Abstract
The midline tissues are important inductive centers of early vertebrate embryos. By signal peptide selection screening, we isolated a secreted factor, Kielin, which contains multiple cys-rich repeats similar to those in chordin (Chd). Expression of Kielin starts at midgastrula stages in the notochord and is detected in the floor plate of neurula embryos. Kielin is induced in mesoderm and in ectoderm by nodal-related genes. Chd is sufficient to activate Kielin expression in mesoderm whereas Shh or HNF-3β in addition to Chd is required for induction in ectoderm. Kielin has a distinct biological activity from that of Chd. Injection of Kielin mRNA causes dorsalization of ventral marginal zone explants and expansion of MyoD expression in neurula embryos. Unlike Chd, Kielin does not efficiently induce neural differentiation of animal cap ectoderm, suggesting that the activity of Kielin is not simply caused by BMP4 blockade. Kielin is a signaling molecule that mediates inductive activities of the embryonic midline.
The Spemann organizer, when grafted to the ventral side of another embryo, not only induces dorsal tissues from presumptive ventral tissues, but also constructs organized structures in the induced tissues (1). As a result, the induced secondary axis contains well-patterned neural and mesodermal organs both in the dorsal-ventral and anterior-posterior directions. The series of inductive events initiated by the organizer can be classified into two categories. First, the organizer itself directly causes initial inductive events (primary induction; e.g., neural induction). Second, more localized inductive activities are suggested to play roles in the fine patterning of tissues (secondary induction). In the latter process, some tissues induced by or derived from the organizer function as an inductive center (secondary organizer).
Over the last several years, a number of secreted signaling molecules acting during primary induction have been isolated in Xenopus (2). The Spemann organizer induces dorsal tissues by emanating organizer factors such as noggin, follistatin, and chordin (Chd) (3–5). These secreted factors dorsalize mesoderm and also induce neural tissues in ectoderm (4, 6, 7). Noggin, follistatin, and Chd bind to and inactivate the ventralizing molecule BMP4 in the extracellular space (8–10). Thus, the dorsal-ventral patterning of gastrula embryos is regulated by a bone morphogenetic protein (BMP) activity gradient (11).
The midline tissues, such as notochord and floor plate, are typical examples of a signaling center of secondary induction. The notochord, which is a major derivative of the Spemann organizer, patterns both the neural tube (12) and somites (ref. 13 and references therein). The notochord induces formation of the overlying floor plate in the neural tube and also promotes polarization of somites. The floor plate provides the ventral neural tube with positional information for fine neuronal patterning (12). A candidate molecule for the patterning activity is Shh, a secreted factor homologous to Drosophila Hedgehog (14–16). In explant experiments using early central nervous system (CNS) tissues, soluble Shh protein can induce markers of motoneuron and floor plate in a dose-dependent manner (17, 18). Shh also mimics the activity of notochord in the patterning of somites (13, 19). However, Shh alone cannot account for all of the activities that the midline tissues have (e.g., refs. 20–23).
To further understand the molecular basis of embryonic patterning, we carried out a cDNA screen for signaling factors involved in secondary induction during early embryogenesis. By using the signal peptide selection method (24), we attempted to isolate secreted factors expressed in possible secondary organizer tissues of the forming head region. In this study, we report the isolation and functional characterization of a secreted factor, Xenopus Kielin. Kielin has a unique domain structure containing multiple Chd-type repeats and is a patterning molecule emanating from the embryonic midline.
Materials and Methods
Molecular Cloning of Kielin cDNA.
Poly(A)+ mRNA was isolated from anterior neural plate regions (with underlying mesodermal and endodermal tissues) of stage 13–14 Xenopus embryos. Double-stranded cDNA was synthesized with random primers and then was fractionated by agarose electrophoresis. cDNA fractions of 300–700 bp were subcloned into the NotI site of the trap-vector lambda RK18 (24). Seventy four pools of 5,000 plaque-forming units of lamba phages were amplified and rescued into plamids (pRK18) by Exassist helper phage and XL-1 Blue bacteria (Stratagene). Each pool of plasmid mixture was used to transform the yeast YT455 (suc2Δ9, ade2–101, and ura3–52; pRK18 contains a wild-type URA gene) by the lithium acetate method (CLONTECH). After selecting transformed YT455 on URA(−) plates, yeast colonies were transferred to sucrose plates with a disposable replica plater (Takara, Osaka). After 7–10 days, growing colonies were picked and plasmids were recovered from the yeasts into Escherichia coli.
Explant Assays and in Situ Hybridization.
The developmental stage of Xenopus embryos was determined by Nieuwkoop and Faber staging. The animal caps and ventral marginal zone (VMZ) (about 60°) were prepared at stage 10.25–10.5 and then cultured in 1× low Ca2+ Mg2+ Ringer's solution (LCMR) supplemented with 0.2% BSA until a given stage. In situ hybridization analyses were performed as described (25) with minor modifications.
Plasmid Construction and mRNA Injection.
