Abstract
The gem turrids (genus Gemmula Weinkauff, 1875) are venomous snails in the family Turridae. A gene superfamily of disulfide-rich peptides expressed in Gemmula venom ducts was characterized. Gemmula speciosa (Reeve, 1843), venom duct cDNA clones revealed two different conotoxin-like prepropeptide precursors, with identical signal sequences, a largely conserved pro region, and a cysteine-rich C-terminal mature peptide region. The conserved signal sequence was used to successfully amplify homologous genes from three other Gemmula species; all had the same pattern of Cys residues in the predicted mature venom peptide. Although the signal sequence and propeptide regions were highly conserved, the mature toxin regions diverged greatly in sequence, except that the Cys residues were conserved. We designate this as the Pg-gene superfamily (Pg-superfamily) of Gemmula venom peptides. Purification of two members of the family directly from G. speciosa venom was achieved; amino acid sequence analysis revealed that these peptides are highly posttranslationally modified. With at least 10-fold as many species of turrids as cone snails, identification of rapidly diversifying gene superfamilies such as the Pg-superfamily of Gemmula is essential before the facile and systematic discovery and characterization of peptide toxins from turrid venoms can be achieved.
Keywords: Conoidea, Venom peptides, Exogene superfamilies, Turrids, Drug discovery
1. Introduction
There are >10,000 species of predatory marine snails that use venom as the primary weapon for prey capture; these are traditionally assigned to a single superfamily, Conoidea (=suborder Toxoglossa) (Kohn, 1998). Based on shell morphology, the superfamily is traditionally divided into three groups, cone snails, augers and turrids. Cone snails and augers are morphologically well defined; species in these groups are easily recognized through their distinctive shells (Fig. 1A). In most molluscan taxonomic treatments, cone snails are generally assigned to a single large genus, Conus, while augers are assigned by most taxonomists to several different genera (Terebra, Hastula, Duplicaria), in the family Terebridae (Bouchet and Rocroi, 2005, Kohn, 1998).
Fig. 1.
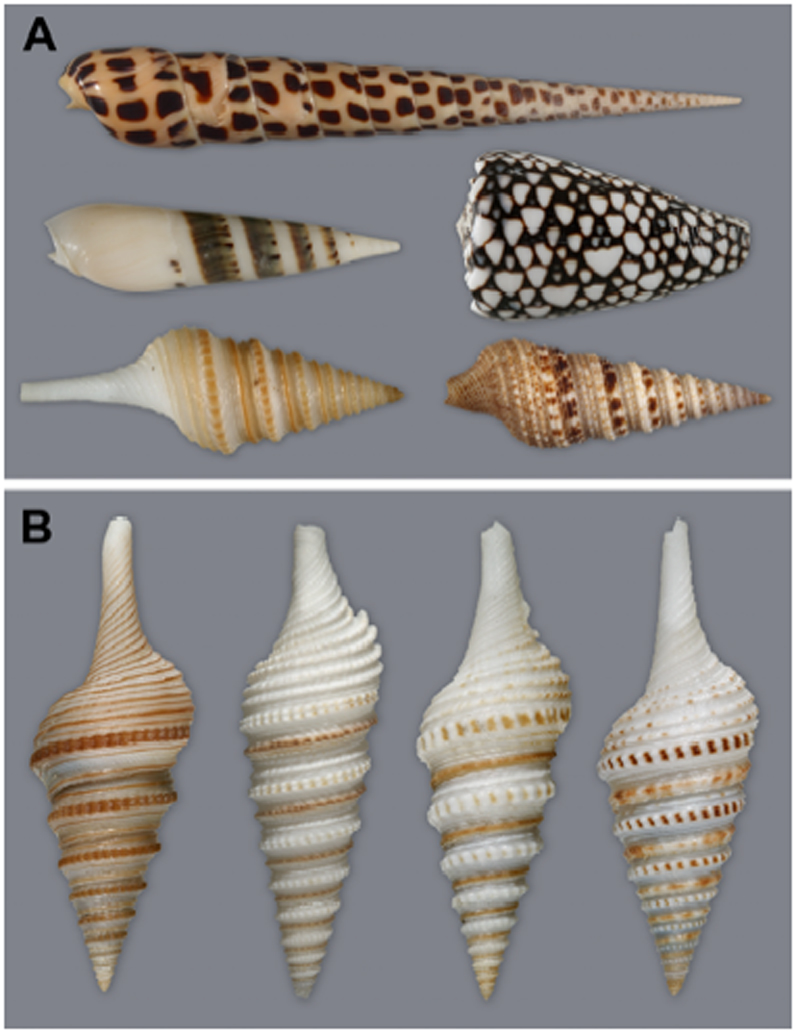
A. Shells of Conus, Terebra, Turrids. Top, Terebra subulata (Terebridae); Middle row: left, Hastula hectica (Terebridae); right, Conus marmoreus; Bottom row: left, Gemmula speciosa; right, Lophiotoma olangoensis (both Turridae).
