Abstract
Islet1 (Isl1) is a LIM homedomain protein that plays a pivotal role in cardiac progenitors of the second heart field. Here, lineage studies with an inducible isl1-cre demonstrated that most Isl1 progenitors have migrated into the heart by E9. Although Isl1 expression is downregulated in most cardiac progenitors as they differentiate, analysis of an isl1-nlacZ mouse and coimmunostaining for Isl1 and lineage markers demonstrated that Isl1 is expressed in distinct subdomains of the heart, and in diverse cardiovascular lineages. Isl1 expression was observed in myocardial lineages of the distal outflow tract, atrial septum, and in sinoatrial and atrioventricular node. The myocardialized septum of the outflow tract was found to derive from Isl1 expressing cells. Isl1 expressing cells also contribute to endothelial and vascular smooth muscle lineages including smooth muscle of the coronary vessels. Our data indicate that Isl1 is a specific marker for a subset of pacemaker cells at developmental stages examined, and suggest genetic heterogeneity within the central conduction system and coronary smooth muscle. Our studies suggest a role for Isl1 in these distinct domains of expression within the heart.
Keywords: Cardiac progenitor, Lineage, Isl1, pacemaker, Coronary smooth muscle, Tamoxifen, Inducible Cre
Introduction
Previous studies of the LIM homeodomain transcription factor Isl1 in cardiac progenitors determined that Isl1 expression both marked and was required within a subset of undifferentiated cardiac progenitors which would substantially contribute to the developing heart (Cai et al., 2003). A majority of cells within the outflow tract, right ventricle and right atria, and a portion of cells within left ventricle and left atria derived from Isl1 expressing progenitors. These results demonstrated that the previously described secondary or anterior heart fields which give rise to outflow tract, or outflow tract and right ventricle, respectively (Abu-Issa et al., 2004; Kelly and Buckingham, 2002; Mjaatvedt et al., 2001; Waldo et al., 2001), were a subset of Isl1 progenitors. Retrospective clonal lineage analysis performed in mouse embryos was consistent with there being two cardiac lineages, the first and second lineage, which derive from the first and second heart fields, respectively (Buckingham et al., 2005; Meilhac et al., 2004). The second heart field corresponds to the domain marked by isl1 expression.
Original isl1 lineage studies were performed with constitutive isl1-cre mouse lines (Park et al., 2006; Srinivas et al., 2001; Yang et al., 2006). Isl1 lineages were observed in myocardium, endocardium and endothelium of the aorta (Cai et al., 2003), suggesting that Isl1 is expressed in a number of distinct cardiovascular lineages. Initial lineage studies with an inducible isl1-cre in postnatal heart demonstrated persistent expression of isl1 in cells within distinct subdomains of the heart (Laugwitz et al., 2005). Analysis of isolated, cultured isl1-expressing cells from postnatal heart demonstrated that these cells did not express markers of cardiomyocyte differentiation, but could be induced to differentiate to myocyte lineages with high efficiency (Laugwitz et al., 2005).
The timing of isl1 progenitor migration into the heart and identification of lineages within the heart exhibiting Isl1 expression remain to be explored. Here, we have performed experiments with an inducible isl1-cre mouse line to investigate the timing of isl1-progenitor migration into the forming heart. We have also examined domains of Isl1 expression utilizing a previously undescribed isl1-nlacZ knock-in mouse line, and performed co-immunostaining for Isl1 and various cardiovascular lineage markers to identify cell types exhibiting Isl1 expression within the heart.