A cDNA fragment containing the full coding sequence of Kielin and a simian virus 40 polyadenylation signal was subcloned into the EcoRI and NotI sites of pBluescript KS. After linearization with NotI, T3 polymerase was used for in vitro transcription (mMessage mMachine Kit, Ambion). A frame-shift mutant of Kielin (for a negative control) was generated by deleting 4 bp at the KpnI site located in the middle of the cys-rich domain. RNAs for Chd, Xnot, goosecoid, and cyc were synthesized as described (5, 26–28). Full coding fragments of HNF-3β (with EcoRI linker at the 5′ end and XhoI at the 3′) and Shh (with EcoRI linker at the 5′ and XbaI at the 3′) were amplified by PCR and subcloned into pSP35 vector. A Xnr-4 coding fragment also was obtained by PCR and subcloned into the ClaI and XbaI sites of pCS2. The plasmids with PCR fragments were sequenced before being used as a template for SP6 polymerase. The HNF-3β plasmid was linearized with SacI and the plasmids for Shh and Xnr-4 with NotI. Synthetic mRNA was injected into blastomeres of eight-cell embryos (1–4 nl) by using a fine glass capillary and a pneumatic pressure injector (Narishige, Tokyo). All of the injection experiments were carried out at least twice and gave reproducible results. The statistical values given in the text were from one representative experiment. In each series of experiments, the total amount of RNA injected was adjusted by adding neutral lacZ mRNA.
Results
Kielin Is a Unique Secreted Factor with Multiple Chd-Type Repeats.
We attempted to systematically isolate genes involved in the complex patterning of the vertebrate head region. We focused on genes coding secreted or cell-surface molecules because extracellular signaling factors are supposed to play roles in intricate cell–cell interactions during brain patterning. An efficient screening method for genes encoding an extracellular factor recently was invented (signal peptide selection) (24). This method uses a mutant yeast that lacks invertase gene. Invertase is an extracellular enzyme that hydrolyzes sucrose and is required for yeast to grow on sucrose plates. By using an expression vector of invertase lacking the signal peptide portion, cDNA that contains a signal peptide sequence can be trapped by nutrient requirement screening (24).
By screening 3.7 × 105 colonies carrying Xenopus cDNAs from anterior neural plate and the underlying tissues (stage 13–14), 119 revertant yeast colonies growing on sucrose plates were obtained. Of these selected cDNA clones, 17 showed a localized expression pattern on whole-mount in situ hybridization. Five cDNA clones had high sequence homology to known extracellular proteins [mucin-like, neural cell adhesion molecule (NCAM)-like, XAG, fibrinogen-related, and plexin]. We focused on a novel protein that showed a mild similarity to a portion of thrombospondin protein, because it had an intriguing expression pattern in the midline of early embryos. As the expression in the CNS resembled a keel of ship, the gene was named Kielin (after Kiel, a German word for keel).
A full-length cDNA coding 8,137 bp then was isolated (DNA Data Base of Japan no. AB026192). The ORF encodes 2,327 amino acid residues (Fig. 1A). Three distinct domains were identified by a homology search (blast). The most striking structural feature was 27 repeats of the cysteine-rich domain that shares homology with that found in Chd protein (Fig. 1B). Chd contains four cys-rich repeats, which are highly conserved between Xenopus and Drosophila counterparts (11). It recently has been shown that these cys-rich repeats are responsible for the conserved activity of BMP antagonism (29). The amino-terminal portion of the Kielin protein shows similarity to the amino-terminal part of thrombospondin, a platelet-derived signaling molecule (18% residue identity) (30). This part of thrombospondin is reported to be involved in binding to the cell surface and matrix (30). The carboxyl-terminal portion of Kielin shares homology with the D domain of von Willebrand factor (vWF) (29% residue identity), an essential hemostasis component attached to the platelet and collagen (31). Interestingly, thrombospondin and vWF also contain one and two cys-rich domains, respectively, which have an arrangement of cys residues similar to that found in the cys-rich domains of Chd (Fig. 1B) (5). The hydropathy profile indicated only one hydrophobic segment at the amino terminus (not shown), indicating that Kielin is a secreted factor. This idea is further strengthened by the fact that the entire protein sequence of Kielin, from the amino terminus to the carboxyl terminus, shares significant homologies with secreted factors (Chd, thrombospondin, and vWF).
Figure 1.
Primary structure of the deduced Kielin protein. (A) Domain structure of Kielin (KLN). A signal sequence (filled box) is located at the N terminus, followed by the thrombospondin (TSP)-like domain, the Chd-type cys-rich repeats, and the vWF-like domain. (B) Sequence comparison of cys-rich (CR) domains of Kielin with those in Xenopus Chd, human TSP2, and human vWF.
Kielin Is Expressed in the Notochord and Floor Plate in a Similar Manner to Shh.
