Abstract
Human SNM1B/Apollo is involved in the cellular response to DNA-damage, however, its precise role is unknown. Recent reports have implicated hSNM1B in the protection of telomeres. We have found hSNM1B to interact with TRF2, a protein which functions in telomere protection and in an early response to ionizing radiation. Here we show that endogenous hSNM1B forms foci which colocalize at telomeres with TRF1 and TRF2. However, we observed that additional hSNM1B foci could be induced upon exposure to ionizing radiation (IR). In live-cell-imaging experiments, hSNM1B localized to photoinduced double-strand breaks (DSBs) within 10s post-induction. Further supporting a role for hSNM1B in the early stages of the cellular response to DSBs, we observed that autophosphorylation of ATM, as well as the phosphorylation of ATM target proteins in response to IR, was attenuated in cells depleted of hSNM1B. These observations suggest an important role for hSNM1B in the response to IR damage, a role that may be, in part, upstream of the central player in maintenance of genome integrity, ATM.
Introduction
All living organisms possess mechanisms which respond to DNA-damage and result in the repair of lesions or the elimination of irreparably damaged cells, thus maintaining genomic integrity. We have recently described hSNM1B as a new gene involved in this cellular response to DNA damage [1]. The hSNM1B protein belongs to the SNM1-family. The common features of the proteins in this group are two domains, a metallo-β-lactamase domain and a β-CASP region, which are characteristic of members of the β-lactamase superfamily of proteins which interact with nucleic acids [2]. The sequence similarity among the SNM1-family members is restricted to these two regions which are conserved from yeast to mammals.
ARTEMIS is the best investigated member of the SNM1-family with an established function in DNA overhang processing and opening of DNA hairpins generated during nonhomologous end joining (NHEJ) and V(D)J recombination [3]. In some cases mutations in the ARTEMIS gene have been shown to be the underlying cause of severe combined immunodeficiency in association with radiosensitivity (RSSCID) [4,5].
Based on its similarity to the S. cerevisiae SNM1(PSO2) gene, we originally identified the human KIAA0086/hSNM1 gene as a potential human DNA-crosslink repair gene with an unusually long 5’UTR [6], a feature which was later shown to play a role in the regulation of hSNM1 translation [7]. Mouse embryonic stem (ES) cells in which mSNM1 is disrupted display a twofold decrease in their survival upon exposure to Mitomycin C (MMC), but not to other DNA crosslinking agents or ionizing radiation (IR) [8]. However, treatment with either IR or MMC does result in an increased number of nuclear hSNM1 foci [9], suggesting that hSNM1 responds in some way to both DNA double strand breaks (DSBs) and interstrand cross links (ICLs). In addition, mammalian SNM1 has been implicated in an early mitotic stress checkpoint, in tumor suppression, and immunity [10,11].
In contrast to the DNA damage response roles identified for Artemis and hSNM1, several groups have recently suggested that hSNM1B functions primarily in telomere protection. Freibaum and Counter found transiently expressed EGFP-hSNM1B colocalized and co-immunoprecipitated with TRF2 [12]. Another group identified this interaction by employing a combination of co-immunoprecipitation and mass spectrometry [13]. Finally, using a fragment of TRF2 as a bait, Lenain and colleagues found hSNM1B as an interactor in a yeast two hybrid screen [14]. These studies showed that transiently expressed hSNM1B fused with GFP or a myc- tag localizes to telomeres. Following hSNM1B knockdown, the phenotype of TRF2 inhibited cells was exacerbated in terms of growth defects, telomere deprotection and increased fusions [14]. Activation of a DNA-damage signal at telomeres was observed as a consequence of hSNM1B knockdown [13]. Altogether these recent findings strongly suggest that hSNM1B cooperates with TRF2 to protect telomeres from being recognized as damaged DNA.
Our own prior studies of hSNM1B have suggested a more general role for the protein in the cellular response to both DNA double-strand breaks or interstrand crosslinks [1]. In the current study, we extend these findings. Using hSNM1B and TRF2 specific antibodies in co-immunoprecipitation and indirect immunofluorescence (IF) experiments we confirm the interaction for the native proteins without transfection and expression of exogenous constructs. We further show that hSNM1B, as with TRF2, accumulate rapidly following photoinduction of DSB at non-telomeric sites, suggesting the cooperation of these two proteins in the early cellular response to DSBs. Moreover, we show that depletion of hSNM1B by treatment with siRNA, attenuates the autophosphorylation of ATM on Serine 1981 resulting in decreased phosphorylation of its target proteins, SMC1, p53 and H2AX. These findings establish hSNM1B as an early DSB-response protein that stimulates ATM and contributes to the maintenance of genomic integrity.