Materials and Methods
Generation of mice
Isl1 nuclear LacZ knock-in mice
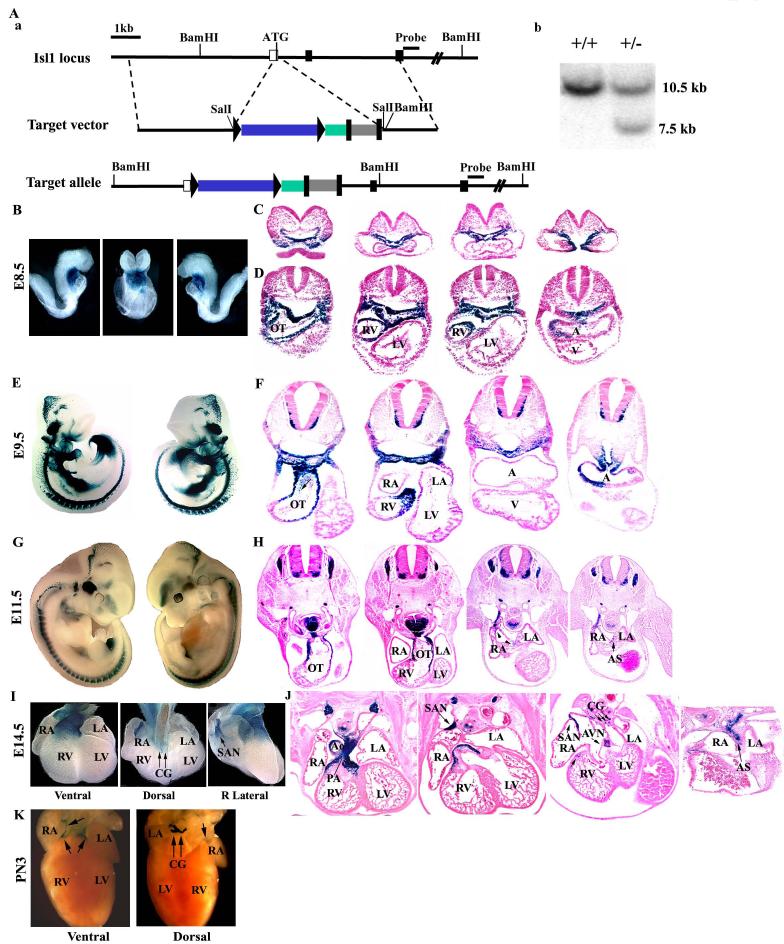
To study the expression pattern and role of Isl1 in mouse development, we generated an Isl1 nuclear LacZ (nLacZ) knock-in mouse line and an isl1 inducible Cre (MerCreMer) mouse line. 129/SV ES cell genomic DNA was used to PCR amplify a 4.9-kb 5' homologous arm containing isl1 untranslated region of exon 1 and a 3.6-kb 3' homologous arm containing exon 2 and part of exon 3. A short coding sequence in exon 1 including ATG would be deleted upon homologous recombination. To construct Isl1 nLacZ knock-in targeting vector, a Sall DNA cassette containing the LoxP-floxed nLacZ followed by hrGFP and FRT-floxed neomycin resistant gene (FRT-mclNeo) were inserted between 5' and 3' homologous arms at Sall site. A BamHl site was introduced at 5' end of 3' arm to discriminate wild type and targeted allele in southern blot. The targeting vector was linearized with Ascl and electroporated into mouse embryonic stem (ES) cells. The DNAs from neomycin resistant ES cell clones were digested with BamHl and screened for recombinant clones by southern analysis using a DNA probe immediately distal to the 3' arm. Wild type allele gave rise to a 10.5kb band and targeted allele gave rise to a 7.5kb band. Two recombinant clones were used for the blastocyst injection and chimera mice were crossed to C57BL/6J females to generate heterozygous Isl1 nLacZ knock-in mice.
Isl1-MerCreMer mice
Isl1 inducible Cre (MerCreMer) mice were generated using the same strategy as that of isl1-nLacZ knock-in, except that a knock-in cassette composed of MerCreMer followed by FRT-mclNeo was introduced at Sall site.
HCN4-H2B-EGFP knock-in mice
Generation and characterization of HCN4-H2B-EGFP knock-in mice will be published elsewhere. Briefly, the targeting cassette containing histone 2B fused EGFP (H2B-EGFP) (a gift from Kat Hadjantonakis) following FRT-mclNeo gene was introduced by homologous recombination into HCN4 locus just before HCN4 translation initiation site. Initial characterization of the mouse line demonstrated that EGFP was expressed in the region of cardiac conduction system and in a pattern similar to that published (Garcia-Frigola et al., 2003).
Tamoxifen injection
To induce MerCreMer translocation to the nucleus, pregnant mice were given a single intraperitoneal injection of tamoxifen (sigma) at a dose of 50-75mg/kg at desired time points (Xu et al., 2005; Zhang et al., 2006). For earlier embryos (E6.5 and E 7.5), we used low dose to avoid tamoxifen toxicity. Embryos were harvested 48 hours after injection and proceeded to X-gal staining or immunostaining.
All the experiments involving mice were carried out according to a protocol reviewed and approved by the Institutional Animal Care and Use Committee of UCSD, in compliance with the USA Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Embryo dissection and X-gal staining
Females with copulation plugs were considered to be at embryonic development day 0.5 (E0.5) of gestation. Pregnant females were sacrificed at different time points of gestation, and embryos were dissected from maternal tissue, examined, photographed. Embryos were harvested in cold PBS and fixed for 1-2 hours in 4% PFA. To optimize tissue fixation and penetrance of X-gal substrate (Roche Molecular, Indianapolis, IN), the chest wall was opened before fixation and in some cases the heart was dissected and incubated in substrate. Whole embryos or hearts were incubated in X-gal solution (5 mM K4Fe (CN)6, 5 mM K3Fe (CN)6, 2 mM MgCl2, 0.01% NP-40, 0.1%deoxycholate, 0.1% X-gal in PBS) at 37°C. For high-resolution analysis of βGal activity, embryos were paraffin embedded, sectioned and counterstained with nuclear Fast Red.