B. Left to right: Gemmula speciosa, G. diomedea, G. sogodensis, and G. kieneri.
In contrast, the term “turrids” has been applied to a group that is much more diverse (Taylor et al., 1993); it has long been recognized that their biodiversity is greater than the two other groups (Powell, 1966, Powell, 1967). However, recent field work of Bouchet and co-workers in New Caledonia and the Philippines has established that the number of “turrid” species has been greatly underestimated and that the “turrids” comprise the vast majority of conoidean biodiversity (Bouchet et al., 2002). The deepwater habitat and/or small size of the majority of turrids has made them a significant component of molluscan biodiversity which has been largely neglected.
We have initiated a broad investigation of the constituents of the venoms of these marine mollusks, and are systematically analyzing representative venoms from each of the major Conoidean genera. Among the turrids, we have concentrated on the subfamily Turrinae (in the family Turridae).
In this work, we report a combined molecular biological/biochemical characterization of venom peptides from several species in the genus Gemmula (Weinkauff, 1875), the gem turrids. Gemmula are characterized by the gemmate sculpture on the peripheral cord of their shells (Fig. 1B), the primary criterion used for taxonomic assignment to the genus (Powell, 1967). In the classical taxonomy of turrids, Gemmula is one of the larger groups; Powell remarked on the extensive fossil record of the group and characterized it as “the most vigorous member of the (subfamily) Turrinae and undoubtedly represents the main stem of the subfamily”. Species in the genus are relatively large compared to most turrids, and like most turrid groups, Gemmula thrive mostly in the deeper waters of the tropics, primarily in the Indo-Pacific. In this study, we address whether Gemmula venoms are as complex as those of the cone snails, and whether the unprecedented rate of interspecific divergence repeatedly noted for cone snail peptides (Olivera, 2006) also occurs in Gemmula.
An important insight into how the impressive diversity of venom peptides was generated during the evolutionary history of the genus Conus is that most peptidic constituents of cone snail venoms are encoded by a relatively small number of rapidly diversifying gene superfamilies (Olivera, 2006). A major question we have addressed is whether similar gene superfamilies, characterized by their accelerated evolution, will also be found in Gemmula venoms.
A final notable characteristic of peptide toxins from Conus venoms is the high frequency of posttranslational modification found in the mature peptidic gene products. It is unknown how widespread these posttranslational modifications are within the superfamily Conoidea; preliminary evidence for the absence of most posttranslational modifications in one major clade of Conoidea has been obtained (Imperial et al., 2007). We demonstrate by purifying and characterizing venom peptides that, as in Conus, the peptide toxins of Gemmula venoms can be highly posttranslationally modified.
2. Materials and Methods
2.1 Specimen collection and RNA isolation
The snail samples were obtained by trawling between 50 –100 m off Manila Bay and through tangle nets between 300–500 m off the seas of Cebu and Bohol, Philippines. Live snails were kept in seawater until processed. Samples were dissected and their venom ducts immediately placed in RNAlater (Ambion, TX, USA) and pooled. The venom ducts (50–100 mg) were homogenized in 1 mL TRIzol reagent (Invitrogen Life Technologies, CA, USA) with a disposable microtissue grinder and the total RNA isolated by phase separation and precipitation following the manufacturer’s standard protocol.
2.2 cDNA preparation and sequencing
One microgram of total RNA was utilized as template for the first-strand cDNA preparation, followed by double-stranded cDNA synthesis and amplification using long-distance polymerase chain reaction and subcloning into pDNR-LIB using the Creator SMART cDNA Library Construction Kit (Clontech Laboratories, CA, USA). Recombinant plasmids were used to transform TOP10 (Invitrogen Life Technologies, CA, USA) electrocompetent cells and plasmid libraries were screened for 0.5–1.4 kb inserts by performing PCR directly on colonies using M13 forward and reverse primers provided by the vendor. Plasmids from selected colonies were purified (Wizprep, Promega, WI, USA) and sequenced using an ABI377 automated sequencer.
