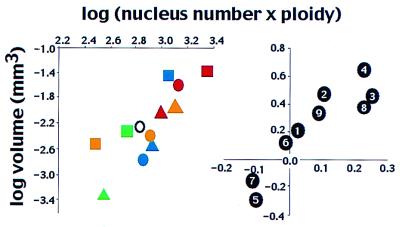
Can haploid genome size or hypodermal ploidy explain more of the variation in body size than nucleus number? At first glance it appears not, for neither is significantly correlated with body size. The correlation between genome size and body size is not significant (across species r = 0.35; P = 0.3; contrasts P = 0.1), nor is that between mean hypodermal ploidy and body size (across species r = 0.5; P = 0.1; contrasts P = 0.1). However, simultaneous analysis of all three variables shows the influence of at least two on body size. A backward stepwise multiple regression of nucleus number, genome size, and ploidy across species shows a significant effect of nucleus number (P = 0.02), genome size (P = 0.03), and ploidy (P = 0.0013); no higher-order interactions are significant. A similar analysis of the contrasts (multiple regression through the origin) shows a significant effect of nucleus number (P = 0.0006), ploidy (P = 0.0002), but not genome size (P = 0.98); of the higher-order interactions only nucleus number × ploidy was significant (P = 0.004). These analyses concur on the importance of nucleus number and ploidy, but not on that of genome size. Another way of assessing the joint importance of nucleus number and ploidy is illustrated in Fig. 4, which shows that body size is strongly correlated with their product (across species, r = 0.79; P = 0.002; across contrasts P = 0.0001). We conclude that body size evolution in nematodes is associated either with alterations in lateral seam cell lineages so as to give various numbers of hypodermal nuclei or else with variation of the ploidy of existing nuclei by varying the number of rounds of endoreduplication, or both.
Figure 4.
The relationship between the product nucleus number × ploidy and body size. (Left) Comparison of species means (log scale). (Right) Phylogenetic contrasts of the same data. See text for regression statistics. Species and contrasts are as in Fig. 1.
The partial association of endoreduplication with body size evolution suggests that variation in the extent of acellular syncytial growth may be an important mechanism of body size evolution. This can be demonstrated directly. The growth curve of any worm can be divided into larval and adult phases. Larval growth is due, at least in part, to cellular proliferation and fusion. But, as in C. elegans, hypodermal cell proliferation in all species studied here ceases at maturity. Three lines of evidence point to this. First, as in C. elegans, all nonneuronal lateral hypodermal cells fuse either with the hypodermal syncytium (hyp 7) or else with the seam syncytium formed at the L4 which, in turn, forms the lateral alae present in all species (data not shown). Second, mitotic chromosomal condensations have never been seen in the adult hypodermis of any species; instead all nuclei undergo at least some endoreduplication, suggesting that they have permanently exited the mitotic cell cycle. Finally, the lateral seam cell lineages (V cells) of 4 of the 12 species studied here have been directly lineaged (12), among them Panagrellus redivivus (15), a species that has a high nucleus number and is only a distant relative of C. elegans. In these 4 species, then, cell proliferation has been directly shown to end before the L4/adult molt. Given an absence of cell proliferation in the adult hypodermis, adult growth must be due entirely to the acellular growth of the syncytium. The percentage contribution of adult growth to final body size is shown in Table 1. Wild-type C. elegans, for example, has a maximum body size of 0.005 mm3, of which 42.8% occurs during embryogenesis and the four larval stages, whereas the remaining 57.2% occurs between maturity and death. In other species, the contribution of adult growth to final body size varies from less than 40% in the small species Acrobeloides nanus to nearly 90% in the large Pellioditis sp. (EM434). In general, the larger the species, the greater the contribution of adult growth to final body size. Regression shows that variation in adult growth explains nearly all of the variation among these species (86% across species; 85% contrast, not log transformed), larval growth explaining the remainder. Body size evolution in these nematodes occurs, then, mainly by altering the extent of adult, acellular, growth.