Immunostaining
Embryos or dissected hearts were fixed overnight in 4% PFA, embedded in OCT, and cut to 5-μm sections. The following antibodies were used: mouse monoclonal anti-Isl1 (1:100, DSHB), Rabbit anti-isl1 (1:1000, a gift from Sam Pfaff), Rabbit anti β-gal (1: 200, Cappel, #55976), PECAM-1 (1:50, BD Pharmingen, cat# 553708), MF-20 (1:50, DSHB), neurofilament (1:50, DSHB), α smooth-muscle-actin (1:200, Abcam, ab7817 and ab15734) and Nkx2.5 (1:50, Santa Cruz, sc14033). αActinin (1:800, Sigma, #A-7811). Sections were incubated with fluorescently labeled antibodies after washing with 0.25% Triton X-100 in PBS. The specimens mounted with Vectashield DAPI medium (Vector Laboratory) were analyzed under a fluorescence microscope.
Results
Isl1 progenitors have migrated into the heart by E9.0
Previous lineage analysis with isl1-cre;R26RlacZ embryos demonstrated a substantial contribution of isl1 expressing progenitors to the heart, and that isl1 mRNA was downregulated as these progenitors enter the heart (Cai et al., 2003). To investigate when migration of isl1 progenitors occurs, we performed lineage analysis with an inducible isl1-cre line, isl1-MerCreMer (Fig. 1A, see Materials and Methods; (Laugwitz et al., 2005). Embryos which were doubly heterozygous for isl1-MerCreMer and R26R-lacZ (Soriano, 1999) and their control littermates, were induced with tamoxifen injected into pregnant dams at E6, E7, E8, and E9. Embryos were harvested 48 hours following tamoxifen injection and stained with X-gal to reveal lineages derived from cells expressing isl1-MerCreMer at the time of injection (Fig. 1B-J). Tamoxifen activity is dependent on the half-life of tamoxifen, 11.9 hours, and persists over a 24-36 hour time period (Robinson et al., 1991; Xu et al., 2005; Zhang et al., 2006).
Fig. 1.
Lineage analysis of embryos doubly heterozygous for isl1-MerCreMer and R26RlacZ by tamoxifen induction. (A) Generation of Isl1 MerCreMer knock-in mouse (a) Targeting strategy. The relevant genomic region of isl1 is shown on top, the targeting construct is shown in the center, and the locus after recombination is shown at the bottom. A knock-in cassette composed of MerCreMer followed by FRT-mclNeo was introduced at SalI site in isl1 5'-untranslated region. Red box represents the MerCerMer cDNA and gray box represents the mcl-neomycin gene. (b) Detection of wild-type and targeted alleles by Southern blot analysis. DNAs from neomycin-resistant ES clones were digested with BamHI and analyzed by Southern blot with probe as shown in A-a. The 10.5- and 7.5-kb bands represent wild-type and targeted alleles, respectively. (B). Embryos injected at E6 and harvested at E8 revealed that labeled cells had already migrated into the early linear heart tube at both anterior and posterior poles, contributing to both myocardium and endocardium. (C) Corresponding sections of ED8.5 (8 somites) embryos are shown, progressing from anterior to posterior and (D) Corresponding sections of ED8.5 (10 somites) embryos are shown, progressing from anterior to posterior. Arrows indicate positive cells in endocardium. (E, F) Embryos injected at E7 and harvested at E9 and (G, H) Embryos injected at E8 and harvested at E10 showed labeled cells in outflow tract, atria, ventricle, and within mesenchymal cells of the atrioventricular (AV) canal (positive cells indicated by arrows and negative cells indicated by arrowheads). (I, J) Inductions performed at E9 revealed positive staining in the most distal cells of the outflow tract and a distinct subset of cells in the atria, in the region of the sinoatrial node and atrioventricular node (indicated by arrows). A: atria; AVN: atrioventricular node; LA: left atria; LV: left ventricle; OT: outflow tract; RA: right atria; LV: left ventricle; SAN: sinoatrial node. The framed areas were shown by high magnification in next photos.
Embryos injected at E6 and harvested at E8 revealed that labeled cells had already migrated into the early linear heart tube at both anterior and posterior poles, contributing to both myocardium and endocardium (Fig. 1B-D). Consistent with earlier isl1-cre lineage studies (Cai et al., 2003), left ventricular and left atrial tissue were relatively less populated by isl1 expressing cells. Continued migration of isl1 progenitors into the heart occurs up to approximately E8.5 (Fig. 1E-H). However, inductions performed at E9 revealed very few cardiac progenitors actively expressing isl1 at this time, with labeled cells observed in the most distal outflow tract, atrial septum, and in the region of the sinoatrial node and atrioventricular nodes (Fig. 1I, J). These data suggest that by E9 most isl1 progenitors have migrated into the heart and have ceased active expression of isl1.