Results
Subcellular localization of endogenous hSNM1B
Previous reports on the subcellular distribution of hSNM1B were based on experiments employing transiently overexpressed and tagged versions of hSNM1B [12,15]. To validate an hSNM1B-antiserum we have shown before to work specifically in immunoprecipitation experiments [1] for indirect immunofluorescence (IF), we expressed Flag-tagged hSNM1B in GM00637 cells and double stained these cells with antibody against the Flag-tag and with the hSNM1B-antiserum. IF analysis with anti-Flag antibody revealed an almost exclusively nuclear localization of hSNM1B with a subset of the transfected cells displaying nuclear foci, a result which is in agreement with the above mentioned reports on hSNM1B localization. In addition, all foci stained with the anti-Flag also stained positive with anti-hSNM1B indicating that the hSNM1B-antiserum is able to recognize hSNM1B in this experimental setting (Figure 1A). We then tested the ability of the anti-hSNM1B antiserum to recognize endogenous hSNM1B foci. The antibody detected bright nuclear foci in a considerable subset of cells of all three cell lines tested. The remaining cells showed a diffuse nuclear staining (Figure 1B). Quantification revealed that ~60% of the GM00637 and HeLa nuclei and ~70% of the U2OS nuclei analysed stained foci positive (Fig. 1B), however, foci positive HeLa cells appeared to have less foci per nucleus.
Figure 1. Subcellular localization of hSNM1B.
A) Transiently expressed Flag-hSNM1B was detected in indirect immunofluorescence experiments by double staining with antibodies against Flag and hSNM1B in GM00637 cells. Flag-hSNM1 foci detected with anti-Flag colocalized with foci detected by anti-hSNM1B (merged) indicating the specificity of the latter.
B) GM00637, U2OS and HeLa cells were fixed and stained with an hSNM1B-antiserum and analyzed under a fluorescence microscope. The anti-hSNM1B antibody detected the endogenous hSNM1B protein with a subset of cells showing nuclear foci (green). DNA was stained with DAPI (blue). The fractions of cells staining hSNM1B foci positive is indicated for each cell line analyzed together with the number of cells, n, evaluated.
Interaction between TRF2 and hSNM1B
We used a full length hSNM1B cDNA as a bait in a yeast two hybrid (Y2H) screen and recovered a single cDNA clone encoding amino acids 40–252 of TRF2 from a HeLa cDNA library. TRF2 is a core component of shelterin, a protein complex involved in chromosome-end regulation and protection [16]. The TRF homology domain of TRF2 mediates homodimerization and interaction with other telomeric proteins and is comprised of amino acids 43–245 of the protein [17]. As shown in Figure 2A, the cDNA identified in the Y2H screen represented almost exclusively the TRF homology domain (TRFH) amino-terminally fused to the vector encoded B42 domain.
Figure 2. Interaction between TRF2 and hSNM1B.
A) Schematic representation of the TRF2 protein and the protein encoded by the cDNA clone identified in the Y2H screen. This cDNA clone contained a partial TRF2 sequence representing the complete TRFH domain of TRF2. The vector encoded B42 domain and the MYB domain of TRF2 are indicated.
B) Endogenous TRF2 co-immunoprecipitates with endogenous hSNM1B and transiently expressed hSNM1B-EGFP. HEK293T cells were transiently transfected with pEGFP-NI-hSNM1B or empty vector. Cells were untreated or irradiated with 20Gy as indicated and harvested 15min post-irradiation. Immunoprecipitations (IPs) were performed using antibodies against the endogenous proteins TRF2 or hSNM1B. Blots were probed with an antibody against GFP or TRF2 to test for interaction. “Input” represents 1.5% of the total lysates used for IPs, (1) from cells transfected with the empty vector and (2) from cells transfected with pEGFP-NI-hSNM1B. Total rabbit IgG antibodies were used as a negative control in a IP.
C) Co-localization of endogenous hSNM1B with TRF1 and TRF2 in GM00637 cells detected by immunofluorescence.
D) Immunoblot analysis of GM00637 cells treated with TRF2 specific siRNAs. Cells were analysed 48h post transfection showing a clear reduction of TRF2 expression following exposure to the TRF2 siRNA when compared to the TRF2 signal from cells treated with hSNM1B or control siRNA.
E) siRNA mediated depletion of TRF2 resulted in a significant reduction of hSNM1B foci in GM00637 and U2OS cells when compared to cells treated with control siRNAs as analyzed by immunofluorescence. For comparison the effect of hSNM1B knockdown on hSNM1B foci formation is indicated.
To further explore the interaction between hSNM1B and TRF2 we performed co-immunoprecipitation (Co-IP) experiments. We and others have so far been unable to detect endogenous hSNM1B in western blots [1,13] presumably because of its low expression level. Therefore HEK293T cells were transiently transfected with hSNM1B-EGFP, or an empty vector control, followed by immunoprecipitation with antibodies against hSNM1B or TRF2. The western blot was probed with antibodies directed against TRF2 and the EGFP tag. Endogenous TRF2 was specifically co-immunoprecipitated along with the endogenous hSNM1B from lysates of cells transfected with the empty vector (con) as well as from lysates with the plasmid encoded hSNM1B-EGFP (Fig. 2B). The reverse IP using the TRF2 antibody did not, however, co-IP the transiently expressed hSNM1B-EGFP (Fig. 2B). In a similar experiment, the monoclonal TRF2 antibody was also unable to co-IP transiently expressed hSNM1B with an amino-terminal Flag-tag (data not shown), suggesting that the tag itself is not disturbing protein interactions. Irradiation of the cells prior to analysis did not change the amount of TRF2 co-immunoprecipitated with hSNM1B.