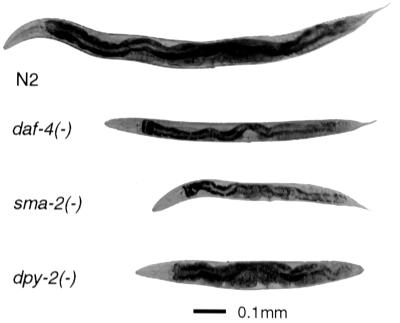
Some Dwarfism Mutants in C. elegans Affect Endoreduplication, but Others Do Not.
We examined three dwarfism mutants in C. elegans to determine the cellular basis of their small body size (Fig. 5). Two of these mutants, daf-4 and sma-2, encode proteins implicated in TGF-β signal transduction, respectively a type II serine/threonine kinase receptor and a Smad (25, 26); dpy-2, encodes a cuticle collagen (27). The sizes of adult daf-4 and sma-2 worms respectively are 27% and 13% of wild-type by volume, whereas dpy-2 is 40% of wild-type (Table 2). The mutants also differ in proportion, daf-4 and sma-2 being both shorter and thinner than wild-type, dpy-2 being mainly shorter. Hypodermal nuclear counts of the mutants showed little or no difference relative to wild-type (Table 2). This observation implies wild-type lateral seam cell lineages and suggests that the severe dwarfism of these mutants must be caused by a deficiency in cell or syncytial growth. We therefore examined hypodermal ploidy in these mutants. dpy-2 had hypodermal nuclei with a final wild-type ploidy of 9.5. daf-4 and sma-2, on the other hand, had mean final ploidies of 5.8 and 7.0. We conclude that dwarfism caused by disruption of the TGF-β pathway in C. elegans is, at least in part, associated with a deficiency of endoreduplication in the hypodermal nuclei, whereas dwarfism caused by a disruption of cuticle structure is not.
Figure 5.
Body size mutants in C. elegans, depicted at 96 h after hatching. daf-4 and sma-2 encode components of a transforming growth factor-β (TGF-β) signaling pathway; dpy-2 encodes a cuticle collagen.
Table 2.
Body size and cellular nature of dwarf C. elegans mutants
| Mutant | Body size, mm3 | Nucleus number | Hypodermal DNA content, Mb |
|---|---|---|---|
| N2 RX008 | 0.0062 (±0.0003) | 76.3 (±0.5) | 917.0 (±18.4) |
| daf-4 (m63) | 0.0017 (±0.0001) | 75.4 (±0.2) | 565.3 (±64.1) |
| sma-2 (e502) | 0.0008 (±0.0001) | 73.6 (±0.7) | 680.3 (±52.8) |
| dpy-2 (e8) | 0.0025 (±0.0002) | 74.2 (±0.8) | 918.0 (±53.5) |
Body size and hypodermal DNA content are taken at age of maximal growth. Results are presented as mean with 95% confidence interval in parentheses.
Discussion
To understand the cellular, and ultimately the molecular, basis of body size evolution we have chosen to study the hypodermis of a group of free-living nematodes closely related to C. elegans. The hypodermis is unusual among metazoan tissues in that it is a syncytium. This syncytium grows in part by the successive fusion of cells produced by the lateral seam. The number of cells that fuse with this syncytium (and which remain visible as embedded nuclei) varies widely among species. We have shown that this number is a poor predictor of adult body size; the remaining variation must be due to either the size of the cells that fuse or differences in syncytial growth independent of cell fusion. As such, our study is one of very few to have demonstrated a role for a process other than cell number in the evolution of body size. Perhaps our results are not surprising. All of the species studied have a considerable amount of postlarval growth, and none of this can be due to cell fusion, but must be simply due to the syncytium's own efforts; we have shown that 86% of the variation in final body size is due to this final phase of growth. The importance of acellular growth in body size evolution is probably not unique to nematodes. The larval tissues of dipterans grow by increases in cell size (37), and the difference in body size between the larva of Drosophila melanogaster and those of the giant Hawaiian fruit flies may well be because of differences in the extent of acellular growth. Indeed, in Drosophila, cell number does not explain all of the body size variation even in cellular organs such as wings (9, 10). Besides dipterans, our findings are also likely to be relevant to many small-bodied taxa such as acoels, rotifers, gastrotrichs, acanthocephalans, and neodermatan platyhelminthes, all of which have syncytial organs, and some of which are thought to grow largely in the absence of cell proliferation (38–40).
How do nematodes accommodate hypodermal evolution in the absence of changes in cell number? We have shown that, as in C. elegans, the hypodermal nuclei of all of the species studied show at least some polyploidization by endoreduplication, that the degree of endoreduplication varies widely among nematodes species, and that the final ploidy of the hypodermal nuclei, together with their number, accounts for most of the variation in final body size. Correlations among traits across divergent populations, species, or higher taxa generally lend themselves to one of two kinds of explanation: hypotheses of correlated selection and hypotheses of pleiotropy (41, 42). Correlated selection hypotheses suppose that the traits in question have no direct mechanistic connection, but have evolved in tandem because of parallel or common selective pressures. For example, it could be that when a population comes under selective pressure for a change in body size, hypodermal volume evolves initially without alteration in its DNA content, and that only later does a functionally optimal nucleocytoplasmic ratio evolve. Pleiotropy hypotheses, on the other hand, suppose a developmental connection either direct or otherwise between hypodermal volume and DNA content such that variant alleles that alter one alter the other and, furthermore, that this connection has been the primary influence in shaping the parallel evolution of these traits.
Because the selective forces that act upon body size and the hypodermal DNA content in these free-living nematodes are unknown and likely to remain so, we cannot provide strong evidence for or against either of these kinds of hypotheses. However, we can ask whether there is any genetic evidence for the kind of mechanistic connections supposed by the pleiotropy hypothesis. We have shown that loss-of-function mutations in two genes in the TGF-β pathway, daf-4 and sma-2, result in both dwarfism and reduced endoreduplication of hypodermal nuclei so that the final hypodermal DNA content of mutant worms is 60–75% of wild-type even as nuclear number is constant or nearly so. In other words, we have identified a signaling pathway of precisely the type supposed by the pleiotropy hypothesis, that is, one that influences both endoreduplication and body size simultaneously. Indeed, among the dozens, perhaps hundreds, of C. elegans genes that influence body size (Z.-Z.S., M. N. Patel, and A.M.L., unpublished data), those of the TGF-β pathway must be considered strong candidates for being causally involved in body size evolution. As the dpy-2 cuticle collagen-defective mutant shows, low hypodermal ploidy is not an inevitable consequence of dwarfism, but is, rather, something unique (so far) to the TGF-β pathway. We have not examined mutations in dbl-1, sma-6, sma-3, or sma-4, respectively a TGF-β-like ligand, type I receptor, and two SMAD proteins which also act in the C. elegans growth control pathway (26, 43–45); they are all grossly similar to daf-4 and sma-2 and presumably are also endoreduplication deficient.
Our finding of low somatic ploidy in daf-4 and sma-2 is of interest for another reason. Although these and other TGF-β pathway mutants have been known for some years (25, 26), beyond the observation that cell numbers of mutant worms are no different from wild type, the cellular basis of their dwarfism has proved, so far, obscure (41–43). Endoreduplication and somatic polyploidy are commonly associated with increases in cell size in many taxa (19). The causal basis of this association remains unclear, but it has also long been noted that germ-line polyploidy also typically leads to increases in cell size (19–23). Insofar that the hypodermis is central to the growth of the worm (covering the entire body and manufacturing the cuticular exoskeleton) it is possible that daf-4 and sma-2 worms are dwarf because they have low somatic ploidy. Against this hypothesis, it should be noted that the daf-4 and sma-2 mutants are probably dwarfed in tissues in which endoreduplication has not been observed [e.g., ventral nerve cord (16)], although this may be a secondary consequence of a primary hypodermal deficiency. In affecting cell (or syncytium) size, the TGF-β-like pathway in C. elegans partly has some similarities to the role of its Drosophila homologue (including dpp, tkv, and punt), which influences the size of the wing imaginal disk partly by cell size (46, 47). The Drosophila pathway is different from that of C. elegans in that its influence on cell size occurs in mitotically active (rather than endoreduplicating) cells, indeed, dpp is best known for influencing cell proliferation (47). In Drosophila, adult organ and cell size are also controlled via an insulin-like pathway (48–50); because this pathway influences the size of larval tissues it would be interesting to know whether it affects their ploidy as well.
Our finding that haploid genome size has at best a weak effect on body size is surprising. Many studies have shown that cell size among species is strongly positively correlated with genome size (24, 51–53), and may, at least in Amphibia, affect organ complexity and size (8, 53–55). But the positive relationship between genome and cell size, although widespread, is not universal. Recent (mutant) polyploid fish strains have large cells relative to their diploid relatives (56, 57), but ancient polyploid fish species frequently do not, apparently having down-regulated cell size in the course of evolution, while maintaining large genomes (57). Perhaps a similar disassociation between haploid genome size and cell size, and hence body size, has occurred in nematode evolution.
We have shown that nematodes may, in evolution, increase the DNA content of their hypodermal syncytia by either increasing the number of nuclei (by evolution of seam cell lineages) or increasing the ploidy of those nuclei (by evolution of endoreduplication). Why should one mechanism be utilized over the other? We suspect that endoreduplication is most commonly used in the evolution of extensive postlarval growth. We have studied only relatively small (<3 mm long) free-living secernentean nematodes. Such worms are thought to resemble the ancestors of mammalian parasites such as Ascaris (32), whose large size is achieved mainly by postlarval growth (18). In ascarids, unlike the nematodes studied here, adult growth is associated with continued cell proliferation as well as massive somatic polyploidy (58). Perhaps the mechanisms that we have identified here contribute to the evolution of nematodes such as Placentonema gigantissima, a parasite of the sperm whale, which at 8 m is, among invertebrates, second in length only to Architeuthis, the giant squid (59).
Acknowledgments
We thank Dave Hall for insight into nematode growth, Ricardo Azevedo for statistical advice, Michael Cammer of the Imaging Facility at Albert Einstein College of Medicine for the confocal reconstructions, and L. Carta, P. De Ley, D. Fitch, B. Robertson, B. Sohlenius, J. Vanfleteren, and W. Wood for kindly providing strains, as well as the Caenorhabditis Genetics Centre, which is funded by the National Institutes of Health National Center for Research Resources. We thank two anonymous referees for useful comments. This work was supported by the National Science Foundation (U.S.A.), the Biotechnology and Biological Sciences Research Council (U.K.), the Natural Environment Research Council (U.K.), the Science and Technology Foundation (Portugal), and the Royal Society (U.K.).
Abbreviations
- Mb
megabase(s)
- TGF-β
transforming growth factor β
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Levi G. Ergeb Anat Entwicklungsgesch. 1925;26:86–352. [Google Scholar]
- 2.Tessier G. Tabulae Biologicae. 1939;19:1–64. [Google Scholar]
- 3.Conklin E G. J Morph. 1912;23:159–188. [Google Scholar]
- 4.Wilson E B. The Cell in Development and Heredity. 3rd Ed. New York: Macmillan; 1925. pp. 97–99. [Google Scholar]
- 5.Thompson D W. In: On Growth and Form; abridged edition. Bonner J T, editor. Cambridge, U.K.: Cambridge Univ. Press; 1969. pp. 39–40. [Google Scholar]
- 6.Bonner J T. The Evolution of Complexity by Means of Natural Selection. Princeton, NJ: Princeton Univ. Press; 1989. p. 134. [Google Scholar]
- 7.Calder W A. Size, Function and Life-History. Mineola, NY: Dover; 1996. p. 87. [Google Scholar]
- 8.Hanken J, Wake D B. Annu Rev Ecol Syst. 1993;24:501–519. [Google Scholar]
- 9.Stevenson R D, Hill M F, Bryant P J. Proc R Soc London B. 1995;259:105–110. doi: 10.1098/rspb.1995.0016. [DOI] [PubMed] [Google Scholar]
- 10.James A C, Azevedo R B R, Partridge L. Genetics. 1997;146:881–890. doi: 10.1093/genetics/146.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sulston J, Horvitz H R. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 12.Sternberg P, Horvitz H R. Dev Biol. 1982;93:181–205. doi: 10.1016/0012-1606(82)90251-2. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V, Fixen W. In: Development as an Evolutionary Process. Raff R A, Raff E C, editors. New York: Liss; 1985. pp. 139–159. [Google Scholar]
- 14.Azevedo, R. B. R., Cunha, A., Emmons, S. W. & Leroi, A. M. (2000) Nematology, in press.