Isl1-nlacZ knock-in mice recapitulate cardiac expression of endogenous isl1 protein and reveal subdomains of active Isl1 expression within the heart
An isl1-nlacZ knock-in mouse line was generated to readily visualize Isl1 expression (Fig. 2A, see Materials and Methods). Analysis of lacZ expression in embryos at E8.5, E9.5, E11.5 and E14.5 (Fig. 2B-J) revealed congruence with previously described expression of the endogenous isl1 gene (Cai et al., 2003; Pfaff et al., 1996). At E8.5, Isl1 is actively expressed in outflow tract and partially in the right atria and right ventricle, but not in the remainder of the myocardium (Fig. 2B-D). Isl1 was asymmetrically expressed in the right atria, and became progressively confined to a subdomain within the right atria, in the region of the cardiac pacemaker (Fig. 2C, D, F, H). At E14.5, isl1-nlacZ expression persisted in subdomains within the outflow tract, aorta, pulmonary artery, right ventricle, venous valves, atrial septum, and in regions corresponding to those of the sinoatrial (SA) and atrioventricular (AV) nodes, and in clusters of cells in the region of cardiac ganglia (Fig 2I, j). At postnatal day 3, Isl1-nlacZ expression was still observed in the region of the sinoatrial node and at the base of the aorta/pulmonary artery, but less extensively, with the exception of cardiac ganglia which still exhibited strong expression (Fig. 2K).
Fig. 2.
Analysis of β-galactosidase expression in isl1-nlacZ knock-in embryos using X-gal staining. (A) Generation of isl1-nlacZ knock-in mice. (a) Targeting strategy is the same as that of isl1-MerCreMer, except that the knock-in cassette is a Sall fragment of Lox-P-nLacZ followed by hrGFP and FRT-mclNeo gene. Colored boxes represent: blue box, nLacZ, which is flanked by Lox-P, green box, hrGFP and gray box,mcl-neomycin resistance gene, which is flanked by FRT. (b) Detection of wild-type and targeted alleles by Southern blot analysis as described in A-a. The 10.5- and 7.5-kb bands represent wild-type and targeted alleles, respectively. (B) Embryos at E8.5 stained with X-gal in left, frontal, and right views revealed active expression of β-gal in foregut endoderm, splanchnic mesoderm. (C) Corresponding sections of ED8.5 (8 somites) embryos are shown, progressing from anterior to posterior, (D) Corresponding sections of ED8.5 (10 somites) embryos are shown, progressing from anterior to posterior, (E) Embryos at E9.5 stained with X-gal in left and right views, and (F) Corresponding sections from anterior to posterior revealed isl1-nlacZ expression within outflow tract, right atria and right ventricle. Arrows indicate positive cells in endocardium. (G) Embryos at E11.5 stained with X-gal in left and right views, and (H) Corresponding sections from anterior to posterior revealed isl1-nlacZ expression within outflow tract, atrial septum, and sinoatrial and atrioventricular nodes. (I) Heart from E14.5 embryos stained with X-gal in ventral, dorsal and lateral dorsal views, and (J) Corresponding sections from ventral to dorsal revealed isl1-nlacZ expression within outflow tract, aorta, pulmonary artery, atrial septum, venous valves, cardiac ganglia and sinoatrial and atrioventricular nodes. (K) At postnatal day 3, Isl1-nlacZ expression was observed in cardiac ganglia, the region of the sinoatrial node and at the base of the aorta/pulmonary artery. A: atria; AS: atria septum; AVN: atrioventricular node; CG: cardiac ganglia, LA: left atria; LV: left ventricle; OT: outflow tract; RA: right atria; SAN: sinoatrial node.
To determine whether isl1-nlacZ expression accurately reflected endogenous Isl1 protein expression, we performed co-immunostaining analysis on cardiac sections, utilizing antibodies to detect β-galactosidase and Isl1. At E11.5 and E13.5, β-galactosidase co-localized with endogenous Isl1 (Fig. 3A-H and I-K). As with X-gal staining, Isl1 was observed in outflow tract (Fig. 3A-D), in the region of the SA and AV nodes (Fig.3E-H), in the atrial septum and within cells in the region of the cardiac ganglia (Fig. 3I-K). At postnatal day 3, Isl1 was observed coincident with β-gal staining, in cardiac ganglia, the region of the sinoatrial node (Fig. 3L, M, P, Q, T, UG), and in scattered cells within myocardium at the base of the aorta/pulmonary artery (Fig. 3N, O, R, S, V). Cardiac ganglia were identified by immunostaining with antibody to neurofilament (see Fig. 7F, G).
Fig. 3.
Expression of isl1-nlacZ mirrors endogenous Isl1 expression. (A-H) Coimmunostaining with antibodies to Isl1 and β-galactosidase (β-gal) on cardiac sections of isl1-nLacZ knock-in embryos at E11.5. (I-K) Co-immunostaining of Isl1 and β–gal antibodies on cardiac sections of isl1-nLacZ knock-in embryos at E13.5. (L-T) Co-immunostaining of Isl1 and β-gal antibodies on postnatal day 3 cardiac sections of isl1-lacZ knock-in mouse.
Fig. 7.
Isl1 expression in cardiac neural crest lineages. (A-D) Co-immunostaining for Isl1 and β-gal on tissue sections from Wnt1-cre;R26RlacZ embryos at E12.5 showed that only a small number of Isl1- and β–gal-positive cells overlapped in the outflow tract (arrows indicate overlapping staining). (E-H) Co-immunostaining for Isl1 and neurofilament on tissue sections from E13.5 embryos revealed Isl1 expression in cardiac ganglia.