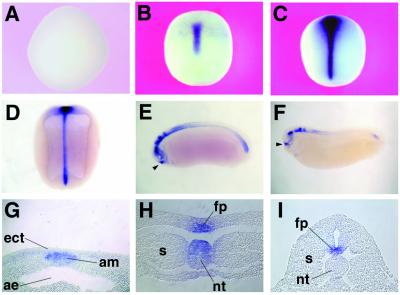
Whole-mount in situ hybridization shows that Kielin expression starts at the midgastrula stage in the dorsal midline (Fig. 2 A and B). The cross-section indicates that Kielin is expressed in the axial mesoderm at this stage (Fig. 2G). At neurula stages, Kielin expression is detected both in the axial mesoderm (notochord and prechordal mesoderm) and in the ventral CNS (floor plate and ventral forebrain) (Fig. 2 C, D, and H). At tailbud stages, Kielin expression is diminished in the notochord and remains in the ventral CNS (Fig. 2 E, F, and I). During and after tailbud stages, other regions start to express Kielin. The epiphyseal placode (adenopituitary anlage; arrowheads in Fig. 2 E and F) has a strong Kielin expression. Additional expression is found in dorsal parts of the CNS, especially in the anterior spinal cord and hindbrain as well as in the tailbud mesoderm (Fig. 2 E and F). At the larval stage, Kielin expression also is found in the forming heart (not shown).
Figure 2.
Whole-mount in situ hybridization analysis of Kielin expression in embryos at stage 10.5 (A; vegetal view), stage 12 (B; dorsal view), stage 14 (C; dorsal view), stage 19 (D; cleared dorsal view), stage 24 (E, cleared lateral view), and stage 28 (F, cleared lateral view). Histological analyses show Kielin expression in the axial mesoderm of a late gastrula (G), in the notochord and presumptive floor plate of a midneurula (H), and in the floor-plate region of an early tailbud embryo (I). Arrowheads, stomodeal-hypophyseal anlage; am, axial mesoderm; ect, ectoderm; ae, archenteron; fp, presumptive floor plate; s, somite; nt, notochord.
The temporal expression of Kielin was compared with that of two midline genes, HNF-3β and Shh, by Northern blot analysis (not shown). As described previously, HNF-3β expression started at the late blastula stage (stage 9) (32, 33). The onset of Kielin and Shh expression occurred at the midgastrula stage (between stages 11 and 12) and both transcripts accumulated at a high level during neurula stages. Collectively, the results above demonstrated that Kielin has a spatial and temporal expression profile similar to that of Shh (34).
Kielin Expression Is Activated by Chd and Xnr-4 in the Marginal Zone Mesoderm.
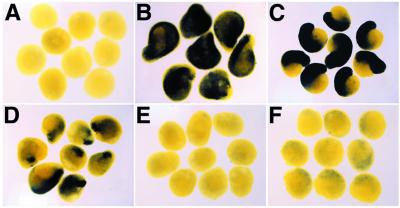
We next studied the regulation of Kielin expression in the axial mesoderm by using VMZ explants. In this experiment, the explants were harvested for analysis during late gastrula stages when Kielin is expressed only in the axial mesoderm. As candidate upstream regulators for Kielin expression, we first tested secreted factors that are accumulated in the axial mesoderm both at the onset of Kielin (stage 11.5) and at late gastrula stages. Chd is expressed in the organizer and the axial mesoderm throughout gastrulation. Microinjection of Chd mRNA resulted in induction of Kielin in the VMZ (100%, n = 32; Fig. 3B) whereas injection of lacZ mRNA did not activate Kielin expression (n = 30; Fig. 3A). nodal-related genes (Xnr-1, -2, and -4), which can promote dorsal mesoderm formation, also are expressed in the gastrula mesoderm (35, 36). Xnr-4 is expressed in the axial mesoderm during gastrula and neurula stages (36), whereas the expression of Xnr-1 and -2 at the RNA level does not overlap significantly with that of Kielin (35), suggesting that Xnr-4 is a candidate regulator of Kielin in the axial mesoderm. Injection of Xnr-4 mRNA efficiently activated Kielin expression in the VMZ (100%, n = 42; Fig. 3C). These data indicate that Kielin expression is positively regulated, at least in part, by two distinct secreted signals: the BMP antagonists (Chd and noggin) and Xnr-4 (or its related factors). Microinjection of Shh mRNA (up to 800 pg) did not significantly activate Kielin under this condition (not shown).
Figure 3.
Regulation of Kielin expression in the marginal zone explant. VMZ explants were prepared from embryos injected with control lacZ mRNA (300 pg; A), Chd mRNA (100 pg; B), Xnr-4 mRNA (200 pg; C), goosecoid mRNA (50 pg; D), HNF-3β mRNA (100 pg; E), and Xnot mRNA (300 pg; F). The explants were prepared at stage 10.5, harvested at stage 12.5, and analyzed by in situ hybridization with a Kielin probe.
To investigate possible intracellular mediators of Kielin activation, the transcription factors expressed in the axial mesoderm were examined in the VMZ assay. Injection of goosecoid (gsc) (37) mRNA induced Kielin expression in the explants (100%, n = 24; Fig. 3D). When mRNA of HNF-3β or Xnot (38, 39) was injected, little Kielin induction, if any, was observed in the VMZ explants from the injected embryos (n = 25 each; Fig. 3 E and F). These results suggest that the activation of Kielin in the axial mesoderm is mediated partly by gsc but unlikely by HNF-3β or Xnot alone.