Toxin cDNA from other snail samples was obtained by PCR using the gene-specific primers, Ptx1gg01_5’ (5’-A(G/T)CGAAG(A/C)GCT(C/G)CATTCG-3’) and Ptx1gg01_3’ (5’-ATC(G/C)A(T/G)(C/T)GAT(C/A)TGTT(T/G)TG-3’) adapted with vector-specific sequences for cloning into pNEB206A linearized plasmid vector following the manufacturer’s standard protocol (USER Friendly Cloning Kit, New England Biolabs, MA, USA). The putative toxin gene was amplified from 2 μL dsDNA product as template in 20 μL PCR reaction mix containing 2 μL PCR buffer (10X), dNTPs (200 µM), primers (0.5 µM each), Taq DNA polymerase (1U) and diethylpyrocarbonate (DEPC)-treated water. The PCR profile used consisted of an initial denaturation of 1 min at 95 ° C, followed by 40 cycles of 20 sec at 95 ° C, 20 sec at 54 ° C and 30 sec at 72 ° C, and a final extension of 5 min at 72 ° C. The amplified product was subcloned into pNEB206A.
2.3 Purification of peptides from G. speciosa venom ducts
Venom ducts were dissected from live specimens of G. speciosa collected from the western Luzon, Philippines. Dissection was done on an ice block; the ducts were transported with dry ice, and stored at −70° C. Frozen venom ducts from 104 specimens were thawed by adding 1 mL of 40% acetonitrile with 0.1% trifluoroacetic acid (TFA), and the mixture homogenized in an Eppendorf tube with a Teflon pestle in 30 strokes. The homogenate was centrifuged in a Beckmann F650 rotor at 20,000 rpm for 15 min at 4 ° C. The supernatant was applied on an analytical C18 high-pressure liquid chromatography (HPLC) column (4.6 mm × 250 mm), and the venom components were separated through sequential elution with gradients of 4.5% to 54% acetonitrile - 0.1% TFA for 55 min, followed by 54% to 90% acetonitrile - 0.1% TFA for 8 min(Fig. 2, Panela). The number and masses of major components of the main peaks were estimated by matrix-assisted laser desorption ionization spectrometry (MALDI) (Peptide Biology Lab, Salk Institute, La Jolla California, USA). Peptidic components within 1–6 kDa were reduced and alkylated.
Fig. 2.
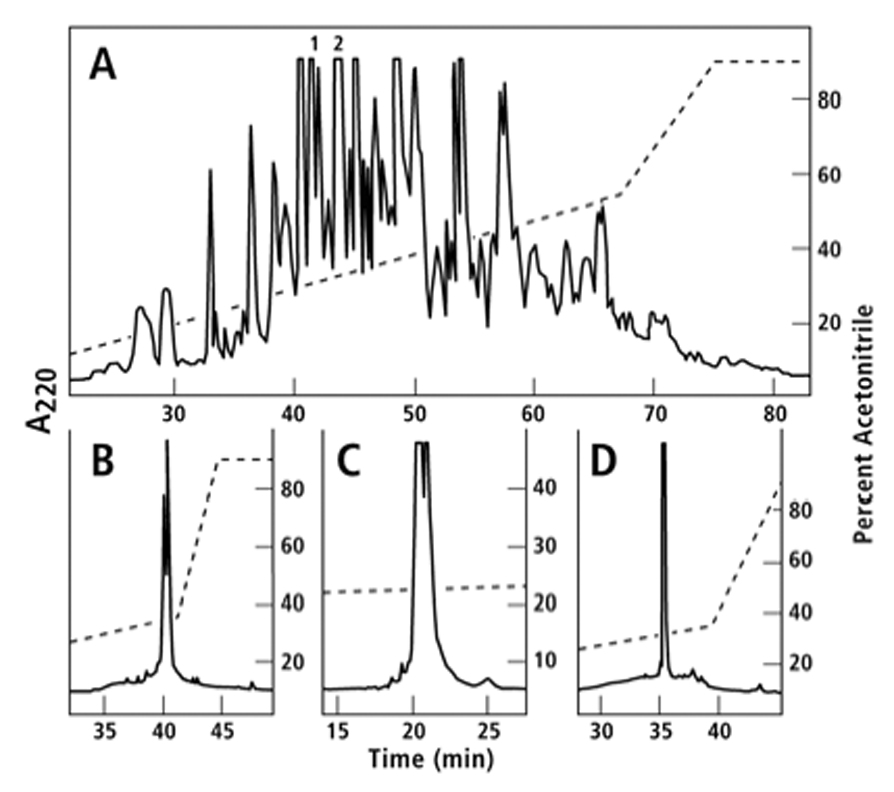
Purification of gsp9a and gsp9b. The components of the G. speciosa venom duct extract prepared as described under Methods were separated by sequential elution with gradients of 4.5% to 54% acetonitrile in 55 min and 54% to 90% acetonitrile in 8 min in the presence of 0.1% TFA. The absorbance profile at 220 nm is shown in Fig. 2, Panel A. An aliquot of Peak 1 in Panel A was reduced and pyridylethylated. The pyridylethylated mixture was fractionated on an analytical C18 column employing a gradient of 0.45% acetonitrile/min in 0/1% TFA, (Panel B). The more hydrophobic peak was determined as described in Methods to be gsp9a and the sequence is shown in Table 2. Peak 2 in Panel A was subfractionated on an analytical C18 HPLC column with a gradient of 0.18% acetonitrile/min in 0.1% TFA (Panel C). The components of the more hydrophobic peak were reduced, pyridylethylated, and then fractionated with a gradient of 0.45% acetonitrile/min in 0.1% TFA on an analytical C18 HPLC column (Panel D). The main peak in Panel D was determined by MALDI and amino acid sequencing to be gsp9b (Table 2).
An aliquot of Peak 1 (Fig. 2, Panel A) was reduced and the peptide components were pyridylethylated following the previously described protocol (Imperial et al., 2003). The pyridylethylated mixture was fractionated by HPLC on an analytical C18 column employing a gradient of 0.45% acetonitrile - 0.1% TFA per minute (Chromatogram is shown in Fig. 2 Panel B). The mass of the native peptide equivalent to the more hydrophobic component in Panel B was measured by MALDI (4071.5 Da) and the amino acid sequence was determined by Edman degradation. The peptide is named gsp9a and its sequence is shown in Table 2.
Table 2.
Predicted Mature Toxin Sequences of the Pg-superfamily (including posttranslational modifications)
 |
Peptides gsp9a and gsp9b were purified from G. speciosa venom; all other sequences are predicted from cDNA clones. All Pro and Glu residues are assumed to be posttranslationally modified to Hyp (4-hydroxyproline) and Gla (γ-carboxyglutamate), which are represented by O and γ respectively. Homologous amino acid residues in the native and predicted sequences are boxed.
Peak 2 (Fig. 2, Panel A) was subfractionated through an analytical C18 HPLC column on a gradient of 0.18% acetonitrile - 0.1% TFA per minute (Chromatogram is shown in Fig 2, Panel C). The components of the more hydrophobic peak in Panel C were reduced and pyridylethylated. The pyridylethylated mixture was eluted through a gradient of 0.45% acetonitrile - 0.1% TFA on an analytical C18 HPLC column (Panel D). The mass of the main peak in Panel D was determined by MALDI (4522.2 Da) and the amino acid sequence of the peptide (referred to as gsp9b), was obtained by Edman degradation.
Posttranslationally modified hydroxyproline residues were identified directly from the characteristic peaks in sequencing chromatograms while the posttranslational modification of glutamic residues to γ-carboxyglutamic acid was deduced when low glutamic acid signals were obtained in the respective cycles and confirmed by the equivalence of calculated and measured molecular masses.
3. Results and Discussion
3.1. Identification of toxin sequences by molecular cloning
Specimens of Gemmula speciosa (Reeve, 1843), the splendid turrid (Fig. 1) were dissected and cDNA from venom ducts prepared as described under Methods. Five out of 40 random clones sequenced from the cDNA gave conotoxin-like sequences; two of these showed considerable sequence identity (Table 1). The translated open reading frame encoded by these two cDNAs exhibit characteristics typical of the venom peptides from Conus: at the N-terminal end there is a signal sequence; at the C-terminal end, after a proteolytic processing at a predictable site (boxed), there is a cysteine-rich mature peptide sequence with an intervening region (the “pro” region) found between the signal sequence and the mature toxin regions. These two cDNA sequences, Gsp9.1 and Gsp9.2 have extensive sequence identity in the signal sequence and propeptide regions (they differ at only one position). However, the predicted mature peptide regions are much more divergent from each other than the N-terminal prepro sequences; although the Cys residues are completely conserved, almost half of the non-Cys residues (14/30) have diverged. This striking juxtaposition of highly conserved and hypervariable regions is characteristic of gene superfamilies encoding the toxin peptides found in Conus venoms, a feature repeatedly noted ever since the first conopeptide gene superfamily was characterized (Conticello et al., 2001, Olivera, 1999, Woodward et al., 1990).