- 15.Cunha A, Azevedo R B R, Emmons S W, Leroi A M. Nature (London) 1999;402:253. doi: 10.1038/46211. [DOI] [PubMed] [Google Scholar]
- 16.Hedgecock E M, White J G. Dev Biol. 1985;107:128–133. doi: 10.1016/0012-1606(85)90381-1. [DOI] [PubMed] [Google Scholar]
- 17.Byerly L, Cassada R C, Russell R L. Dev Biol. 1976;51:23–33. doi: 10.1016/0012-1606(76)90119-6. [DOI] [PubMed] [Google Scholar]
- 18.Malakhov V V. Nematodes. Structure, Development, Classification and Phylogeny. Washington, DC: Smithsonian Inst. Press; 1994. [Google Scholar]
- 19.Brodsky V Y, Uryvaeva I V. Genome Multiplication in Growth and Development. Cambridge, U.K.: Cambridge Univ. Press; 1985. [Google Scholar]
- 21.Fankhauser G. Q Rev Biol. 1945;20:20–78. [Google Scholar]
- 22.Guo X, Allen S K. Genetics. 1994;138:1199–1206. doi: 10.1093/genetics/138.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galitski T, Sladanha A J, Styles C A, Lander E S, Fink G R. Science. 1999;285:251–253. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- 24.Szarski H. Int Rev Cytol. 1976;44:93–111. doi: 10.1016/s0074-7696(08)61648-4. [DOI] [PubMed] [Google Scholar]
- 25.Estevez M, Attisano L, Wrana J L, Albert P S, Massagué J, Riddle D L. Nature (London) 1993;365:644–649. doi: 10.1038/365644a0. [DOI] [PubMed] [Google Scholar]
- 26.Savage C, Das P, Finelli A L, Townsend S R, Sun C Y, Baird S E, Padgett R W. Proc Natl Acad Sci USA. 1996;93:790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy A D, Yang J, Kramer J M. Mol Biol Cell. 1993;8:803–817. doi: 10.1091/mbc.4.8.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sulston J, Hodgkin J. In: The Nematode, Caenorhabditis elegans. Wood W, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 587–606. [Google Scholar]
- 29.Emmons S W. In: The Nematode, Caenorhabditis elegans. Wood W, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. p. 53. [Google Scholar]
- 30.Hauser-Urfer I, Leemann U, Ruch F. Exp Cell Res. 1982;142:455–459. doi: 10.1016/0014-4827(82)90388-3. [DOI] [PubMed] [Google Scholar]
- 31.Madl J E, Herman R K. Genetics. 1979;93:393–402. doi: 10.1093/genetics/93.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blaxter M L, De Ley P, Garey J R, Liu L X, Scheldeman P, Vierstraete A, Vanfleteren J R, Mackey L Y, Dorris M, Frisse L M, et al. Nature (London) 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 33.Fitch D J, Bugaj-Gaweda B, Emmons S W. Mol Biol Evol. 1995;12:346–358. doi: 10.1093/oxfordjournals.molbev.a040207. [DOI] [PubMed] [Google Scholar]
- 34.Felsenstein J. Am Nat. 1985;125:1–15. [Google Scholar]
- 35.Purvis A, Rambaut A. Comput Appl Biosci. 1995;11:247–251. doi: 10.1093/bioinformatics/11.3.247. [DOI] [PubMed] [Google Scholar]