As shown above, the anti-hSNM1B antibodies were able to detect hSNM1B in IF experiments which allowed us to determine whether endogenous hSNM1B localizes to telomeres, as suggested by the yeast-two-hybrid and co-IP results and previously published results on ectopic overexpressed hSNM1B [12–15]. Double staining of hSNM1B and either of the telomere markers, TRF1 or TRF2, demonstrated a high degree of co-localization of these proteins (Figure 2C) and showing, for the first time, that the majority of endogenous hSNM1B foci are localized at telomers.
We next explored the ability of cells to form nuclear hSNM1B or TRF2 foci following siRNA mediated knockdown of either of the proteins. The hSNM1B-siRNA used here was validated before in various assays [1] and hSNM1B knockdown was tracked by counting hSNM1B foci positive cells in indirect IF for each experiment. The fraction of foci positive cells (cells containing at least one focus) was typically reduced by 60–70% when compared to cells treated with a control siRNA (Fig. 2E). Knockdown of TRF2 reduced the protein amount to less than 20% when compared to control cells by western blot (Fig 2D). In U2OS cells, the TRF2 knockdown resulted in a significant reduction of hSNM1B foci positive cells from ~73% in controls to ~50% after treatment with TRF2-siRNA (p <0.0001) (Fig. 2E). We observed an even more pronounced reduction of hSNM1B foci positive cells in another cell line, GM00637 (Fig. 2E).
In order to analyse the impact of hSNM1B knockdown on TRF2 foci formation, we counted the number of TRF2 foci per cell. No significant difference in TRF2 foci formation was observed between hSNM1B-siRNA treated cells and controls when nuclei with > 20 TRF2 foci were counted (data not shown).
hSNM1B functions in early DNA-damage responses
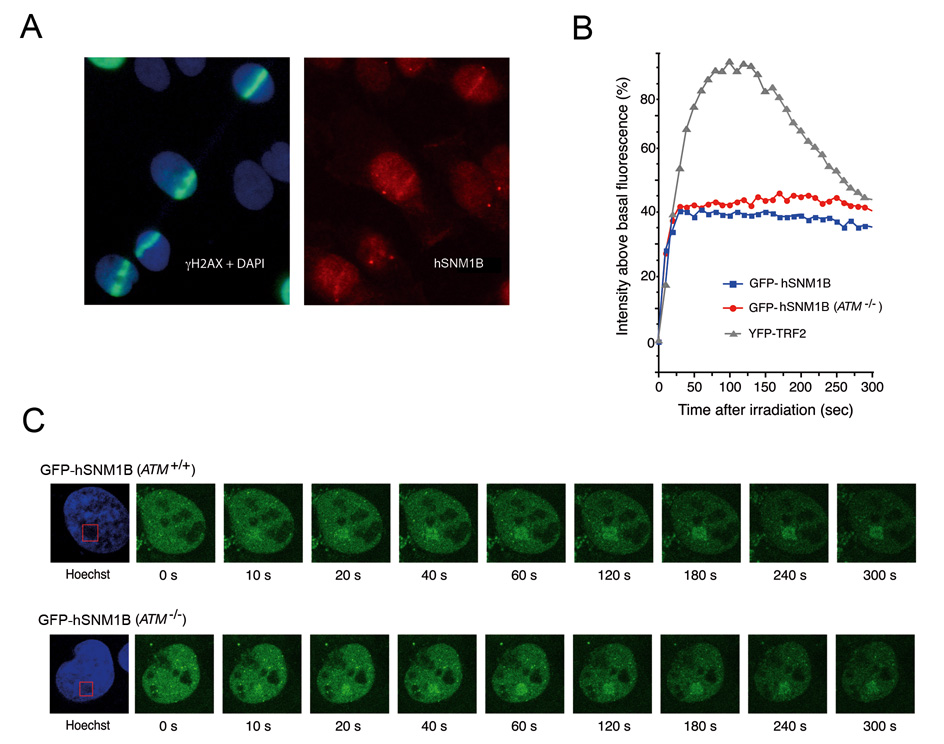
TRF2 has been reported to accumulate at the sites of DSBs in non-telomere DNA within seconds following photo-induction [18]. Given the interaction between TRF2 and hSNM1B that we and others have observed, we sought to determine hSNM1B underwent similar relocalization in response to DNA damage. We first examined the nuclear dynamics of endogenous hSNM1B following induction of DNA breaks by laser micro-irradiation of GM00639 human fibroblasts photo-sensitized by a brief exposure to the intercalating agent, Hoechst 33258. This technique generates DNA breaks only in those sub nuclear regions exposed to the 355 nm high-intensity laser. The location of induced DNA breaks was monitored by indirect immunofluorescence of γH2AX, a phosphorylated histone that forms foci in DSB-containing chromatin. Using this technique, we detected accumulation of hSNM1B at sites of DNA damage 10 minutes post-irradiation, the time of the first measurement (Figure 3A).
Figure 3. Endogenous hSNM1B forms foci in regions of photo-induced DNA damage in SV40-transformed human fibroblasts.
A) γH2AX (green) and hSNM1B (red) form overlapping tracks in DAPI-stained nuclei of GM00639 cells fixed 10 minutes post induction of DNA breaks by laser-irradiation. (B) and (C) Live cell imaging of fluorescently-tagged hSNM1B protein following photo-induction of DNA breaks in either GM00639 (ATM+/+) or GM05849 (ATM−/−) fibroblasts demonstrates rapid association of GFP-hSNM1B protein with regions of photo-induced DNA breaks in individual nuclei. Confocal images were captured at 10 sec intervals up to 300 s post-irradiation. Quantitative kinetics of GFP-hSNM1B association with photo-induced DNA breaks in GM00639 and GM05849 fibroblasts are plotted in (B), along with previously determined kinetics of YFP-TRF2 association with photo-induced DNA breaks in GM00639 cells [18]. In (C), nuclear regions containing photo-induced DSBs appear as dark areas in the Hoechst images due to photo-bleaching of the Hoechst dye by the laser pulses.
To further study the kinetics of hSNM1B localisation to DNA breaks, we carried out live cell imaging of ATM+/+ and ATM−/− human fibroblasts expressing GFP-hSNM1B. DNA breaks were induced in pre-defined areas of the nucleus by laser irradiation followed by image capture at 10 second intervals for 300 seconds after induction of damage (Fig. 3B and 3C). On average, GFP-hSNMB1 localisation to regions of induced DNA breaks was observable by 10 seconds post-irradiation, with a peak accumulation of 40% above baseline levels at 40 seconds post-irradiation. The magnitude of this association with photo-induced DNA damage was not as great as that previously reported for YFP-TRF2 [18] (Fig. 3B) or for GFP-ATM (unpublished results). From 1 to 5 minutes post-irradiation, GFP-hSNM1B concentrations in the DNA break-containing nuclear regions remain constant. In contrast, concentrations of YFP-TRF2 in these regions begin to decline after 2 minutes (Fig. 3B). The association of GFP-hSNM1B with induced DNA damage was not dependent on ATM, as the absence of functional ATM protein in GM05849 cells did not significantly affect the association of GFP-hSNM1B with photo-induced DNA damage (Fig. 3B and 3C).
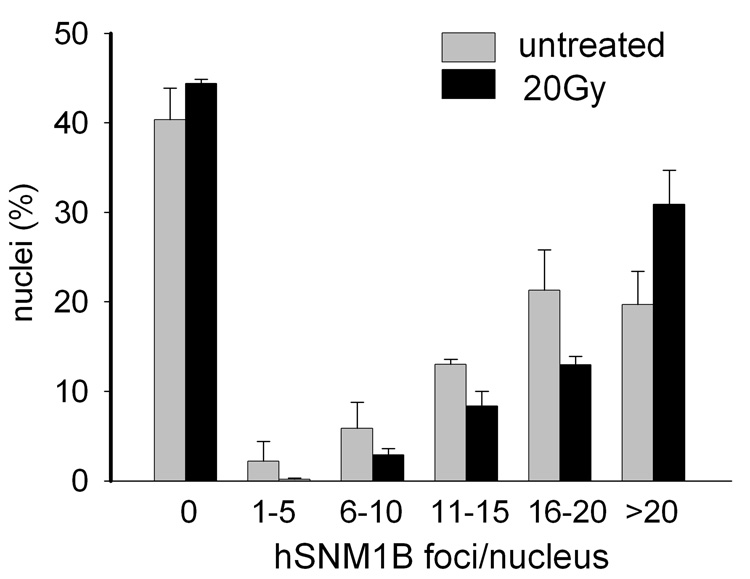
To further examine hSNM1B in the cellular response to DNA-damage we analysed irradiated (IR) and non-irradiated GM00637 cells in IF experiments by counting the number of foci per nucleus. As illustrated in Fig. 4, the proportion of cells containing hSNM1B foci did not change significantly 15 minutes after irradiation with 20Gy when compared to untreated cells. However, the average number of hSNM1B foci per cell was significantly increased after radiation exposure; ~31% of the nuclei contained more than 20 foci compared to ~20% in unirradiated control cells (p < 0.001).
Figure 4. hSNM1B foci formation induced by IR.
Quantitative analysis of hSNM1B foci induced by ionizing radiation. GM00637 cells were irradiated with 20Gy and analysed for hSNM1B foci 15 min later by indirect immunofluorescence. Whereas no significant difference in the proportions of cells containing foci and cells with a diffuse stained nucleus was detected, there was a clear difference in the number of hSNM1B foci per nucleus. Cells were grouped according to the number of foci, as indicated. The number of nuclei with more than 20 hSNM1B-foci increases in response to IR (p< 0.0001) and the number of nuclei in the other groups also increases. Results from at least two independent experiments are shown (mean counts per nucleus are given together with error bars indicating the standard error of the mean).
hSNM1B stimulates the damage-induced phosphorylation of ATM
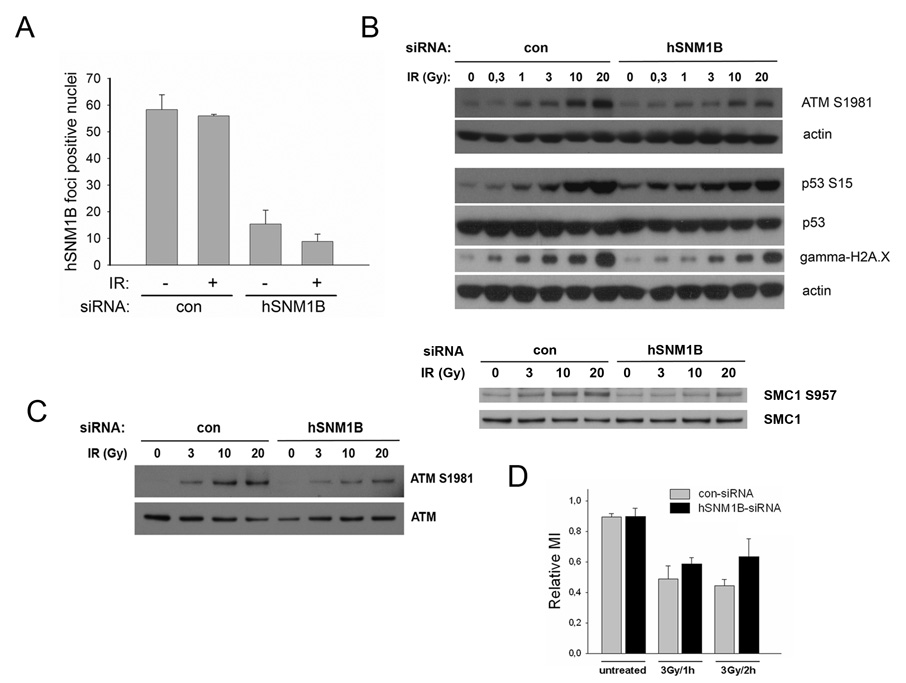
Karlseder and colleagues have shown that overexpression of TRF2 inhibits the phosphorylation of several targets of the ATM kinase, including nibrin and p53, in response to ionizing radiation exposure. In addition, they found ATM autophosphorylation itself attenuated in cells overexpressing TRF2 [19]. The interaction between hSNM1B and TRF2 and the co-localization of both proteins in IR induced nuclear foci raised the possibility that hSNM1B might similarly be involved in the ATM phosphorylation process. In order to test whether hSNM1B was also involved in this early step of ATM activation, we transfected GM00637 cells with hSNM1B-siRNAs and evaluated the ATM phosphorylation status in immunoblots following increasing doses of IR. Functionality of the hSNM1B-siRNAs was shown previously [1] and the extent of hSNM1B knockdown was tracked for each experiment by indirect IF using anti-hSNM1B antibodies. In a typical experiment, the proportion of cells with hSNM1B foci was reduced to 10–20% compared to ~60% in cells transfected with control-siRNAs (Fig. 5A).
Figure 5. hSNM1B depleted cells (siRNA) show reduced ATM activation and a compromised G2/M checkpoint in response to IR.
A) GM00637 cells were treated with control or hSNM1B siRNAs and were irradiated with 20 Gy 66h post transfection and subsequently stained for indirect immunofluorescent analysis using anti-hSNM1B antibodies. Results of quantitative analysis of hSNM1B foci without or 15 min after IR are shown. Results from two independent experiments are shown (mean counts of foci positive nuclei are given together with error bars indicating the standard error of the mean).
B) siRNA treated cells were irradiated with the indicated dose and cells were analysed 15 min later by immunoblotting. Samples were separated on 4–12% Bis-Tris gels and immunoblotted to detect ATM phospho-serine1981, p53, p53-phospho-serine15, gamma-H2A.X and SMC1, SMC1-phospho-serine 957. Actin was detected as a loading control.
C) GM00637 cells were treated with control or hSNM1B siRNAs and were irradiated with the indicated dose 66h post-transfection. The cells were harvested and ATM was immunoprecipitated from whole cell lysates 15 min post-irradiation. Samples were separated on a 3–8% Tris-acetate gel and immunoblotted to detect ATM phosphorylated at serine1981 and total ATM.
D) GM00637 cells were transfected with siRNAs as described in C). Triplicates of each sample were irradiated with 3Gy and the relative mitotic index was determined 1h or 2h later.
As shown in Fig. 5B, siRNA-mediated knockdown of hSNM1B affected the autophosphorylation of ATM at serine1981 in response to IR with a clear reduction in phosphorylated ATM following IR between 3Gy and 20Gy. The relative level of ATM phosphorylated at serine 1981 in hSNM1B depleted cells (normalised to total ATM) at 20Gy was 72% (±2, n=2) of the control-siRNA treated cells. In order to rule out non-specific effects related to the anti-phospho-ATM antibody, we also analysed ATM phosphorylation status on immunoprecipitated ATM from siRNA treated and irradiated cells. This confirmed the result of an attenuated ATM phosphorylation at serine1981 (Fig. 5C).
Since phosphorylation of ATM serine1981 is generally considered a marker of its activation, the reduction in phosphorylated ATM in hSNM1B depleted cells detected here might be expected to result in reduced phosphorylation of ATM target molecules. To test this, we evaluated cells treated with hSNM1B-siRNAs and irradiated with increasing doses of IR for their ability to phosphorylate different ATM targets. The tumor suppressor, p53, is phosphorylated and stabilized in response to DNA damage by the ATM kinase (reviewed in [20]). Both phosphorylation and stabilization of p53 were affected in hSNM1B depleted cells as revealed by immunoblotting with antibodies specific for p53 phosphorylated at serine15 and antibodies detecting total p53 levels (Figure 5B). Interestingly, there was considerable induction of p53 already in untreated and low-dose irradiated hSNM1B depleted cells. However, when irradiated at higher doses, p53 induction was clearly reduced in hSNM1B depleted cells when compared to cells treated with control-siRNAs (Fig. 5B). One of the earliest detectable events in cells responding to DNA damage is the ATM-mediated phosphorylation of the histone variant, H2A.X [21]. By immunoblotting with an antibody specifically recognizing the phosphorylated form of H2A.X, γ-H2A.X, we found that modification of this ATM target was also affected following siRNA treatment. In the case of γ-H2A.X, a reduced signal was detected over the whole range of applied IR dose (Fig. 5B). Similar results were obtained for another ATM substrate, SMC1, whose phosphorylation at serines 957 and 966 is required for S-phase checkpoint activation in response to IR [22,23] (Fig. 5B).
hSNM1B depleted cells display a G2/M checkpoint defect
The activation of cell cycle checkpoints is disturbed in cells from AT patients and in cells mutated in genes whose products participate in the ATM-mediated signalling cascade, e.g. the NBS1 gene [24–26]. To explore the role of hSNM1B in cell cycle checkpoint activation, we determined the mitotic index of irradiated GM00637 cells transfected with a control or hSNM1B-siRNA. Irradiation of the control-siRNA treated cells resulted in an approximately 50% reduction of mitotic cells. As shown in Figure 5D, cells depleted for hSNM1B responded with a less pronounced reduction in mitotic index 2h after IR (70%; p = 0.0458).
Discussion
We have previously identified hSNM1B as a gene involved in the cellular DNA-damage response on the basis of the increased sensitivity of hSNM1B depleted (siRNA) cells to treatment with ionizing radiation, Mitomycin C and Cisplatin. While we had interpreted our prior results as indicative of a general role for hSNM1B in the cellular response to DNA damage, recent published studies reporting a role for hSNM1B in telomere protection raise the possibility that hSNM1B might function predominantly or entirely at telomeres. In the current study we address this issue and demonstrate that hSNM1B plays a significant role in the cellular response to DNA DSBs; a role that is not confined to telomeres.
A major limitation to prior investigations of the hSNM1B function was that we, and others, had been unable to detect endogenous hSNM1B either in western blots [1,13] or in indirect immunofluorescent analysis [13], a fact that was interpreted to reflect the low abundance of the protein.
Here we demonstrate that an hSNM1B-antiserum, which we have previously successfully used in detecting ectopic overexpressed Flag-hSNM1B in immunoblots following IP [1], recognizes endogenous hSNM1B in IF experiments. This allowed us, for the first time, to explore the subcellular localization of the endogenous hSNM1B protein.
Between 60% and 70% of the cells from three different cell lines analysed stained positive for hSNM1B foci with the remaining cells displaying diffuse nuclear staining. Further IF studies revealed that the majority of hSNM1B-foci co-localized with the telomere core protein, TRF1, and are therefore localized at telomeres. These findings substantiate previous reports on the localization of ectopic expressed hSNM1B at telomeres [12,14,16]. The observation that only a fraction of cells contained hSNM1B foci suggests a transient, cell cycle dependent function for hSNM1B at telomeres consistent with reports that hSNM1B functions in repressing the “DNA-damage” signal at telomeres during or after their replication [13].
As previously reported, we observed that hSNM1B associated with TRF2, and that, like TRF2, it accumulated at sites of DSB induction. hSNM1B localized to tracks of photoinduced DSBs where it co-localized with γH2A.X. Interestingly, at the early timepoint after IR analysed here, the fraction of cells displaying hSNM1B foci did not change, while the number of hSNM1B foci per nucleus increased significantly. This may reflect the low expression level of hSNM1B which only crosses the threshold for detection by fluorescence microscopy in a fraction of cells.
GFP-hSNM1B could be found at sites of DSB at the earliest timepoint analyzed, 10s after photo-induction, with the maximal accumulation of GFP-hSNM1B after 40s. This initial fast response of GFP-hSNM1B is similar to that observed for TRF2 [18] and precedes accumulation of YFP-NBS1 and γH2A.X [21,27]. The association of hSNM1B with induced breaks appeared to be stable over the next few minutes, which differs from the more transient YFP-TRF2 response which decreases after reaching a maximum 100-120s post-induction [18].
Autophosphorylation of the protein kinase ATM at serine 1981 and subsequent monomerization is an early event in the cellular response to IR [28]. Activated ATM monomers phosphorylate a variety of downstream transducer and effector molecules, e.g. H2A.X, nibrin, p53, SMC1, CHK2, 53BP1 and FANCD2, involved in regulating cell cycle checkpoints, DNA-repair and/or apoptosis (reviewed in [29]). The association between hSNM1B and TRF2, the formation of hSNM1B foci as an early and ATM independent IR-response, and the known role of TRF2 in ATM activation/inhibition [19] prompted us to analyse hSNM1B function with respect to ATM phosphorylation. We observed that ATM autophosphorylation was attenuated across a broad range of IR doses. This result differs from the attenuation of ATM autophosphorylation observed with depletion of MRN complex components which is only observed at low doses of IR. As expected, hSNM1B knockdown also resulted in a reduction in damage-induced phosphorylation of ATM substrates such as SMC1, p53 and H2A.X. In addition, we observed an increased stabilization and phosphorylation of p53 serine15 in non-irradiated cells depleted for hSNM1B which, together with the finding of upregulated expression of p21 (which is tightly regulated by p53) in hSNM1B knockdown cells [13] suggests that depletion of hSNM1B induces an ATM independent response mediated, at least in part, through p53.
The involvement of hSNM1B in ATM phosphorylation in response to IR, as described here, provides a novel insight into its cellular role. It has been proposed that the primary function of hSNM1B might be to protect telomeres from DNA repair acting inappropriately on chromosome ends [13,14]. However, the data presented here indicate that hSNM1B plays a role in the early response to DSBs occurring in non-telomeric DNA, as shown by its role in ATM phosphorylation, the formation of IR-induced foci, the reduced activation of the G2/M checkpoint in hSNM1B knockdown cells and our prior demonstration of IR sensitivity in cells depleted of hSNM1B by siRNA [1]. We speculate that protection from DNA repair at chromosome ends is not a role of hSNM1B but a task performed by TRF2 which binds hSNM1B at telomeres and thereby prevents hSNM1B from activating ATM. However, we can not rule out the possibility that hSNM1B is involved in an other aspect of ATM phosphorylation status regulation early after IR such as ATM-dephosphorylation.
Cells depleted for hSNM1B also show hypersensitivity to ICL inducing agents in colony forming assays as well as in chromosome breakage analysis [1]. ATM is not known to play any significant role in the response to ICLs, suggesting that another phosphatidylinositol 3-kinase-related protein kinase, such as ATR, might also be affected by hSNM1B knockdown.
While our knowledge about the downstream effects of ATM has grown considerably during the past years, much less is known about the initial events leading to the detection of DSBs and initiating the signal cascade by activating ATM. Our data presented here establish hSNM1B as a new factor acting early in the DSB response at the stage of ATM activation. Further studies are necessary to identify the exact role of hSNM1B and TRF2 within the growing network of molecules involved in the early DNA damage response of the cell.
Materials and methods
Cell lines and culture
HEK293T, GM00637, U2OS, HeLa, GM00639 and GM05849 (ATM−/−) human fibroblasts were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin (Life Technologies). Cells were grown in a humidified 5% CO2 incubator at 37°C.
hSNM1B-expression constructs
Generation of the plasmid pCMV-Tag2B-hSNM1B, allowing the expression of hSNM1B with an N-terminal fused Flag-tag, was previously described [1]. The previously described plasmid pT7T319U-hSNM1B [1] was used as a PCR template to amplify the hSNM1B ORF with oligonucleotides designed to introduce PstI and XmaI sites at the 5’-terminus and the 3’-terminus respectively and to remove the stop codon. This fragment was cloned into the expression plasmid pEGFP-NI (Clonetech) in frame with EGFP at its 3’-end.
The pEG202-hSNM1B plasmid was constructed by subcloning of the blunted PstI insert of pCMV-Tag2B-hSNM1B followed by sequence verification of the vector-insert borders.
siRNAs, transfections
siRNAs specific for hSNM1B, TRF2 or for luciferase GL2 (control siRNA) were purchased from Dharmacon Research (Lafayette, CO) and have been described before [1,30].
GM00637 cells, 1.5×105 cells in 800µl DMEM without antibiotics, were plated 24 hours before transfection into the wells of a 6-well plate. For immunofluorescence analysis, cells were grown on coverslips. 7.4µl of the siRNA-duplexes (20µM) were diluted in Opti-MEM1 medium (Life Technologies) to a final volume of 185µl. In a separate tube, 3µl Oligofectamine transfection reagent (Invitrogen) were mixed with 12µl Opti-MEM and incubated for 5 min at room temperature. The diluted siRNAs were combined with the oligofectamine mixture, incubated for 20min at room temperature and then added to the cells without changing the media. After 6 hours incubation at 37°C, the transfection medium was replaced by DMEM without antibiotics. Immunoblotting and immunofluorescence analysis were performed 66h after transfection as described below.
Photo-induction of DNA breaks
Laser micro-beam irradiation was performed using minor modifications of the method of Bradshaw et al. This technique is believed to induce predominantly DSBs although, as with IR, other damage will also be produced [18,31,32]. In brief, human fibroblasts were grown in DMEM media with 10% FCS on 25 mm round glass coverslips. Nearly confluent cells were exposed to 10 ng/ml of Hoechst 33258 dye in media for 10 minutes, then irradiated on a heated stage in DMEM without Hoechst using a 355 nm MMI Cell Cut microdissection laser coupled to the epifluorescence path of a Zeiss Axiovert microscope. Irradiation was undertaken in predefined regions of the coverslip using a 63x 1.4 NA objective, scan speed of 10% and power output of 85%. Following irradiation, cells were fixed and stained as previously described [18].
Live image analysis
GM00639 (ATM+/+) and GM05849 (ATM−/−) human fibroblasts were transfected with pEGFP-N1/hSNM1B using the FUGENE (Roche) transfection reagent following the manufacturer’s protocol. The following day the cells were subcultured onto 25 mm2 coverslips in the same media. Cells then were exposed to 10 ng/ml of Hoechst 33258 dye in media for 10 minutes, placed in fresh media and mounted on the heated stage (37°C) of a Zeiss LSM510 confocal microscope fitted with a 2-photon tunable laser module. DSBs were introduced using a 790 nm laser beam focused through a 63× 1.4 NA objective and set for a 90% power, 200 ms pulse. Quantitative analyses of captured images were carried out using Openlab v3.01 software (Improvision) as described [18].
Immunoblotting and Immunofluorescence
siRNA-transfected GM00637 cells from three 6-well plates were resuspended in 6ml PBS and aliquots of 1ml were irradiated with the indicated dose. Total cell extracts were prepared 15 min after IR as described [33] and were electrophoresed using the NuPage system (Invitrogen) in 4–12% gradient Bis-Tris or 3–8% Tris-Acetate gradient gels. Following electrophoresis, proteins were transferred to Invitrolon PVDF membranes (Invitrogen). Membranes were blocked for at least 1h in 10% non-fat milk in Tris-buffered saline, pH 7.6, with 0.1% Tween 20 (TBS-T). Incubation with primary and secondary antibodies was performed in 5% non-fat milk in TBS-T. All washing steps were carried out using TBS-T. Immunoblots were probed with the following primary antibodies: ATM phospho-serine 1981 (Rockland), ATM, SMC1, actin, GFP (abcam), p53 phospho-serine15, H2A.X phospho-serine139, p53 (Santa Cruz Biotechnology), SMC1 phospho-serine 957 (Bethyl Laboratories), TRF2 (Imgenex). Primary antibodies were detected with horseradish peroxidase-conjugated goat anti-rabbit IgG, donkey anti-goat IgG or goat anti-mouse IgG (BD Pharmingen, San Diego, CA). Chemiluminescence was developed using Western Lightning (PerkinElmer Life Sciences, Boston, MA). To quantify signals, band intensities were determined using ImageJ software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2005).
Immunoprecipitates were prepared by lysing transfected cells in 50mM Tris-HCl, pH 7.5, 150mM NaCl, 5mM EDTA, 0.3% Triton X-100 containing a protease inhibitor mixture (Roche Applied Science). Lysates were immunoprecipitated with ATM-antibody (Novus), TRF2-antibody (Imgenex) or hSNM1B antibody (Ab-1, [1]) and Dynabeads Protein G (Dynal Biotech ASA, Oslo, Norway) for 3h. Immunoprecipitates were washed four times with lysis buffer and proteins eluted from the beads by boiling for 5min. Immunoblotting was performed as described above.
For indirect immunofluorescence analysis, cells were grown overnight on glass coverslips and exposed to 0 or 20 Gy of irradiation. Cells were fixed after 15min with 4% paraformaldehyde–0.1% Triton X-100 and were blocked overnight in 10% fetal calf serum in phosphate-buffered saline. Cells were stained to detect hSNM1B, TRF2 and TRF1 according to the indicated combinations. The primary antibodies were detected with goat anti-rabbit IgG coupled to Alexa 568 or Alexa 488 and goat anti-mouse IgG coupled to Alexa 488 (Molecular Probes, Eugene, OR) and analysis was performed using the Zeiss Axiophot microscope equipped with a CCD camera (SensiCam) and using the Zeiss filter set 13 (excitation 470, emission 505–530) for Alexa-488 stains and filter set 20 (excitation 546, emission 575–640) for Alexa-568 stains. