Abstract
Chikungunya virus (CHIKV) is an emerging arbovirus and is an important human pathogen. Infection of humans by CHIKV can cause a syndrome characterized by fever, headache, rash, nausea, vomiting, myalgia, arthralgia and occasionally neurological manifestations such as acute limb weakness. It is also associated with a fatal haemorrhagic condition. CHIKV is geographically distributed from Africa through Southeast Asia and South America, and its transmission to humans is mainly through the Aedes aegypti species mosquitoes. The frequency of recent epidemics in the Indian Ocean and La Reunion islands suggests that a new vector perhaps is carrying the virus, as Aedes aegypti are not found there. In fact, a relative the Asian tiger mosquito, Aedes albopictus, may be the culprit which and has raised concerns in the world health community regarding the potential for a CHIK virus pandemic. Accordingly steps should be taken to develop methods for control of CHIKV. Unfortunately, currently is no specific treatment for Chikungunya virus and there is no vaccine currently available. Here we present data of a novel consensus-based approach to vaccine design for CHIKV, employing a DNA vaccine strategy. The vaccine cassette was designed based on CHIKV Capsid and Envelope specific consensus sequences with several modifications, including codon optimization, RNA optimization, the addition of a Kozak sequence, and a substituted immunoglobulin E leader sequence. The expression of Capsid, envelope E1 and E1 was evaluated using T7-coupled transcription/translation and immunoblot analysis. A recently developed, adaptive constant-current electroporation technique was used to immunize C57BL/6 mice with an intramuscular injection of plasmid coding for the CHIK-Capsid, E1 and E2. Analysis of cellular immune responses, including epitope mapping, demonstrates that electroporation of these constructs induces both potent and broad cellular immunity. In addition, antibody ELISAs demonstrate that these synthetic immunogens are capable of inducing high titer antibodies capable of recognizing native antigen. Taken together, these data support further study of the use of consensus CHIK antigens in a potential vaccine cocktail.
Keywords: Chikungunya virus (CHIKV), DNA vaccination, Envelope, Capsid, Electroporation
INTRODUCTION
Chikungunya virus (CHIKV) is an alphavirus indigenous to tropical Africa and Asia, where it is transmitted to humans by the bite of infected mosquitoes, usually of the genus Aedes [1]. Chikungunya fever, the disease caused by CHIKV, was first recognized in epidemic form in East Africa during 1952–1953 [2]. The word "chikungunya" is thought to derive from description in local dialect of the contorted posture of patients afflicted with the severe joint pain associated with this disease [1–3]. Because CHIK fever epidemics are sustained by human-mosquito-human transmission, the epidemic cycle is similar to those of dengue and urban yellow fever. Large outbreaks of CHIK fever have been reported recently on several islands in the Indian Ocean and in India [4–7]. In 2006, CHIK fever cases also have been reported in travelers returning from known outbreak areas to Europe, Canada, the Caribbean (Martinique), and South America (French Guyana) [5–9]. During 2005–2006, 12 cases of CHIK fever were diagnosed serologically and virologically at CDC (USA) in travelers who arrived in the United States from areas known to be epidemic or endemic for CHIK fever [10]. CHIKV is spread by the bite of an infected mosquito. Mosquitoes become infected when they feed on CHIKV infected individuals. Monkeys, and possibly other wild animals, may also get infected but their role as reservoirs of the CHIKV is not yet documented. Infected mosquitoes can then spread the virus to other humans when they bite. □ Aedes aegypti (the yellow fever mosquito), a household container breeder and aggressive daytime biter which is attracted to humans, is the primary vector of CHIKV to humans. Aedes albopictus (the Asian tiger mosquito) may also play a role in human transmission is Asia, and various forest-dwelling mosquito species in Africa have been found to be infected with the virus [11–17]. Recently studies have demonstrated that an envelope E1-A226V mutation is directly responsible for a significant increase in CHIKV infectivity for Aedes albopictus, and further confirmed that a single amino acid substitution can influence vector specificity. This finding provides a plausible explanation of how this mutant virus caused an epidemic in a region lacking the typical insect vector [18]. There is no specific vaccine or specific antiviral treatment for Chikungunya. Live attenuated vaccine trials were carried out in 2000, but funding for the project was discontinued and there is no vaccine currently available. However, several adverse events associated with this prior vaccine are well documented, accordingly, new vaccine strategies must be developed [3,5]. Here we describe construction of and immunization of mice with a new prototypic DNA vaccine for CHIKV. This is the first such study we are aware of. The vaccine induces both humoral and cellular immunity in vivo.
2. MATERIALS AND METHODS
2.1. CELLS AND ANIMALS
The BHK-21 cell line obtained from the ATCC was grown and maintained in DMEM medium supplemented with 10% fetal calf serum. The mammalian plasmid expression vector, pVax1, was purchased from Invitrogen (Carlsbad, CA). Three- to 4-week-old female C57BL/6 mice (Jackson laboratories, Indianapolis, IN) were used in these experiments and divided into three experimental groups (n□□4). All animals were housed in a temperature-controlled, light-cycled facility in accordance with the guidelines of the National Institutes of Health (Bethesda, MD, USA) and the University of Pennsylvania (Philadelphia, PA, USA) Institutional Animal Care and Use Committee (IACUC).