Isl1 is actively expressed in some myocardial lineages, including those of the sinoatrial and atrioventricular nodes
To identify cardiac lineages which express Isl1, we performed co-immunostaining on tissue from wildtype or isl1-nlacZ embryos, utilizing antibodies to specific cardiac lineages and to Isl1 or β-galactosidase. At E11.5, co-immunostaining with Isl1 antibody and a monoclonal antibody to muscle specific myosin heavy chain, MF20 (Bader et al., 1982; Han et al., 1992), revealed co-expression of those two markers in outflow tract myocardium (Fig. 4A-D). Outflow tract myocardium also expresses α-smooth muscle actin (Beall and Rosenquist, 1990; Kruithof et al., 2003), which also co-localizes with Isl1 expression (Fig. 4E-H). At E14.5, β-galactosidase expressing cells at the base of the outflow tract which derived from Isl1 expressing cells, co-express α-smooth muscle actin, and MF20 (Fig. 4I-L and M-P). This population represents the “myocardialized” proximal septum of the outflow tract which provides continuity with the ventricular septum to divide the heart (Kruithof et al., 2003).
Fig. 4.
Isl1 expression in myocardial lineages and cardiac conduction system. (A-D) Coimmunostaining of isl1 or β-gal with a monoclonal antibody to muscle specific myosin heavy chain, MF20, on sections from E11.5 (A-D) and E14.5 (I-L) embryos revealed expression of isl1 in outflow tract myocardium. Co-immunostaining of isl1 or β-gal and α-smooth muscle actin on sections from E11.5 (E-H) and E14.5 (M-P) embryos revealed expression of isl1 in outflow tract myocardium. (Q-T) Co-immunostaining for isl1 on sections obtained from an HCN4-GFP knock-in mouse revealed expression of isl1 in subsets of HCN4+ populations in the regions of SA node, AV node and atrial septum (arrow). Co-immunostaining of isl1 on E13.5 sections demonstrated that Isl1 expressing cells in atrial septum co-expressed Nkx2.5 (U-X), and α-actinin (Y-B′), the latter a marker for differentiated myocytes.
To determine whether observed expression of Isl1 in the region of the SA and AV nodes reflected expression of Isl1 in pacemaker cells, we performed immunostaining for Isl1 on sections obtained from an HCN4-H2B-GFP knock-in mouse (YS, SE, unpublished) (Fig. 4Q-T). HCN4 is a hyperpolarization activated cyclic nucleotide gated cation channel which marks developing SA and AV nodes in mouse embryos, and the HCN4-H2B-GFP knock-in recapitulates expression of the endogenous gene (Garcia-Frigola et al., 2003); YS, SE, unpublished observations). We observed colocalization of Isl1 with some GFP expressing cells at E11.5 (Fig. 4Q-T), which demonstrated that Isl1 is expressed in at least a subset of pacemaker cells.
We also observed Isl1 expression in the atrial septum. Co-immunostaining on E13.5 sections demonstrated that Isl1 expressing cells in this region co-expressed Nkx2.5 (Fig. 4U-X), and α-actinin (Fig. 4Y-B′), the latter a marker for differentiated myocytes.
Isl1 is expressed in some endothelial lineages
Previous lineage studies with isl1-cre suggested a contribution of isl1 progenitors to endothelial cells within the outflow tract, a subset of endocardial cells, and to aortic endothelium (Cai et al., 2003). Here, we observed active Isl1 expression in the endothelial populations evidenced by co-expression of Isl1 and PECAM in cells within the outflow tract at E11.5 (Fig. 5A-E) and within the aorta and pulmonary artery at E12.5 (Fig. 5F-I), indicating that Isl1 is actively expressed in these endothelial populations at this time.
Fig. 5.
Isl1 expression in endothelial lineages. Co-immunostaining for Isl1 and PECAM-1 showed expression of Isl1 protein in PECAM positive endothelial cells within outflow tract at E11.5 (A-E) and in endothelial cells within aorta and pulmonary artery at E12.5 (F-I).