Table 1.
Comparison of Pg and P-conotoxin superfamilies
| Designation | Precursor Sequence |
|---|---|
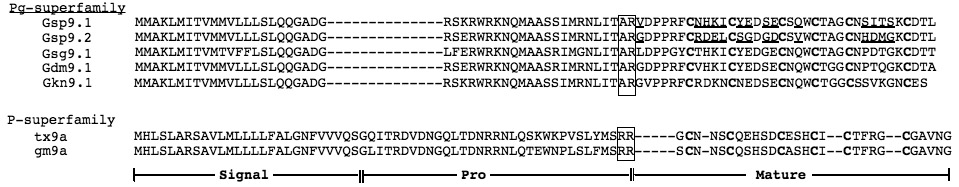 | |
Predicted proteolytic cleavage signals are boxed. Differences in the amino acid sequence between Gsp9.1 and 9.2 are underlined. The 6 Cys residues in the mature toxin presumably involved in disulfide cross-links are in bold type.
In the last 15 years, since the first Conus peptide was characterized directly from its gene sequence (Hillyard et al., 1992), the presence of conserved sequences in Conus gene superfamilies has led to a strategy for new peptide discovery using PCR. We explored using the same strategy for venom peptide discovery from Gemmula. We used conserved sequences from Gsp9.1 and 9.2 to design PCR primers that were used to scan for related venom peptide cDNAs from other Gemmula species. The results of the PCR analysis of mRNA isolated from three other Gemmula species, Gemmula sogodensis (Olivera, 2004), Gemmula diomedea (Powell, 1964), and Gemmula kieneri (Doumet, 1840) are shown in Table 1. An alignment of all five sequences shows a striking conservation of signal sequences and the same pattern of cysteine residues in all five mature toxin regions. Thus, as in Conus, once one member of a gene superfamily has been identified, the conserved signal sequence can be used to identify related toxin sequences that belong to the same superfamily from other species through the use of PCR primers based on the conserved sequences. A notable feature of the sequences in Table 1 is that the two sequences obtained from Gemmula speciosa (Gsp9.1 and Gsp9.2) are more divergent from each other than Gsp9.1 is from the sequences obtained from two of the other Gemmula species, Gsg9.1 (G. sogodensis) and Gdm9.1 (G. diomedea).
The results presented above provide the first compelling evidence for a rapidly diversifying gene superfamily that encodes peptide toxins in venom ducts of Gemmula, the gem turrids. Gemmula is one of the well-known genera in a major family of venomous molluscs, the Turridae. Gem turrids occur in deeper waters of tropical oceans, with the bulk of species found in the Indo-Pacific. A variety of criteria, including both morphological and molecular data, suggest that Gemmula and Conus are on branches of the Conoidean superfamily tree that diverged early in the evolutionary history of Conoidea; thus, Conus uses a hypodermic needle-like radular tooth to inject venom while Gemmula apparently stabs its prey and injects venom at the wound site (Gemmula sp. do not have hollow, hypodermic-type teeth). However, little is known regarding the detailed biology of any Gemmula.
This study demonstrates that the biosynthesis of Gemmula venom peptides is very similar to the pattern established for Conus: each toxin gene is translated as a prepropeptide precursor and proteolytic cleavage of the precursor releases the mature peptide toxin.
3.2 Biochemical purification of venom peptides
The venom ducts of Gemmula speciosa are extremely fine, and it was a challenge to obtain enough specimens to perform biochemical purification (a detailed account of the field collecting that had to be carried out is presented elsewhere – Heralde, et al. manuscript in preparation). We purified major components of the venom, and two peptides that are encoded by the gene superfamily characterized by molecular cloning described in the preceding section were identified. The purification of these two peptides is detailed in Fig. 2.