- 36.Tobler H, Etter A, Müller F. Trends Genet. 1992;8:427–432. doi: 10.1016/0168-9525(92)90326-y. [DOI] [PubMed] [Google Scholar]
- 37.Trager W. J Exp Zool. 1934;71:489–508. [Google Scholar]
- 38.van Cleave H. Q Rev Biol. 1932;7:59–67. [Google Scholar]
- 39.Brusca R C, Brusca G J. Invertebrates. Sunderland, MA: Sinauer; 1990. pp. 336–368. [Google Scholar]
- 40.Winnepenninckx B M H, Backeljau T, Kristensen R M. Nature (London) 1998;393:636–638. [Google Scholar]
- 41.Lauder G V, Leroi A M, Rose M R. Trends Ecol Evol. 1993;8:294–297. doi: 10.1016/0169-5347(93)90258-Q. [DOI] [PubMed] [Google Scholar]
- 42.Leroi A M, Rose M R, Lauder G V. Am Nat. 1994;143:381–402. [Google Scholar]
- 43.Krishna S, Maduzia L L, Padgett R W. Development (Cambridge, UK) 1999;126:251–260. doi: 10.1242/dev.126.2.251. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki Y, Yandell M D, Roy P J, Krishna S, Savage-Dunn C, Ross R M, Padgett R W, Wood W B. Development (Cambridge, UK) 1999;126:241–250. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- 45.Morita K, Chow K L, Ueno N. Development (Cambridge, UK) 1999;126:1337–1347. doi: 10.1242/dev.126.6.1337. [DOI] [PubMed] [Google Scholar]
- 46.Burke R, Basler K. Development (Cambridge, UK) 1996;122:2261–2269. doi: 10.1242/dev.122.7.2261. [DOI] [PubMed] [Google Scholar]
- 47.Su T T, O'Farrell P H. Curr Biol. 1998;19:R687–R689. doi: 10.1016/s0960-9822(98)70436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Böhni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss B F, Beckingham K, Hafen E. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- 49.Weinkove D, Neufeld T P, Twardzik T, Waterfield M D, Leevers S J. Curr Biol. 1999;9:1019–1027. doi: 10.1016/s0960-9822(99)80450-3. [DOI] [PubMed] [Google Scholar]
- 50.Conlon I, Raff M. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 51.Olmo E. Basic Appl Histochem. 1983;27:227–256. [PubMed] [Google Scholar]
- 52.Hughes A, Hughes M K. Nature (London) 1995;377:391. doi: 10.1038/377391a0. [DOI] [PubMed] [Google Scholar]
- 53.Roth G, Blanke J, Wake D B. Proc Natl Acad Sci USA. 1994;91:4796–4800. doi: 10.1073/pnas.91.11.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth G, Blanke J, Ohle M. Brain Behav Evol. 1995;45:84–95. doi: 10.1159/000113542. [DOI] [PubMed] [Google Scholar]
- 55.Roth G, Nishikawa K C, Wake D B. Brain Behav Evol. 1997;50:50–59. doi: 10.1159/000113321. [DOI] [PubMed] [Google Scholar]
- 56.Benfey T J. Rev Fish Sci. 1999;7:39–67. [Google Scholar]
- 57.Leipoldt M. Hum Genet. 1983;65:11–18. doi: 10.1007/BF00285022. [DOI] [PubMed] [Google Scholar]
- 58.Anisimov A P, Usheva L N. Soviet J Dev Biol. 1973;4:379–383. [PubMed] [Google Scholar]
- 59.Gubanov N M. C R Acad Sci URSS. 1951;77:1123. [PubMed] [Google Scholar]