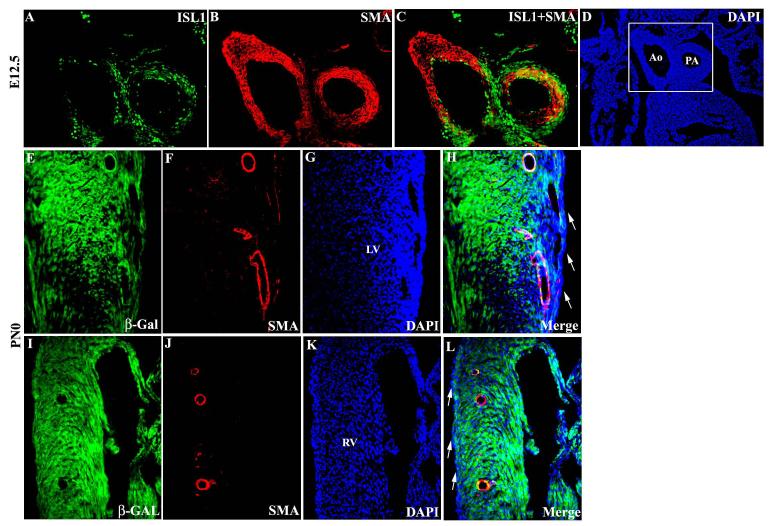
Isl1 is expressed in smooth muscle lineages, including those of the proximal outflow tract, and the coronary vasculature
At E12.5, Isl1 co-localized with smooth muscle at the proximal portion of the aorta and pulmonary arteries (Fig 6A-D). Expression of Isl1 in vascular smooth muscle prompted us to examine whether smooth muscle of the coronary vessels might also derive, at least in part, from isl1-expressing cells. Examination of cardiac sections from postnatal isl1-cre;R26R-lacZ mice revealed co-localization of β-galactosidase and α-smooth muscle actin within some smooth muscle cells of the coronary vessels (Fig. 6E-L). Isl1 lineage-traced cells, as indicated by β-galactosidase expression, were observed sporadically within smooth muscle of the coronary vasculature both within the left (Fig. 6E-H) and right (Fig. 6I-L) ventricles. Isl1 lineage-traced cells were not observed in the epicardium (Fig. 6H and L).
Fig. 6.
Isl1 expression in smooth muscle lineages. (A-D) Coimmunostaining for Isl1 and α-smooth muscle actin in sections from E12.5 embryos showed colocalization in proximal aorta and pulmonary artery trunk. (E-L) In the heart from postnatal day 0 (PN0) isl1-cre;R26RlacZ mouse, β-gal expressing cells co-expressed α-smooth muscle actin in coronary vasculature in Left (E-H) and right (I-L) ventricles but not in eipcardium (arrow in H and L) .
Isl1 is expressed in very few cells within outflow tract derived from cardiac neural crest, but is expressed throughout cardiac ganglia
Cardiac neural crest cells migrate into the aortic sac and outflow tract and contribute smooth muscle to the great vessels, and associated aortic arch arteries (Creazzo et al., 1998; Waldo et al., 1998). As Isl1 is expressed in a number of neural crest derivatives that give rise to sensory neurons in central and peripheral sensory ganglia and neurons of the autonomic nervous system, including cardiac ganglia (Kirby and Stewart, 1983; Thor et al., 1991) , we investigated whether Isl1 was expressed in cardiac neural crest cells within the outflow tract. To visualize cells derived from cardiac neural crest, lineage studies were performed utilizing wnt1-cre;R26RlacZ mice (Jiang et al., 2002). Co-immunostaining for β-galactosidase and Isl1 was performed on tissue sections from Wnt1-cre;R26RlacZ embryos. No substantial overlap was observed between Isl1 and β-galactosidase in outflow tract, with the exception of a few scattered cells, suggesting that Isl1 is not expressed in the majority of cardiac neural crest derived cells within cardiac outflow tract (Fig. 7A-D).
Cardiac ganglia also derive from the cardiac neural crest (Kirby and Stewart, 1983). Coimmunostaining for Isl1 and neurofilament demonstrated that Isl1 is expressed in cardiac ganglia (Fig. 7E-H).
Discussion
Lineage studies performed here with inducible isl1-MerCreMer;R26RlacZ embryos, and previous studies with Isl1-cre;floxed fgf8-GFP mice (Park et al., 2006) demonstrate that isl1 expressing progenitors migrate into the heart shortly following fusion of cardiac primordia. Tamoxifen induction of Isl1-MerCreMer;R26RlacZ embryos revealed that by E9, most isl1 progenitors have migrated into the heart, and have downregulated isl1 expression.
From E9 to postnatal day 3, Isl1 expression is observed in select subdomains of the heart, including the outflow tract and pacemaker cells of the sinoatrial and atrioventricular nodes. Consistent with Isl1 expression in outflow tract myocardium, expression of a MEF2c anterior heart field enhancer in the outflow tract requires consensus elements recognized by Isl1 and GATA transcription factors (Dodou et al., 2004). Isl1 is expressed in outflow tract myocardium and endocardium during the time at which extensive outflow tract remodelling is occuring (Kruithof et al., 2003), suggesting a potential role for Isl1 in this process.
Although retroviral lineage studies have demonstrated a myocytic lineage origin for His-Purkinje fibers (Gourdie et al., 2003), no lineage studies have been performed which address the origin of pacemaker lineages in the central conduction system. Observed Isl1 expression in the SA and AV nodes suggests that pacemaking cells at stages examined may arise, at least in part, from the second heart field. HCN4 is an early marker for nodal lineages, and we observed Isl1 expression in a subset of the HCN4 domain, suggesting that Isl1 is actively expressed in, and is a marker for, a subset of HCN4 positive cells. The cardiac pacemaker is comprised of heterogeneous cells (Anderson and Ho, 1998), and it will be of future interest to address the properties of the subset of pacemaker cells actively expressing Isl1. Pacemaker cells of the central conduction system in developing heart are relatively undifferentiated (Moorman et al., 1998), and it is interesting that this correlates with Isl1 expression.
Previous lineage studies with a constitutive isl1-cre (Cai et al., 2003; Yang et al., 2006) suggested that Isl1 expressing cells contributed to endothelial cells of the aorta, endothelial cells within the outflow tract, and a substantial number of endocardial cells within chamber myocardium. Consistent with these results, our data with the inducible isl1-cre demonstrated a substantial contribution of isl1 cells to endothelial lineages, including mesenchymal cells within outflow tract and atrioventricular cushions. We also observed active expression of Isl1 in endothelial cells of the outflow tract, aorta and pulmonary artery.
Isl1 was actively expressed in several smooth muscle populations, including that of the proximal portion of the aorta and pulmonary artery, consistent with previous results which demonstrated that this smooth muscle population derives from the secondary or anterior heart field (Verzi et al., 2005; Waldo et al., 2005). During outflow tract remodelling, a process called myocardialization has been described, whereby the lower portion of the outflow tract septum expresses smooth muscle actin and myosin heavy chain and contributes to the final septation of the heart (Christoffels et al., 2004). The source of the myocardialized septum has not been identified. Analysis of Isl1-lacZ expression revealed that these cells derive at least in part from Isl1 expressing cells.
The observation that Isl1 was expressed in smooth muscle contributing to the aorta and pulmonary artery, and previously described expression of Isl1 in neural crest derivatives (Thor et al., 1991), led us to examine potential overlap of Isl1-expressing cells with those derived from the cardiac neural crest (Kirby and Stewart, 1983). Very little overlap was observed in the outflow tract domain. However, Isl1 was expressed in cardiac ganglia which also derive from the cardiac neural crest.
Our studies also reveal a contribution of Isl1 expressing cells to the smooth muscle of the coronary vessels. Previous lineage studies in chick embryos have demonstrated that smooth muscle of the coronary vessels derives from the proepicardium/epicardium (Gittenberger-de Groot et al., 1999; Mikawa and Gourdie, 1996; Olivey et al., 2004). In mouse, coronary vessels are invested with smooth muscle at approximately E15, well past the time at which the majority of isl1 progenitors of the second heart field have migrated into the heart. We did not observe isl1-lineage traced cells in the epicardium. The origin of the coronary smooth muscle cells which were labelled by isl1-cre;R26R lineage tracing is currently unknown. These cells may represent a subset of epicardially derived cells which activate isl1 expression, may have been conscripted from surrounding isl1-derived myocardium, or may derive from another source. In any case, the observation that a subset of smooth muscle cells derives from an isl1 expressing lineage demonstrates genetic heterogeneity of coronary vascular smooth muscle, which may have implications for coronary vessel disease.
The observation that Isl1 expressing cells contribute to multiple cardiovascular cells of distinct lineages, including myocyte, conduction system, endothelial and smooth muscle lineages, raises the question as to the role of Isl1 in specification of each of these lineages. Is Isl1 expressed in a pluripotential cardiovascular progenitor? Or is Isl1 expressed independently in each of the lineage-restricted precursors? The early onset of Isl1 expression is consistent with the former, and will be a subject for future investigation.
Contribution of Isl1 expressing cells to multiple cardiovascular lineages also has implications for studies utilizing isl1-cre lines to ablate genes which may be expressed in more than one of these lineages.