The purified peptides were analyzed utilizing Edman sequencing and mass spectrometry. The identity of the posttranslationally modified amino acids was established as outlined in Methods; the final sequence assignments are shown in Table 2. These sequences were compared to the mature sequences predicted from the putative precursors obtained by molecular cloning (Table 1).
The two peptides characterized from Gemmula speciosa venom, gsp9a and gsp9b appear to be polymorphic variants of the venom peptides encoded by the two clones from Gemmula speciosa, Gsp9.1 and Gsp9.2. A comparison of the peptide sequences of the components purified from venom with the predicted amino acid sequences from the clones reveals that the purified venom peptides are highly posttranslationally modified. Apparently, in peptides of this gene superfamily, proline is posttranslationally modified to 4-transhydroxyproline and glutamate residues are posttranslationally carboxylated to γ-carboxyglutamate. Both the final posttranslationally modified mature toxin sequences predicted from the clones, as well as the directly sequenced venom peptides are summarized in Table 2.
The sequences fall into two classes, those that display greater homology to gsp9a comprise a major class, and two peptides, gsp9b and its polymorphic variant encoded by Gsp9.2. The sequence from Gemmula kieneri, Gkn9.1, is quite divergent from the others. Although the physiological function of these peptides is unknown, a reasonable working hypothesis is that the different groups are differentially targeted (possibly to different molecular isoforms of homologous targets encoded by the same gene family), and that the predicted venom peptides from Gemmula sogodensis and Gemmula diomedea will have a physiological function homologous to that of gsp9a, but divergent from that of gsp9b. The results suggest that Gemmula venom peptides have good potential to be subtype-specific ligands, and be useful as are conotoxins to functionally characterize different ion channels and receptors in nervous systems.
Members of the gene superfamily directly purified from venom have proven to be highly posttranslationally modified. Two characteristic Conus posttranslational modifications, the hydroxylation of proline residues and the γ-carboxylation of glutamate (Buczek et al., 2005) were found in the Gemmula speciosa peptides directly isolated from venom — all of the proline and glutamate residues in gsp9a and gsp9b were posttranslationally modified. Thus, these unusual posttranslational modifications are not restricted to Conus and may be widely distributed across the superfamily Conoidea.
The arrangement of Cys residues in the primary sequence of the venom peptides gsp9a and gsp9b, is similar to that of the peptides from Conus in the P-superfamily. However, as is shown in Table 1, there is no obvious homology between the sequence of P-superfamily conopeptides and the presently characterized superfamily members from Gemmula. We propose to call this peptide superfamily the Pg-superfamily, in accordance with the nomenclature described in the Appendix. This superfamily of venom peptides was detected in all four Gemmula species investigated; how widely distributed the gene superfamily is within the family Turridae and across the superfamily Conoidea remains to be established.
As is evident from Fig. 2, the venom of Gemmula speciosa is comparable in complexity to Conus venoms. A much more extensive characterization of this venom is in progress (J. Imperial and B. Olivera unpublished results). Gemmula venoms have many of the characteristics now well established for Conus: venom constituents that are disulfide rich peptides, many with extensive posttranslational modifications. Initial mass measurements of native and reduced major peak components demonstrated the presence of peptides within the 1–6 kDa, the size range of most conotoxins. The results of this study suggest that the different constituents of these complex Gemmula venoms are likely to be generated by the rapid diversification of gene superfamilies such as the Pg-superfamily, the first gene superfamily shown to be expressed in Gemmula venom ducts. We note that the only previous study of any Gemmula venom yielded a peptide toxin that was strikingly different in its biochemical properties (Lopez-Vera et al., 2004).
The larger significance of establishing for the first time in the presence of rapidly diversifying gene superfamilies in the turrid genus Gemmula is that the general strategy used to discover, identify and characterize peptides from the venoms of Conus has been demonstrated to be applicable to turrids. An important component of the discovery strategy developed for Conus is the systematic examination of venom peptide sequences obtained by PCR amplification of gene superfamily homologs from different Conus species. We have provided proof-of-principle that this strategy is effective for the Gemmula Pg-superfamily; not only were we able to obtain five sequences by PCR amplification, but the availability of the gsp9a and gsp9b peptide sequences allows us to predict with some confidence which amino acid residues are posttranslationally modified.
Conus peptides have proven not only to provide ligands that are important tools in neuroscience, but compounds that are used for diagnostic and therapeutic purposes as well. One of the peptides found in Conus venoms is now Prialt, a commercial drug for intractable pain. There are ~ 70,000 venom peptides in the venoms of the 500–700 living Conus species; the total biodiversity of turrids is still undetermined, but present estimates indicate that there should be an order of magnitude more turrid species, thereby increasing the total pharmacological resource that can theoretically be accessed from the superfamily Conoidea by at least tenfold. The problem is that unlike Conus, turrids tend to be smaller, live in deeper water habitats (such as the Gemmula species described in this report) and have proportionally smaller venom ducts. However, the exogene-based discovery strategy developed for Conus (Olivera, 2006) makes such differences less of a barrier to discovery.
It would be virtually impossible to access enough venom from most turrid species to do traditional biochemical characterization of venom components. However, by defining gene superfamilies as has been accomplished here, even tiny and rare Gemmula species can now theoretically be examined for the members of the Pg-gene superfamily expressed in their venoms. Thus, the definition of venom constituents can bypass the tedious purification procedures involved in traditional discovery. Clearly, the Pg-superfamily will be only the first of the gene superfamilies that generate the chemical diversity found in turrid venoms.
Acknowledgements
This work was supported by grant GM48677 from the U.S. National Institute of General Medical Sciences. FMH would like to acknowledge PCASTRD-DOST and Cavite State University for his PhD Scholarship. We thank Carlo Lapid for his assistance in the preparation of G. speciosa cDNA library and Maren Watkins for annotating the cDNA library sequences.
Appendix
A proposal for the nomenclature of Conoidean venom peptides
In this manuscript, we adopt a nomenclature that we propose be applied to all Conoidean venoms. It builds on the widely used nomenclature for Conus peptides. We propose that the term conopeptide be generalized for peptidic constituents in all Conoidean venoms, not just for Conus.
In Conus, when the mechanism of action for a peptide is unknown, it is referred to by one or two lower case letters followed by an Arabic number and a small letter. Thus, the peptides might be initially called rg1a or r11c: the letters refer to the Conus species, while the Arabic number provides information about the pattern of cysteine residues in the primary sequence. Thus, rg refers to Conus regius, r to Conus radiatus; and the number 1 indicates the characteristic Cys pattern of the A-superfamily (CC---C---C) while 11 suggests the Cys pattern of the I-superfamily (C---C---CC---CC---C---C). The last lower case letter following the number simply refers to the order in which the peptides have been either characterized from the venom or synthesized. Thus, r11a, r11b and r11c all come from Conus radiatus with r11c the third peptide to be characterized in that series, (different in sequence from r11a and r11b, but sharing the same cysteine pattern).
We propose that basically the same nomenclature be used for all conoidean venoms, excepts that peptides that come from taxa other than Conus will have either three or four letters. In the present work, we use three letters, with the first letter always being G or g (for Gemmula), and the two following derived from the species name. Thus, we refer to the two peptides isolated from Gemmula speciosa venom as gsp9a and gsp9b.
Similarly, references to specific clones should follow the Conus nomenclature; in Conus, the first letter is capitalized and instead of using an Arabic number followed by a letter, the clones use the same number with the designation 1.1, 1.2, 1.3, etc. for those that encode peptides with Cys pattern 1 and 11.1, 11.2, 11.3, etc. for clones encoding peptides with Cys pattern 11. Thus, in the present work, we use this nomenclature, with the first letter capitalized, when clones are being referred to. The clones from Gemmula speciosa are designated Gsp9.1, Gsp9.2, while the clone from Gemmula sogodensis is Gsg9.1 and from Gemmula diomedea, Gdm9.1.
We further propose that superfamilies be referred to in the same manner as in Conus except that the name of the superfamily will now have a capital letter and a small letter with the small letter referring to the genus from which the superfamily was first characterized. Thus, in the present work, since the superfamily has a Cys pattern similar to that of the P-superfamily of Conus peptides, but it has been characterized from Gemmula, we refer to the gene superfamily as the Pg-superfamily.
We propose that the physiological mechanisms of actions, once clarified, use the same Greek letters as in the Conus nomenclature. Thus, if peptide gsp9a in the Pg-superfamily proves to be nicotinic receptor antagonist, then this will be referred to as α-conopeptide GspIXA (abbreviated as α-GspIXA). This reveals a nicotinic antagonist (“α”) from Gemmula speciosa (“Gsp”) that has the Cys pattern “C--C--C--C--C--C” (“1X”) characteristic of the P-like superfamilies. A second nicotinic antagonist in the same superfamily would be α-GspIXB; note that αGspIXA and αGspIXB may target different subtypes of nicotinic receptors.
Thus, all the conopeptides will have the same basic nomenclature, and the 106 diverse peptides from multiple genetic families with diverse structure, functional mechanisms and biological origins will be named to provide information about the species the conopeptide was derived from, structural information based on the Cys pattern, and a general mechanism of action (based on the Greek letter designation).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bouchet P, Lozouet P, Maestrati P, Heros V. Assessing the magnitude of species richness in tropical marine environments: high numbers of molluscs at a New Caledonia site. Biol. J. Linnean Soc. 2002;75:421–436. [Google Scholar]
- Bouchet P, Rocroi JP. Malacologia: International Journal of Malacology, Classification and nomenclature of gastropod families. ConchBooks; 2005. [Google Scholar]
- Buczek O, Bulaj G, Olivera BM. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 2005;62:3067–3079. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Gilad Y, Avidan N, Ben-Asher E, Levy Z, Fainzilber M. Mechanisms for evolving hypervariability: the case of conopeptides. Mol. Biol. Evol. 2001;18:120–131. doi: 10.1093/oxfordjournals.molbev.a003786. [DOI] [PubMed] [Google Scholar]
- Hillyard DR, Monje VD, Mintz IM, Bean BP, Nadasdi L, Ramachandran J, Miljanich G, Azimi-Zoonooz A, Mcintosh JM, Cruz LJ, Imperial JS, Olivera BM. A new Conus peptide ligand for mammalian presynaptic Ca2+ channels. Neuron. 1992;9:69–77. doi: 10.1016/0896-6273(92)90221-x. [DOI] [PubMed] [Google Scholar]
- Imperial J, Kantor Y, Watkins M, Heralde F, Stevenson BJ, Chen P, Ownby J-P, Bouchet P, Olivera BM. The Venomous Auger Snail Hastula (impages) hectica (Linnaeus, 1758): Molecular Phylogeny, Foregut Anatomy and Comparative Toxinology. Journal of Experimental Zoology, Part B. 2007 doi: 10.1002/jez.b.21195. In press. [DOI] [PubMed] [Google Scholar]
- Imperial JS, Watkins M, Chen P, Hillyard DR, Cruz LJ, Olivera BM. The augertoxins: biochemical characterization of venom components from the toxoglossate gastropod Terebra subulata. Toxicon. 2003;41:391–398. doi: 10.1016/s0041-0101(03)00169-7. [DOI] [PubMed] [Google Scholar]
- Kohn AJ. Superfamily Conoidea. In: Beesley PL, Ross GJB, Wells A, editors. Mollusca: Southern Synthesis. Fauna of Australia. Vol. 5. Vol. 5. Melbourne: CSIRO Publishing; 1998. pp. 846–854. Part A xvi 563 pp. [Google Scholar]
- Lopez-Vera E, Heimer de la Cotera EP, Maillo M, Riesgo-Escovar JR, Olivera BM, Aguilar MB. A novel structural class of toxins: the methionine-rich peptides from the venoms of turrid marine snails (Mollusca, Conoidea) Toxicon. 2004;43:365–374. doi: 10.1016/j.toxicon.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Olivera BM. Conus venom peptides: correlating chemistry and behavior. J Comp Physiol [A] 1999;185:353–359. doi: 10.1007/s003590050394. [DOI] [PubMed] [Google Scholar]
- Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281:31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- Powell AWB. The Molluscan Families Speightiidae and Turridae: An evaluation of the valid taxa, both Recent and fossil, with lists of characteristic species. Auckland, New Zealand: Unity Press Limited; 1966. [Google Scholar]
- Powell AWB. The family Turridae in the Indo-Pacific, Part 1a. The Turrinae concluded. Indo-Pacific Moll. 1967;1:409–444. [Google Scholar]
- Taylor JD, Kantor YI, Sysoev AV. Foregut anatomy, feeding mechanisms, relationships and classification of the Conoidea (=Toxoglossa) (Gastropoda) Bull. nat. Hist. Mus. Lond. (Zool.) 1993;59:125–170. [Google Scholar]
- Woodward SR, Cruz LJ, Olivera BM, Hillyard DR. Constant and hypervariable regions in conotoxin propeptides. EMBO J. 1990;1:1015–1020. doi: 10.1002/j.1460-2075.1990.tb08204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