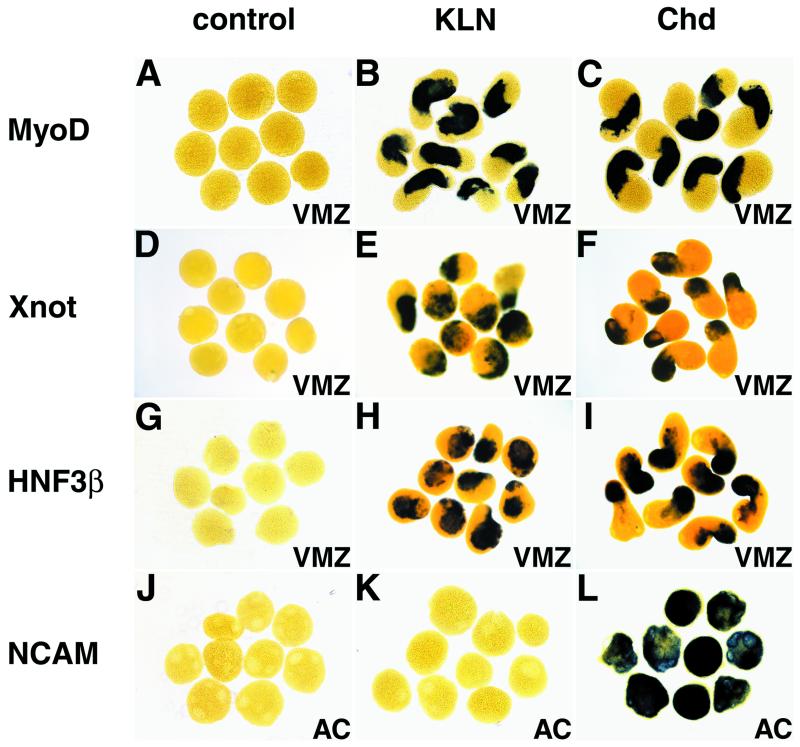
Kielin Is Induced in the Ectoderm by Chd+Shh and by nodal-Related Genes.
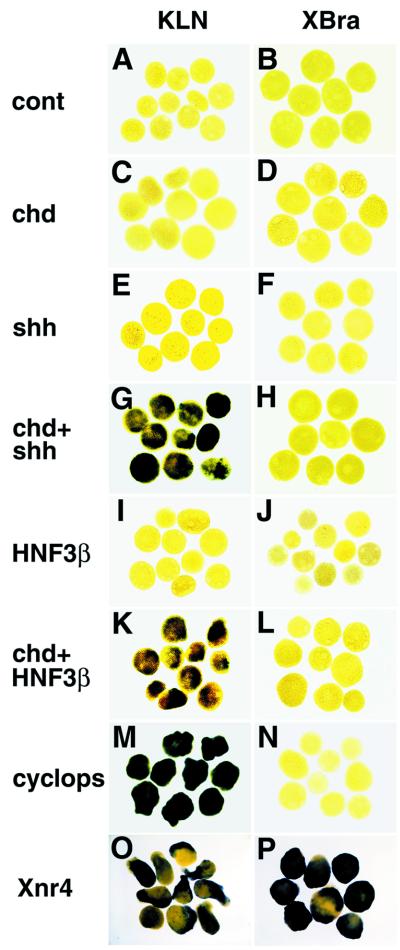
To examine possible regulators of Kielin expression in the ectoderm, we next conducted animal cap assays. In this study, the explants were analyzed at stage 20 when the ventral CNS is the main ectodermal tissue expressing Kielin. Unlike in VMZ, Chd alone did not induce Kielin expression in animal cap ectoderm (n = 35, Fig. 4 A and C). Next, we tested two midline genes, Shh and HNF-3β, which are essential for the midline formation (21, 40–42). Kielin was not induced by mRNA injection of Shh (n = 30) or HNF-3β (n = 40) alone (Fig. 4 E and I). When Shh or HNF-3β was combined with Chd, Kielin transcription was activated in animal cap ectoderm (100%, n = 25; Fig. 4G) (90%, n = 30, Fig. 4K). As the mesodermal marker Xbra was not induced in the explants (n = 20 each; Fig. 4 B, D, F, H, J, and L), the effects of the factors above were direct on the ectoderm rather than secondary because of mesoderm induction. These results demonstrate that a combination of the neural inducer (Chd) and the midline specifier (HNF-3β or Shh) is sufficient to induce Kielin expression in the floor plate.
Figure 4.
Regulation of Kielin expression in the animal cap ectoderm. Animal caps were explanted at stage 10.5 from embryos injected with control mRNA (1 ng; A and B), Chd mRNA (100 pg; C and D), Shh mRNA (900 pg; E and F), Chd and Shh mRNAs (G and H), HNF-3β mRNA (50 pg; I and J), Chd and HNF-3β mRNAs (K and L), cyc mRNA (200 pg; M and N), and Xnr4 mRNA (200 pg; O and P). The explants were harvested at stage 20 and analyzed with a Kielin (KLN) probe (A, C, E, G, I, K, M, and O) or harvested at stage 12 and hybridized with a Xbra probe (B, D, F, H, J, L, N, and P).