2.2. CHIKV DNA CONSTRUCT AND SYNTHESIS
The CHIKV core and envelope genes were designed by synthetic primer synthesis followed by DNA-PCR amplification using the consensus strain predicted of sequences collected from the NCBI database of all CHIKV viruses. Consensus sequences were optimized for expression, including codon and RNA optimization (GeneArt, Regensburg, Germany) and inserted into the pVax1 expression vector (Invitrogen).
2.3. IN VITRO AND IN VIVO EXPRESSION
Construct expression was confirmed by utilizing a T7 promoter in the pVax1 backbone and T7-based coupled transcription/translation system (Promega, Madison, WI) containing S35-methionine CHIKV genes. The synthesized protein was immunoprecipitated using anti-E1, anti-E2 or anti-Cap antibodies. The immunoprecipitated protein was electrophoresed on a 12% NuPage SDS-PAGE gel (Invitrogen, CA) and subsequently fixed and dried. Autoradiography was performed to detect an incorporated S35-labeled gene product. In vivo expression, BHK-21 cells (1×106) were transfected with CHIKV constructs using Fugene transfection method (Roche, NJ). Seventy-two hours after transfection, proteins (50µg) were fractioned on SDS–PAGE (12%) and transferred to a PVDF membrane (Bio-Rad, Hercules, CA). Immunoblot analyses were performed with specific antiserum, which was raised in mice and the expressed protein s were visualized with horseradish peroxidase conjugated goat anti-mouse IgG using an ECL detection system (Amersham Pharmacia Biotech, Piscataway, NJ) [19].
2.4. IMMUNIZATION AND ELECTROPHORATION
A standard protocol was used to prime animals with plasmid DNA [20]. Groups of four mice were immunized twice with pCHIKV genes (25µg) 2–3 times, 2 weeks apart, and sacrificed 1 week following the final immunization. All immunizations were delivered into the quadriceps muscles in a total volume of 100µl by in vivo electroporation (EP) (VGX Pharmaceuticals Inc, Blue Bell, PA). The animals were sacrificed 7 days after the last immunization, whereupon serum and the spleen were collected for immunology assays. Blood from both control and immunized mice was obtained 1 week after the second and third immunizations, respectively.
Square-wave pulses were used in all experiments and delivered with the constant-current EKD that was designed and tested in our laboratory [20–22]. A three electrode array (3-EA) was used in the mouse experiments. The 3-EA consists of three 26-gauge solid stainless steel electrodes in an isosceles triangle formation, with the two long sides 0.5mm in length and short side 0.3mm in length, held together with a nonconductive plastic. Specific EP conditions for the mouse experiments were using constant current, 0.1Amps, three pulses, 52 msec/pulse, 4 sec between pulses. The lag time between plasmid injection and EP was about 20sec. The sequence of events for plasmid administration/EP was as follows: Place a disposable electrode assembly in the receptacle of the handle, press initiation button on handle and enter animal experimental group number, inject 50µl of DNA construct (25µg total DNA) plasmid using insulin syringe, immediately place needles into area surrounding the injection site, press initiation button on handle, and after 4 second countdown, pulses will be delivered. After 5 seconds following electroporation, the array is gently removed from muscle. All electrodes were completely inserted into the muscle during all treatments [21, 22]. All DNA was made using endotoxin-free Qiagen columns. All animals were housed in a temperature-controlled, light-cycled facility at the University of Pennsylvania, and their care was under the guidelines of the National Institutes of Health and the University of Pennsylvania.
2.5. CELLULAR RESPONSE: ELISPOT ASSAY
An ELISPOT assay was conducted as previously described [23]. Briefly, ELISpot 96-well plates (Millipore) were coated with anti-mouse IFN-γ capture Ab and incubated for 24h at 4 °C (R&D Systems). The following day, plates were washed and blocked for 2h with 1% BSA. Two hundred thousand splenocytes from the immunized mice were added to each well and stimulated overnight at 37 °C in 5% CO2 in the presence of RPMI 1640 (negative control), Con A (positive control), or specific peptide Ags (10µg/ml; Invitrogen). Peptide pools consist of 15-mer peptides overlapping by 11 amino acids. After 24h of stimulation, the cells were washed and incubated for 24 h at 4°C with biotinylated anti-mouse IFN-γ Ab (R&D Systems). The plates were washed, and streptavidin–alkaline phosphatase (R&D Systems) was added to each well and incubated for 2h at room temperature. The plate was washed, and 5-bromo-4-chloro-3′-indolylphosphate p-toluidine salt and nitro blue tetrazolium chloride (chromogen color reagent; R&D Systems) were added to each well. The plate was then rinsed with distilled water and dried at room temperature. Spots were counted by an automated ELISPOT reader (CTL Limited) [21–23].
2.6. HUMORAL IMMUNE RESPONSE: ANTIBODY ELISA
The antibody levels following each DNA priming injection and the humoral immune response to vaccination was determined to each CHIKV DNA constructs. Briefly, 96-well high-binding polystyrene plates (Corning, NY) plates were coated overnight at 4 °C with synthesized specific peptides (2µg/ml), which was diluted in PBS. The next day, plates were washed with PBST (PBS, 0.05% Tween 20), blocked for 1h with 3% BSA in PBST, and incubated with 1;100 dilutions of serum from immunized and naïve mice for 1h at 37 °C. Bound IgG was detected using goat anti-mouse IgG-HRP (Research Diagnostics, NJ) at a dilution of 1:5,000. Bound enzyme was detected by the addition of the chromogen substrate solution TMB (R&D Systems), and read at 450 nm on a Biotek EL312e Bio-Kinetics reader. All serum samples were tested in duplicate [22].
3. RESULTS
3.1. CONSTRUCTION OF NOVEL ENVELOPE AND CAPSID DERIVED EXPRESSION CASSETTES
CHIKV is a variable pathogen and is an arthropod-borne virus transmitted to human beings by Aedes spp mosquitoes. Chikungunya virus illness is associated with fever, severe arthralgias, rash, headache, and malaise. Other symptoms include muscle aches and retro-orbital pains. Chikungunya disease is rarely fatal but is associated with significant morbidity. Chikungunya illness has an approximate incubation period of 1–2 weeks. At the beginning of 2006, after a period of lower transmission during the winter and with the arrival of the Southern Hemisphere summer, Reunion Island suffered an explosive outbreak. An estimate of 266,000 residents (population 770,000) infected with CHIKV was reported, and 248 death certificates gave CHIKV as the possible cause of death [10,12]. Evidence for intrauterine infection in pregnant women and vertical transmission has been documented [12,13,17]. Sequence analysis has revealed the existence of geographically clustered lineages of the virus. Phylogenetic analyses based on partial E1 sequences revealed the existence of three distinct phylogroups for CHIKV: one with the West African isolates, another including the Asian isolates and one regrouping the Eastern, Central and South African isolates [15,17]. To develop an immunogen with the ability to induce cross-reactive immune responses against Chikungunya virus (CHIKV), we designed consensus constructs against CHIKV viral envelope E1, E2 and the core protein Capsid. For these constructs 21 sequences were chosen from chikungunya viruses isolated between 1952 and 2006 that caused infection and death in humans. The DNA sequences chosen for each gene were from S27 strain (first isolate) and the strains across various countries including the La Reunion island outbreak isolates in order to avoid sampling bias. The DNA sequences were aligned and the most common nucleotide at each position was chosen for the synthetic sequence. Deduced amino acid sequences were used to guide the introduction of alignment gaps so that they were introduced between codons maintaining the reading frame. After generation of the consensus sequences, an IgE leader sequence was added to N-terminus to enhance expression and the secretion, the construct was optimized by the codon optimization, and replacement of the existing Kozak sequence with a stronger sequence (GCCGCCACC) (Fig. 1A). For analysis, a His tag was added to the C-terminus of both E1 and Capsid for expression verification. These constructs were then produced in bacterial and purified as previously described for analysis, expression and immunogenicity studies [21]. Fig. 1B depict the agrose gel electrophoresis of the constructs encoding envelope E1, E2 and Capsid DNA.
Figure 1.
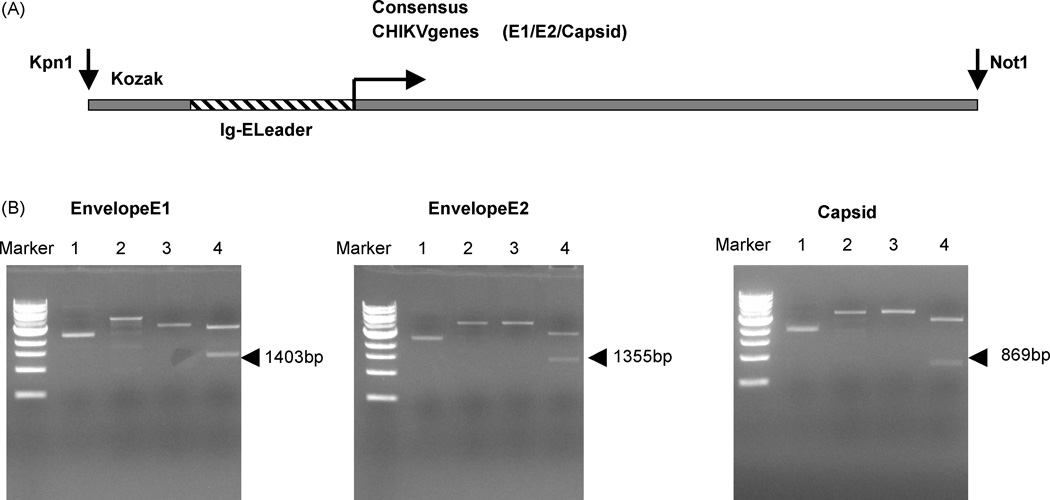
(A) Schematic representation of the strategy for cloning the IgE–leader CHIKV fusion gene into the pVax1 vector. (B) Agarose gel photograph showing the CHIKV plasmid (Envelope E1, E2 and Capsid) linear specific band indicated (lane 4) with Kpn1 and Not1 double digestion which produced the size of 1403bp, 1355bp and 869bp size, respectively.
3.2. CONSENSUS CONSTRUCT EXPRESSION
The expressions of the three CHIKV consensus constructs were verified using multiple techniques. To visualize the proteins produced in vitro S35-labeled In vitro T7-coupled transcription and translation assay was performed. The translation products were immunoprecipitated using the His tag antibody and gel analysis was performed. SDS-PAGE and radiographic analysis showed that each construct (Envelope E1, E2 and Capsid) runs at its theoretically predicted molecular weight (Fig. 2A). We next sought to examine the expression of these constructs in mammalian cells vivo. Following transfection into BHK-21 cells the proteins were extracted after three days and expression was detected using specific polyclonal antibodies by Western blot analysis (Fig. 2B). In envelope E1 construct transfected cells a 52-kDa protein and a 36-kDa protein was observed in Capsid construct transfected cells upon immunoblotting with specific antibodies.
Figure 2.
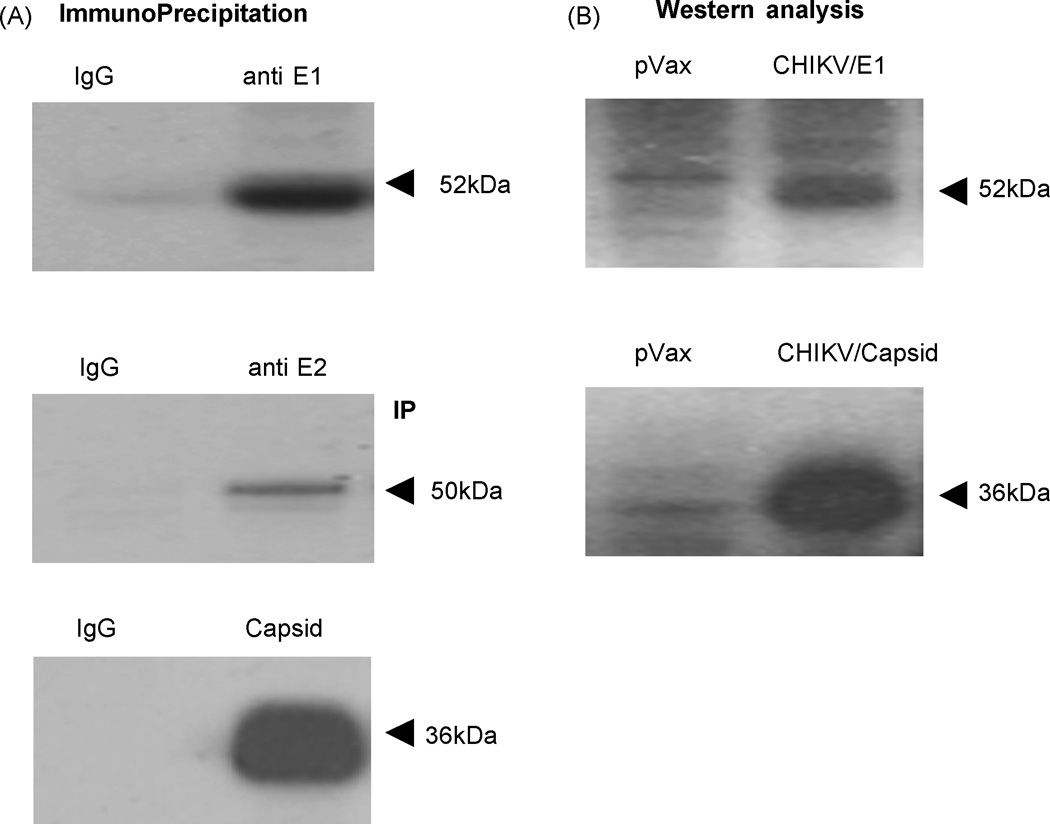
Characterization of CHIKV constructs. (A) Shows S35-labeled in vitro translation of the synthesized construct. The antigens CHIKV-E1, CHIKV-E2 and CHIKV-Capsid were translated and was immunoprecipitated using the specific E1, E2 and Caspsid antibodies respectively and run on a 12% SDS gel, and subject to radiographic analysis. The antigen runs at its predicted molecular weight, confirming expression. (B) Western blot analysis of CHIKV-E1 and CHIKV-Capsid construct in BHK-21 cells. Two days post transfection, the transfected cell lysates were prepared and immunoblot with polyclonal CHIKV-E1 antiserum, which was raised in mice shows the expression of 52kDa E1 protein and 36kDa for Capsid protein.
3.3. HUMORAL IMMUNOGENICITY
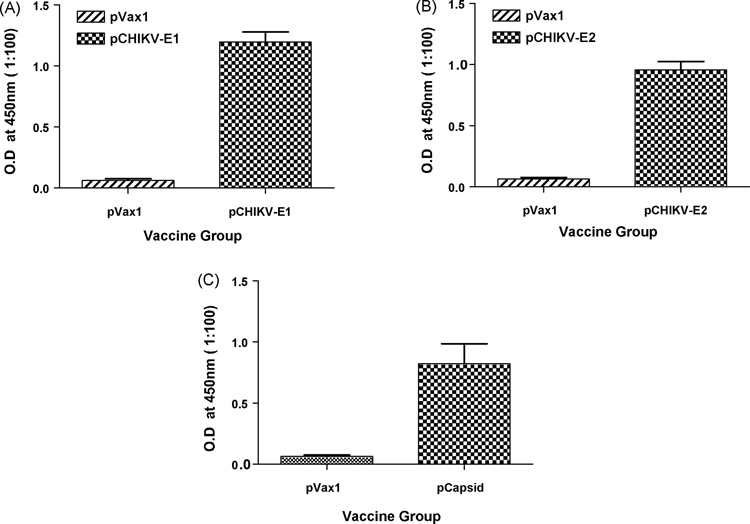
We hypothesize that the strength of our consensus immunogens to protect from lethal CHIK virus will lie with the cellular arm of the immune system. Furthermore, cross-reactive but non-neutralizing antibodies can provide a certain degree of protection against disease severity. In order to determine if our constructs induce antibody responses, we performed an antibody ELISA on CHIK-immunized mouse serum to determine antibody titer from the sera obtained after the DNA immunizations were tested for antibody response by ELISA. Anti E1 specific IgG antibody in the sera of mice immunized with envelope E1 were significantly higher than in the sera of mice immunized with vector control (Fig. 3A). Similarly anti E2 specific IgG antibody and capsid specific IgG antibody in the sera of mice immunized with envelope E2 and capsid constructs respectively were significantly higher than in the sera of mice immunized with vector control (Fig. 3B&C). These results further supported the alternative means of plasmid delivery, specifically electroporation increased the antibody production response to the DNA vaccine immunogen.
Figure 3.
Anitbody ELISA. (A) C57BL/6 mice were immunized two times, each 2 weeks apart, with 25µg 25µg pVax1 vector or CHIKV plasmids as indicated and sacrificed 1 week later. Serum was collected and subject to analysis for Total IgG production. Serum was incubated for 1h at 37 °C on 96-well plates coated with 2µg/ml of respective CHIKV peptides, and antibody was detected using anti-mouse IgG-HRP. Values represent the mean (±S.D.) of duplicate wells.
3.4. CELLULAR IMMUNOGENICITY
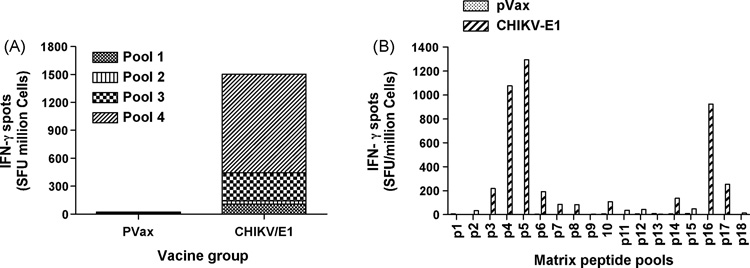
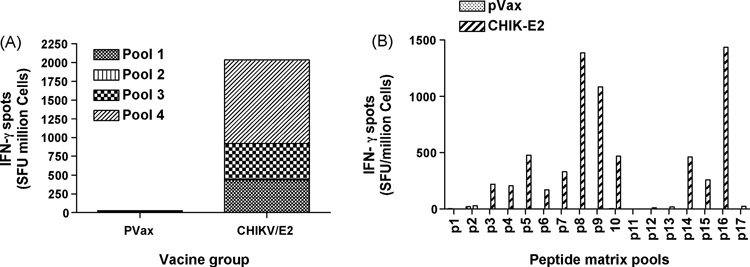
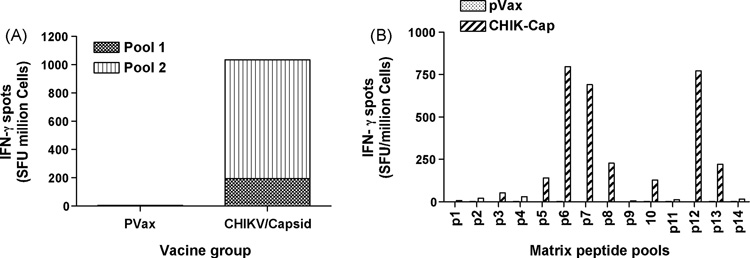
The ability of E1, E2 and Capsid constructs to induce CD8+ CTL responses was determined next by IFN-γ ELISpot assays. The consensus envelope constructs E1, E2 as well as and the Capsid vaccines were able to induce strong IFN-γ responses in C57BL/6 mice after three immunizations (Fig. 4A, 5A and 6A). For molecular characterization of the cellular immune responses induced by the envelope E1, the ELISpot assay was performed against a library of peptides spanning entire envelope E1. Seventy four 15-mer peptides with 9 amino acid overlaps between them, which span residues 1– 435 of E1 protein and peptides which span 1–423 of E2 protein were used. The envelopes induced a dominant epitope HSMTNAVTI in the E1 protein (Fig. 4B) and IILYYYELY in the E2 protein (Fig. 5B). Similarly for Capsid protein, the ELIspot assay was performed against a library of peptides spanning entire Capsid protein. Forty five 15-mer peptides with 9 amino acid overlaps between them, which span residues 1– 261 of Capsid protein, were used. The dominant epitope ACLVGDKVM was induced by the construct Capsid (Fig. 5B). Interestingly the dominant epitope which is induced by the construct CHIKV-E1 carries the 226A–V mutation which suggests that the construct can also effectively induce immune response against the newly emerged mutant virus. This finding may suggest that the immune response may be capable of driving the evolution of the virus through at cell selection process.
Figure 4.
Interferon-γ response to envelope E1 measured by ELISpot. C57BL/6 mice were immunized two times, each 2 weeks apart, with 25µg pVax1 vector or pCHIKV-E1 and sacrificed 1 week later. (A) Splenocytes were harvested and cultured overnight in the presence of R10 (negative control) or 10µg/ml of one of four peptide pools, made up of 15-mer peptides overlapping by 9 amino acids, spanning the length of the E1 protein. Responses to CHIKV-E1 are shown as stacked group mean responses. (B) Splenocytes were harvested and cultured overnight in the presence of R10 (negative control) or 10µg/ml of one of eighteen peptide pools, made up of 15-mer peptides overlapping by 9 amino acids, spanning the length of the matrix E1 protein. Spot forming units (SFU) were quantified by an automated ELISpot reader, and the raw values were normalized to SFU per million splenocytes. Values represent the mean of triplicate wells.
Figure 5.
Interferon-γ response to CHIKV envelope E2 measured by ELISpot. C57BL/6 mice were immunized two times, each 2 weeks apart, with 25 µg pVax1 vector or pCHIKV-E2 and sacrificed 1 week later. (A) Splenocytes were harvested and cultured overnight in the presence of R10 (negative control) or 10µg/ml of one of four peptide pools, made up of 15-mer peptides overlapping by 9 amino acids, spanning the length of the E2 protein. Responses to CHIKV-E2 are shown as stacked group mean responses. (B) Splenocytes were harvested and cultured overnight in the presence of R10 (negative control) or 10µg/ml of one of eighteen peptide pools, made up of 15-mer peptides overlapping by 9 amino acids, spanning the length of the matrix E2 protein. Spot forming units (SFU) were quantified by an automated ELISpot reader, and the raw values were normalized to SFU per million splenocytes. Values represent the mean of triplicate wells.
Figure 6.
Interferon-γ response CHIKV-Capsid measured by ELISpot. C57BL/6 mice were immunized two times, each 2 weeks apart, with 25µg pVax1 vector or pCHIKV-Capsid and sacrificed 1 week later. (A) Splenocytes were harvested and cultured overnight in the presence of R10 (negative control) or 10µg/ml of one of four peptide pools, made up of 15-mer peptides overlapping by 9 amino acids, spanning the length of the capsid protein. Responses to CHIKV-Capsid are shown as stacked group mean responses. (B) Splenocytes were harvested and cultured overnight in the presence of R10 (negative control) or 10µg/ml of one of eighteen peptide pools, made up of 15-mer peptides overlapping by 9 amino acids, spanning the length of the matrix capsid protein. Spot forming units (SFU) were quantified by an automated ELISpot reader, and the raw values were normalized to SFU per million splenocytes. Values represent the mean of triplicate wells.
4. DISCUSSION
Chikungunya, an arbo virus with epidemic potential that can produce a sudden debilitating disease, its potential as a threat to the developing world, the developed world based on its continued spread and as military threat due to soldier deployment in newly emerging endemic region conflict zones is considerable. CHIKV infections have a significant impact economically as, local businesses are affected by absenteeism in endemic areas due to the incapacitating symptoms of this infection on employees. This economic effect is highest on the individual family members who are unable to work for weeks or months. Due to the debilitating infection sequelai, the lack of specific antiviral treatment and any current usable vaccine to prevent the disease is a major impediment to managing or controlling new CHIKV outbreaks. Since late 2004, the Chikungunya virus has reemerged with large outbreaks in various parts of the world predominantly in the Indian Ocean islands. These have caused public health crisises and grabbed the attention of the researchers worldwide. Importantly, most Chikungunya virus infections completely resolve within weeks or months however, there have been documented cases of CHIKV-induced arthralgia persisting for several years developing chronic joint problems. The fact that infection resolves after a long period supports that the immune system can rally to control this infection eventually. Furthermore, such a clearance phenotype supports a role in clearance for the T cell response. Earlier attempts to develop vaccines against Chikungunya such as formalin killed vaccine [3], Tween ether inactivated virus vaccine and live attenuated vaccines were moderately successful however, they were discontinued for various reasons [3].
Moreover all these vaccines were reported to produce only a serological response without induction of useful cellular immunity. The sheer magnitude of the 2005–2007, Chikungunya outbreaks underscores the need for a safe and effective vaccine against CHIKV [6]. One approach that may have benefit here is DNA immunization which induce both humoral and cellular responses and or protective immunity in some animal systems against viral infections such as, HIV, Hepatitis B, Hepatitis C viruses, influenza virus and arbo viruses such as JEV [22–24].
In this study we designed a vaccine cassette based on Capsid (Cap) and Envelope (E1) and Envelope (E2) specific consensus sequences with several modifications, including codon optimization, RNA optimization, the addition of a Kozak sequence, and a substituted immunoglobulin E leader sequence. The vaccine cassette was introduced into the DNA vaccine vector pVax1 at the specific site. The vaccine constructs were checked for the inserts using specific restriction digestion and by sequencing with the primer of the T7 promoter. The final constructs were efficiently expressed based on both in vitro expression as well as using in vivo western blot analysis. This confirmed that the viral constructs were correctly expressed and processed for further immunogenicity studies.
Recently, there has been much interest in the use of EP for the delivery of DNA vaccines. Recent studies of IM immunization + EP in small animal models and non-human primates have consistently reported increases in cellular and in particular, antibody responses [20–22]. Evaluation of the immune response induced in the C57BL/6 mice showed that the constructs were highly immunogenic and elicited T-cell immune response in the terms of an IFN-γ response and proliferation. ELISpot data from the present study suggests the magnitude of IFN-γ response was broad based in terms of the number of spots obtained. Although the envelope protein and the capsid in other related alpha viruses are known to be immunogenic, there is little knowledge about the immunogenicity of Chikungunya envelope and Capsid proteins. Elicitation of IFN-γ production from splenocytes by matrix peptide pools from different regions of the envelope E1 and Capsid identified the T cell dominant epitopes HSMTNAVTI and ACLVGDKVM in E1 and Capsid proteins respectively. Total IgG levels in vaccinated mice were found to be increased compared to that of unvaccinated controls suggesting the induction of strong humoral immune response. Subsequent studies further analyzing the type of antibody responses induced, in addition to the ability of these vaccines to drive protection against a wide range of Chikungunya virus challenges, is currently in progress.
This study suggests that these constructs could be studied further as vaccine candidates. Nonetheless, as this study is limited to the demonstration of efficient expression as well as immunogenicity after intramuscular injection of vaccine constructs followed by electrophoration in mice, an elaborate evaluation of its immunogenicity in more models including the nonhuman primate model is important. The synthetic cassette constructs described appear to be a convenient tool to investigate the immunobiology of Chikungunya virus further.
ACKNOWLEDGEMENT
This work was supported in part by grants from the NIH to D.B.W. and VGX Pharmaceuticals Inc, Philadelphia, PA, to D.B.W. and K.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Strauss JH, Strauss EG. The Alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson MC. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53. I. Clinical features. Trans R Soc Trop Med Hyg. 1955;49:28–32. doi: 10.1016/0035-9203(55)90080-8. [DOI] [PubMed] [Google Scholar]
- 3.Powers AM, Logue CH. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. Journal of General Virology. 2007;vol. 88(part 9):2363–2377. doi: 10.1099/vir.0.82858-0. [DOI] [PubMed] [Google Scholar]
- 4.Porterfield JH. Antigenic characteristics and classification of the Togaviridae. In: Schlesinger R, editor. The Togaviruses. New York: Academic Press; 1980. pp. 13–46. [Google Scholar]
- 5.Weaver SC, Barrett DT. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2004;2:789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chevillon C, Briant L, Renaud F, Devaux C. The Chikungunya threat: an ecological and evolutionary perspective. Trends Microbiol. 2008;16(2):80–88. doi: 10.1016/j.tim.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Lahariya C, Pradhan SK. Emergence of chikungunya virus in Indian subcontinent after 32 years: a review. J. Vector Borne Dis. 2006;43:151–160. [PubMed] [Google Scholar]
- 8.Vanlandingham DL, Hong C, Klingler K, Tsetsarkin K, McElroy KL, Powers AM, Lehane MJ, Higgs S. "Differential infectivities of o'nyong-nyong and chikungunya virus isolates in Anopheles gambiae and Aedes aegypti mosquitoes". Am J Trop Med Hyg. 2005;72(5):616–621. [PubMed] [Google Scholar]
- 9.Yergolkar PN, et al. Chikungunya outbreaks caused by African genotype, India. Emerg. Infect. Dis. 2006;12:1580–1583. doi: 10.3201/eid1210.060529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warner E, Garcia-Diaz J, Balsamo G, Shranatan S, Bergmann A, Blauwet L, Sohail M, Baddour L, Reed C, et al. Chikungunya fever diagnosed among international travelers – United States, 2005–2006. MMWR Morb Mortal Wkly Rep. 2006;55:1040–1042. [PubMed] [Google Scholar]
- 11.Turell MJ, Beaman JR, Tammariello RF. Susceptibility of selected strains of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) to chikungunya virus. J Med Entomol. 1992;29:49–53. doi: 10.1093/jmedent/29.1.49. [DOI] [PubMed] [Google Scholar]
- 12.Reiter P, Fontenille D, Paupy C. Aedes albopictus as an epidemic vector of chikungunya virus: another emerging problem? Lancet Infect Dis. 2006;6:463–464. doi: 10.1016/S1473-3099(06)70531-X. [DOI] [PubMed] [Google Scholar]
- 13.Johnston RE, Peters CJ. Alphaviruses associated primarily with fever and polyarthritis. In: Fields BN, Knipe DM, Howley PM, editors. Fields virology. Philadelphia: Lippincott-Raven Publishers; 1996. pp. 843–898. [Google Scholar]
- 14.Savarino A, Boelaert JR, Cassone A, Majori G, Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases? Lancet Infect Dis. 2003;3(11):722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A Single Mutation in Chikungunya Virus Affects Vector Specificity and Epidemic Potential. PLoS Pathog. 2007;7(312):e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grivard P, Le Roux K, Laurent P, Fianu A, Perrau J, Gigan J, Hoarau G, Grondin N, Staikowsky F, Favier F, Michault A. Molecular and serological diagnosis of Chikungunya virus infection. Pathol Biol (Paris) 2007;55(10):490–494. doi: 10.1016/j.patbio.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Vazeille-Falcoz M, Mousson L, Rodhain F, Chungue E, Failloux AB. Variation in oral susceptibility to dengue type 2 virus of populations of Aedes .aegypti from the islands of Tahiti and Moorea, French Polynesia. Am J Trop Med Hyg. 1999;60:292–299. doi: 10.4269/ajtmh.1999.60.292. [DOI] [PubMed] [Google Scholar]
- 18.Marie Vazeille, et al. Two Chikungunya Isolates from the Outbreak of La Reunion (Indian Ocean) Exhibit Different Patterns of Infection in the Mosquito, Aedes albopictus. PLoS ONE. 2007;11:1–9. doi: 10.1371/journal.pone.0001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muthumani K, Choo AY, Zong WX, Madesh M, Hwang DS, Premkumar A, Thieu KP, Emmanuel J, Kumar S, Thompson CB, Weiner DB. The HIV-1 Vpr and glucocorticoid receptor complex is a gain-of-function interaction that prevents the nuclear localization of PARP-1. Nat Cell Biol. 2006;8:170–179. doi: 10.1038/ncb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan AS, Smith LC, Abruzzese RV, Cummings KK, Pope MA, Brown PA, Draghia-Akli R. Optimization of electroporation parameters for the intramuscular delivery of plasmids in pigs. DNA Cell Biol. 2003;22(12):807–811. doi: 10.1089/104454903322625019. [DOI] [PubMed] [Google Scholar]
- 21.Lauren LA, Hirao LA, Wu L, Khan AS, Satishchandran A, Draghia-Akli R, Weiner DB. Intradermal/subcutaneous immunization by electrophoration improves plasmid vaccine delivery and potency in pigs and rhesus macaques. Vaccine. 2008;17(263):440–448. doi: 10.1016/j.vaccine.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 22.Laddy DJ, Yan J, Corbitt N, Kobasa D, Kobinger GP, Weiner DB. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine. 2007;25(16):2984–2989. doi: 10.1016/j.vaccine.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyer JD, Robinson TM, Kutzler MA, Vansant G, Hokey DA, Kumar S, Parkinson R, Wu L, Sidhu MK, Pavlakis GN, Felber BK, Brown C, Silvera P, Lewis MG, Monforte J, Waldmann TA, Eldridge J, Weiner DB. Protection against simian/human immunodeficiency virus (SHIV) 89.6P in macaques after coimmunization with SHIV antigen and IL-15 plasmid. Proc Natl Acad Sci., USA. 2007;20(10447):18648–18653. doi: 10.1073/pnas.0709198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng GH, Liu N, Zhou Y, Zhai YZ, Li XM, Dou XG. Immunologic analysis induced by DNA vaccine encoding E protein of Beijing-1 strain derived from Japanese encephalitis virus. Intervirology. 2007;50(2):93–98. doi: 10.1159/000097395. [DOI] [PubMed] [Google Scholar]