Acknowledgments
We thank Sam Pfaff for providing Isl1 antibody, and Kat Hadjantonakis for providing H2B-EGFP vector. This work was supported by NIH RO1 HL074066 to SE and American Heart Association/Western States Affiliate Postdoc fellowship Award # 0525226Y to YFS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Issa R, Waldo K, Kirby ML. Heart fields: one, two or more? Dev Biol. 2004;272:281–5. doi: 10.1016/j.ydbio.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Anderson RH, Ho SY. The architecture of the sinus node, the atrioventricular conduction axis, and the internodal atrial myocardium. J Cardiovasc Electrophysiol. 1998;9:1233–48. doi: 10.1111/j.1540-8167.1998.tb00097.x. [DOI] [PubMed] [Google Scholar]
- Bader D, Masaki T, Fischman DA. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J Cell Biol. 1982;95:763–70. doi: 10.1083/jcb.95.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall AC, Rosenquist TH. Smooth muscle cells of neural crest origin form the aorticopulmonary septum in the avian embryo. Anat Rec. 1990;226:360–6. doi: 10.1002/ar.1092260313. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–35. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–89. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Burch JB, Moorman AF. Architectural plan for the heart: early patterning and delineation of the chambers and the nodes. Trends Cardiovasc Med. 2004;14:301–7. doi: 10.1016/j.tcm.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Creazzo TL, Godt RE, Leatherbury L, Conway SJ, Kirby ML. Role of cardiac neural crest cells in cardiovascular development. Annu Rev Physiol. 1998;60:267–86. doi: 10.1146/annurev.physiol.60.1.267. [DOI] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–42. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Garcia-Frigola C, Shi Y, Evans SM. Expression of the hyperpolarization-activated cyclic nucleotide-gated cation channel HCN4 during mouse heart development. Gene Expr Patterns. 2003;3:777–83. doi: 10.1016/s1567-133x(03)00125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, DeRuiter MC, Bergwerff M, Poelmann RE. Smooth muscle cell origin and its relation to heterogeneity in development and disease. Arterioscler Thromb Vasc Biol. 1999;19:1589–94. doi: 10.1161/01.atv.19.7.1589. [DOI] [PubMed] [Google Scholar]
- Gourdie RG, Harris BS, Bond J, Edmondson AM, Cheng G, Sedmera D, O'Brien TX, Mikawa T, Thompson RP. His-Purkinje lineages and development. Novartis Found Symp. 2003;250:110–22. discussion 122-4, 276-9. [PubMed] [Google Scholar]
- Han Y, Dennis JE, Cohen-Gould L, Bader DM, Fischman DA. Expression of sarcomeric myosin in the presumptive myocardium of chicken embryos occurs within six hours of myocyte commitment. Dev Dyn. 1992;193:257–65. doi: 10.1002/aja.1001930306. [DOI] [PubMed] [Google Scholar]
- Jiang X, Choudhary B, Merki E, Chien KR, Maxson RE, Sucov HM. Normal fate and altered function of the cardiac neural crest cell lineage in retinoic acid receptor mutant embryos. Mech Dev. 2002;117:115–22. doi: 10.1016/s0925-4773(02)00206-x. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Buckingham ME. The anterior heart-forming field: voyage to the arterial pole of the heart. Trends Genet. 2002;18:210–6. doi: 10.1016/s0168-9525(02)02642-2. [DOI] [PubMed] [Google Scholar]
- Kirby ML, Stewart DE. Neural crest origin of cardiac ganglion cells in the chick embryo: identification and extirpation. Dev Biol. 1983;97:433–43. doi: 10.1016/0012-1606(83)90100-8. [DOI] [PubMed] [Google Scholar]
- Kruithof BP, Van Den Hoff MJ, Tesink-Taekema S, Moorman AF. Recruitment of intra- and extracardiac cells into the myocardial lineage during mouse development. Anat Rec A Discov Mol Cell Evol Biol. 2003;271:303–14. doi: 10.1002/ar.a.10033. [DOI] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–53. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell. 2004;6:685–98. doi: 10.1016/s1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–32. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- Moorman AF, de Jong F, Denyn MM, Lamers WH. Development of the cardiac conduction system. Circ Res. 1998;82:629–44. doi: 10.1161/01.res.82.6.629. [DOI] [PubMed] [Google Scholar]
- Olivey HE, Compton LA, Barnett JV. Coronary vessel development: the epicardium delivers. Trends Cardiovasc Med. 2004;14:247–51. doi: 10.1016/j.tcm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–33. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–20. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Robinson SP, Langan-Fahey SM, Johnson DA, Jordan VC. Metabolites, pharmacodynamics, and pharmacokinetics of tamoxifen in rats and mice compared to the breast cancer patient. Drug Metab Dispos. 1991;19:36–43. [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor S, Ericson J, Brannstrom T, Edlund T. The homeodomain LIM protein Isl-1 is expressed in subsets of neurons and endocrine cells in the adult rat. Neuron. 1991;7:881–9. doi: 10.1016/0896-6273(91)90334-v. [DOI] [PubMed] [Google Scholar]
- Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:134–45. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- Waldo K, Miyagawa-Tomita S, Kumiski D, Kirby ML. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: aortic sac to ventricular septal closure. Dev Biol. 1998;196:129–44. doi: 10.1006/dbio.1998.8860. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Hutson MR, Ward CC, Zdanowicz M, Stadt HA, Kumiski D, AbuIssa R, Kirby ML. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev Biol. 2005;281:78–90. doi: 10.1016/j.ydbio.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–88. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- Xu H, Cerrato F, Baldini A. Timed mutation and cell-fate mapping reveal reiterated roles of Tbx1 during embryogenesis, and a crucial function during segmentation of the pharyngeal system via regulation of endoderm expansion. Development. 2005;132:4387–95. doi: 10.1242/dev.02018. [DOI] [PubMed] [Google Scholar]
- Yang L, Cai CL, Lin L, Qyang Y, Chung C, Monteiro RM, Mummery CL, Fishman GI, Cogen A, Evans S. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 2006;133:1575–85. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Huynh T, Baldini A. Mesodermal expression of Tbx1 is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development. 2006;133:3587–95. doi: 10.1242/dev.02539. [DOI] [PMC free article] [PubMed] [Google Scholar]