Recently, cyclops (cyc), a zebrafish mutant with floor plate defects (43), has been attributed to a nodal-related gene expressed in the axial mesoderm (28, 44–46). Overexpression of wild-type cyc mRNA activated Kielin transcription efficiently (100%, n = 50; Fig. 4M) without inducing the mesodermal marker Xba (n = 37; Fig. 4N). This indicates that a nodal-related gene (a Xenopus cyc homologue) is likely involved in the induction of the floor plate expression of Kielin. The expression pattern suggests that Xnr-4 is the most appropriate candidate for the Xenopus counterpart of cyc in the view of axial patterning (28, 36, 46). Injection of Xnr-4 mRNA induced Kielin in the explants (100%, n = 19; Fig. 4O). Unlike cyc, Xnr-4 induced the mesoderm marker (100%, n = 18; Fig. 4P) as previously reported (36), which makes it difficult to tell whether the activation of Kielin by Xnr-4 was direct or secondary because of mesoderm induction.
Kielin mRNA Injection Causes Up-Regulation of MyoD Expression in Vivo.
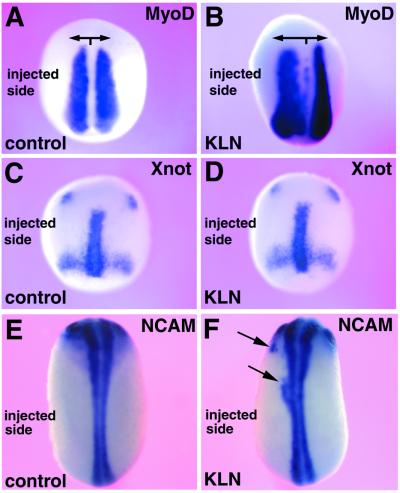
We next tested activities of Kielin by misexpressing it in the developing embryo. When Kielin mRNA was injected into left vegetal blastomeres of eight-cell embryos, expansion of the MyoD-expressing region was observed on the injected side of neurula embryos (56%, n = 25; Fig. 5B). Injection of frame-shift Kielin did not affect MyoD expression (n = 30; Fig. 5A). Ectopic or enlarged expression of the notochordal marker Xnot was not detected (n = 30; Fig. 5 C and D). Unlike Chd injection (5), Kielin injection in vivo did not induce strong secondary axes (n = 50, not shown). Small patches of ectopic NCAM expression were observed at a low frequency in Kielin-injected embryos (19%, n = 77; Fig. 5F), but not significantly in embryos injected with frame-shift Kielin (2%, n = 47; Fig. 5E) (see Materials and Methods).
Figure 5.
Overexpression of Kielin expands the MyoD expression region in vivo. Two left vegetal blastomeres of eight-cell embryos were injected with frame-shift Kielin (control) mRNA (600 pg; A, C, and E) and wild-type Kielin (KLN) mRNA (600 pg; B, D, and F). The injected embryos were harvested at stage 16 (A and B), stage 13 (C and D), or stage 19 (E and F) and analyzed by whole-mount in situ hybridization with a MyoD probe (A and B), a Xnot probe (C and D), or a NCAM probe (E and F).
Kielin Overexpression Dorsalizes Mesoderm But Does Not Neuralize Ectoderm in Explant Assays.
The in vivo data showed that Kielin injection caused expansion of the dorso-lateral mesodermal marker (MyoD) and, less efficiently, ectopic expression of the neural marker. To test whether the in vivo activities reflected direct effects of Kielin on differentiation or not, we next used isolated mesodermal and ectodermal explants. Kielin mRNA was injected into all vegetal blastomeres of eight-cell embryos, from which VMZ explants were prepared at stage 10.5. When the siblings reached the tailbud stage, Kielin-injected VMZ were elongated (83%, n = 24) whereas control VMZ injected with frame-shift Kielin mRNA were round (100%, n = 29)(not shown). Kielin injection caused induction of MyoD (muscle marker; 96%, n = 23; Fig. 6B), HNF-3β and Xnot (dorsal axial mesoderm; 100%, n = 31 and 100%, n = 55, respectively; Fig. 6 E and H) in the VMZ analyzed at stage 13. The efficiency of induction of these dorsal markers by Kielin was comparable to that by the typical dorsalizing factor Chd (100%, n = 24 each; Fig. 6 C, F, and I). Injection of frame-shift Kielin mRNA did not induce any of the dorsal markers (n = 20 each; Fig. 6 A, D, and G). These results show that Kielin overexpression causes dorsalization of mesoderm in VMZ explants. This explains the in vivo effects of Kielin on mesodermal patterning at least in part. On the other hand, the notochordal marker Xnot was induced by Kielin in vitro (Fig. 6E) but not in vivo (Fig. 5D). In this respect, the dorsalizing activity of Kielin is not completely the same as that of Chd, which induces Xnot both in vitro and in vivo (5, 26). This may indicate that the effects of Kielin are partially compensated in vivo by certain regulatory mechanisms.
Figure 6.
Kielin has a mesoderm-dorsalizing activity in VMZ assays, but does not induce neural differentiation in the animal cap. VMZ explants were prepared from embryos injected with control frame-shift Kielin (control) mRNA (300 pg; A, D, G, and J), wild-type Kielin (KLN) mRNA (300 pg; B, E, H, and K), and Chd mRNA (100 pg; C, F, I, and L). As Kilein mRNA is three times larger in size than Chd mRNA, the injected mounts of mRNAs are similar in terms of molar ratio. (A–I) VMZ assays. The VMZ explants harvested at stage 13 were analyzed by in situ hybridization with a MyoD probe (A–C), Xnot probe (D–F), or a HNF-3β probe (G–I). (J–L) Animal cap assays. Animal caps were explanted at stage 10.25 from embryos injected with control frame-shift Kielin mRNA (J), wild-type Kielin mRNA (K), and Chd mRNA (L). The animal cap explants were harvested at stage 20 and hybridized with a NCAM probe.
Finally, Kielin activity was examined in animal cap assays. The mesodermal marker Xbra was not induced in the animal caps injected with control, Kielin, or Chd mRNA (n = 20 each; not shown). Unlike Chd injection (100%, n = 20; Fig. 6L), Kielin mRNA injection (100, 300, 800, 1,200, or 1,600 pg/animal blastomere of eight-cell embryo; n = 24 each) did not induce neural differentiation of animal caps (Fig. 6 J and K and data not shown). Note that strong mesodermal dorsalization was induced at 300 pg Kielin mRNA per cell in VMZ whereas injection of a 5-fold higher amount did not cause neuralization in the animal cap. Therefore, it is unlikely that Kielin is an efficient neural inducer. This suggests that NCAM expansion in Kielin-injected embryos was caused by secondary effects such as mesodermal dorsalization.
Discussion
Kielin Is a Unique Secreted Signaling Molecule.
By using the signal peptide selection screen (24), a unique secreted signaling molecule, Kielin, was isolated. Kielin contains multiple Chd-type repeats and has a partially overlapping distribution with Chd in the axial mesoderm (5). Furthermore, both Chd and Kielin dorsalize VMZ mesoderm to a similar extent in vitro (Fig. 6). Because Kielin contains multiple Chd-type repeats, one explanation for the dorsalizing activity of Kielin was that Kielin antagonizes the ventralizing factor BMP4 just as Chd does. However, the animal cap assay demonstrated that Kielin injection did not have a direct neuralizing effect. Animal caps differentiate into neural tissues when BMP signaling is blocked (11, 47) and is shown to provide a sensitive assay system for attenuation of BMP. These results show that Kielin has a dorsalizing activity, which is not simply explained by blockade of BMP signaling. As a recent report demonstrated that Chd-type cys-rich modules bind directly to BMP (29), it remains possible that Kielin may function through binding to other transforming growth factor beta family molecules that have differential effects on mesoderm and ectoderm. So far, biochemical data on physical interaction with BMP are not available because Kielin protein has not yet been successfully overproduced. We infer that the large protein size (2,327 aa long) and a high cys content in the Chd repeats may have been hindrances to overproduction.
Another signaling event that leads to mesoderm dorsalization in the VMZ assay is blockade of the Wnt signaling. Wnt signaling has two contrasting activities depending on the time of action. Injection of Wnt8 DNA constructs, which reflects the zygotic effects, ventralizes mesoderm, whereas injection of Wnt8 mRNA, which mimics the effects of maternal factors, induces double-axis formation (48). Wnt-binding antagonists such as WIF dorsalize mesoderm when injected alone whereas they suppress Wnt8-induced double axis formation when coinjected with Wnt8 mRNA (49). In our preliminary experiments, we did not observe suppressing effects of Kielin on Wnt8 mRNA-induced double-axis formation. This may indicate that dorsalization of Kielin involves a novel mode of action rather than simply antagonizing and binding to BMP and Wnt in the extracellular space. In the future, it will be intriguing to examine the possibility that the activity of Kielin is mediated through the amino-terminal (thrombospondin-like) and carboxyl-terminal (vWF-like) domains.
Possible Roles of Kielin in Embryonic Patterning.
The dorsalizing activity of Kielin is consistent with its expression in the axial mesoderm, which patterns neighboring mesodermal tissues. In Xenopus, the dorsal-ventral polarity of the mesodermal mantle is roughly determined during early gastrulation by the antagonistic BMP and noggin/Chd signals (2). Because the onset of Kielin expression occurs at the midgastrula stage, Kielin is likely to be involved in the late events, such as fine patterning and maintenance, rather than in the initiation of the dorsal-ventral axis formation. For instance, the dorsalizing activity of Kielin may play a role in somite patterning by inducing MyoD and in the maintenance of axial mesoderm itself by keeping up the expression of HNF-3β and Xnot (Fig. 6).
On the other hand, Kielin also is expressed in the floor plate. However, it remains to be elucidated what function Kielin products have in the CNS. Although we tested a number of regional neural markers, we have not yet observed apparent effects of Kielin on the CNS patterning. For instance, injection of Kielin mRNA in vivo or in the animal cap does not induce floor plate markers (Shh, HNF-3β, F-spondin, or Kielin itself) or the motoneuron marker HB9. It is possible that the lack of apparent neural phenotypes is caused by the time course of overexpression. Kielin expression in the ventral CNS starts at neurula stages. In Xenopus, RNA is injected at early cleavage stages, and an efficient accumulation of the translated products is observed between the midblastula transition and midgastrula stages. This period is just right for mesodermal patterning but may be too early for neural patterning in which the floor plate-derived factors are involved. To further investigate the roles of Kielin in Xenopus neural patterning, it is essential to drive Kielin expression after gastrulation by using the frog transgenic technique and appropriate promoters (50). Transgenic and gene disruption studies using a mouse homologue also may provide complementary information on roles of Kielin in the CNS. Considering the unique biological properties of Kielin described above, further studies on Kielin should reveal new aspects of inductive functions of the embryonic midline.
Acknowledgments
We thank Drs. Robert Klein and Arnon Rosenthal (Genentech, Inc.) for their generous gift of SPS materials; Drs. Igor Dawid, Eddy De Robertis, and Herbert Steinbeisser for plasmids; and Drs. Siew-Lan Ang, Chris Wright, Matthias Hammerschmidt, and Hiroshi Kawasaki for valuable comments on this work. M.M. is grateful to Professor Jun Kimura for providing the opportunity to work on this project. This work was supported by grants from the Ministry of Education, the Ministry of Health and Welfare, the Organization of Pharmaceutical Safety and Research, the Precursory Research for Embryonic Science and Technology (PRESTO) program, Human Frontier Science Program Organization, Mochida Foundation, Yamanouchi Foundation Naitou Foundation, and Nissan Foundation.
Abbreviations
- Chd
chordin
- BMP
bone morphogenetic protein
- CNS
central nervous system
- VMZ
ventral marginal zone
- vWM
von Willebrand factor
- NCAM
neural cell adhesion molecule
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB026192).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090020497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090020497
References
- 1.Spemann H, Mangold H. Roux's Arch Entw Mech. 1924;100:599–638. [Google Scholar]
- 2.De Robertis E M, Kim S, Leyns L, Piccolo S, Bachiller D, Agius E, Belo J A, Yamamoto A, Hainski-Brousseau A, Brizuela B, et al. Cold Spring Harbor Symp Quant Biol. 1997;XLII:169–175. [PubMed] [Google Scholar]
- 3.Smith W C, Harland R M. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- 4.Hemmati-Brivanlou A, Kelly O G, Melton D A. Cell. 1994;77:283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 5.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont L K, De Robertis E M. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb T M, Knecht A K, Smith W C, Stachel S E, Economides A N, Stahl N, Yancopolous G D, Harland R M. Science. 1993;262:713–718. doi: 10.1126/science.8235591. [DOI] [PubMed] [Google Scholar]
- 7.Sasai Y, Lu B, Steinbeisser H, De Robertis E M. Nature (London) 1995;376:333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- 8.Piccolo S, Sasai Y, Lu B, De Robertis E M. Cell. 1996;86:589–598. doi: 10.1016/s0092-8674(00)80132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmerman L B, De Jesus-Escobar J M, Harland R M. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 10.Fainsod A, Deissler K, Yelin R, Marom K, Epstein M, Pillemer G, Steinbeisser H, Blum M. Mech Dev. 1997;63:39–50. doi: 10.1016/s0925-4773(97)00673-4. [DOI] [PubMed] [Google Scholar]
- 11.Sasai Y, De Robertis E M. Dev Biol. 1997;182:5–20. doi: 10.1006/dbio.1996.8445. [DOI] [PubMed] [Google Scholar]
- 12.Yamada T, Pfaff S L, Edlund T, Jessell T M. Cell. 1993;73:673–686. doi: 10.1016/0092-8674(93)90248-o. [DOI] [PubMed] [Google Scholar]
- 13.Fan C-M, Tessier-Lavigne M. Cell. 1994;79:1175–1186. doi: 10.1016/0092-8674(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 14.Echelard Y, Epstein D J, St-Jacques B, Shen L, Mohler J, McMahon J A, McMahon A P. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 15.Krauss S, Concordet J-P, Ingham P W. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- 16.Riddle R D, Johnson R L, Laufer E, Tabin C. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 17.Roelink H, Augsburger A, Heemskerk J, Korzh V, Norlin S, Ruiz i Altaba A, Tanabe Y, Placzek M, Edlund T, Jessell T M, et al. Cell. 1994;76:761–775. doi: 10.1016/0092-8674(94)90514-2. [DOI] [PubMed] [Google Scholar]
- 18.Tanabe Y, Roelink H, Jessell T M. Curr Biol. 1995;5:651–658. doi: 10.1016/s0960-9822(95)00130-8. [DOI] [PubMed] [Google Scholar]
- 19.Johnson R L, Laufer E, Riddle R D, Tabin C. Cell. 1994;79:1165–1173. doi: 10.1016/0092-8674(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 20.Hemmati-Brivanlou A, Stewart R M, Harland R M. Science. 1990;250:800–802. doi: 10.1126/science.1978411. [DOI] [PubMed] [Google Scholar]
- 21.Chiang C, Litingtung Y, Lee E, Young K E, Corden J L, Westphal H, Beachy P A. Nature (London) 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 22.Schauerte H E, van Eeden F J, Fricke C, Odenthal J, Strahle U, Haffter P. Development (Cambridge, UK) 1998;125:2983–2993. doi: 10.1242/dev.125.15.2983. [DOI] [PubMed] [Google Scholar]
- 23.Chandrasekhar A, Warren J T, Jr, Takahashi K, Schauerte H E, van Eeden F J, Haffter P, Kuwada J Y. Mech Dev. 1998;76:101–115. doi: 10.1016/s0925-4773(98)00101-4. [DOI] [PubMed] [Google Scholar]
- 24.Klein R D, Gu Q, Goddard A, Rosenthal A. Proc Natl Acad Sci USA. 1996;93:7108–7113. doi: 10.1073/pnas.93.14.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sive H, Grainger R M, Harland R M. In: Early Development of Xenopus laevis. Sive H, Grainger R M, Harland R M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 2000. pp. 249–274. [Google Scholar]
- 26.Gont L K, Fainsod A, Kim S H, De Robertis E M. Dev Biol. 1996;174:174–178. doi: 10.1006/dbio.1996.0061. [DOI] [PubMed] [Google Scholar]
- 27.Steinbeisser H, Fainsod A, Niehrs C, Sasai Y, De Robertis E M. EMBO J. 1995;14:5230–5243. doi: 10.1002/j.1460-2075.1995.tb00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebagliati M R, Toyama R, Haffter P, Dawid I B. Proc Natl Acad Sci USA. 1998;95:9932–9937. doi: 10.1073/pnas.95.17.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larraín J, Bachiller D, Lu B, Agius E, Piccolo S, De Robertis E M. Development (Cambridge, UK) 2000;127:821–830. doi: 10.1242/dev.127.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bornstein P. FASEB J. 1992;6:3290–3299. doi: 10.1096/fasebj.6.14.1426766. [DOI] [PubMed] [Google Scholar]
- 31.Hunt L T, Barker W C. Biochem Biophys Res Commun. 1987;144:876–882. doi: 10.1016/s0006-291x(87)80046-3. [DOI] [PubMed] [Google Scholar]
- 32.Dirksen M L, Jamrich M. Genes Dev. 1992;6:599–608. doi: 10.1101/gad.6.4.599. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz i Altaba A, Jessell T M. Development (Cambridge, UK) 1992;116:81–93. doi: 10.1242/dev.116.Supplement.81. [DOI] [PubMed] [Google Scholar]
- 34.Ekker S C, McGrew L L, Lai C-J, Lee J J, von Kessler D P, Moon R T, Beachy P A. Development (Cambridge, UK) 1995;121:2337–2347. doi: 10.1242/dev.121.8.2337. [DOI] [PubMed] [Google Scholar]
- 35.Jones C M, Kuehn M R, Hogan B L, Smith J C, Wright C V. Development (Cambridge, UK) 1995;121:3651–3662. doi: 10.1242/dev.121.11.3651. [DOI] [PubMed] [Google Scholar]
- 36.Joseph E M, Melton D A. Dev Biol. 1997;184:367–372. doi: 10.1006/dbio.1997.8510. [DOI] [PubMed] [Google Scholar]
- 37.Cho K W, Blumberg B, Steinbeisser H, De Robertis E M. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Dassow G, Schmidt J E, Kimelman D. Genes Dev. 1993;7:355–366. doi: 10.1101/gad.7.3.355. [DOI] [PubMed] [Google Scholar]
- 39.Gont L K, Steinbeisser H, Blumberg B, De Robertis E M. Development (Cambridge, UK) 1993;119:991–1004. doi: 10.1242/dev.119.4.991. [DOI] [PubMed] [Google Scholar]
- 40.Ang S-L, Rossant J. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 41.Weinstein D C, Ruiz i Altaba A, Chen W S, Hoodless P, Prezioso V R, Jessell T M, Darnell J E., Jr Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- 42.Sasaki H, Hogan B L M. Cell. 1994;76:103–115. doi: 10.1016/0092-8674(94)90176-7. [DOI] [PubMed] [Google Scholar]
- 43.Hatta K, Kimmel C B, Ho R K, Walker C. Nature (London) 1991;350:339–341. doi: 10.1038/350339a0. [DOI] [PubMed] [Google Scholar]
- 44.Feldman B, Gates M A, Egan E S, Dougan S T, Rennebeck G, Sirotkin H I, Schier A F, Talbot W S. Nature (London) 1998;395:181–185. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- 45.Sampath K, Rubinstein A L, Cheng A M, Liang J O, Fekany K, Solnica-Krezel L, Korzh V, Halpern M E, Wright C V. Nature (London) 1998;395:185–189. doi: 10.1038/26020. [DOI] [PubMed] [Google Scholar]
- 46.Rebagliati M R, Toyama R, Fricke C, Haffter P, Dawid I B. Dev Biol. 1998;199:261–272. doi: 10.1006/dbio.1998.8935. [DOI] [PubMed] [Google Scholar]
- 47.Hemmati-Brivanlou A, Melton D A. Cell. 1997;88:13–17. doi: 10.1016/s0092-8674(00)81853-x. [DOI] [PubMed] [Google Scholar]
- 48.Christian J L, Moon R T. Genes Dev. 1993;7:13–28. doi: 10.1101/gad.7.1.13. [DOI] [PubMed] [Google Scholar]
- 49.Hsieh J C, Kodjabachian L, Rebbert M L, Rattner A, Smallwood P M, Samos C H, Nusse R, Dawid I B, Nathans J. Nature (London) 1999;398:431–436. doi: 10.1038/18899. [DOI] [PubMed] [Google Scholar]
- 50.Kroll K L, Amaya E. Development (Cambridge, UK) 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]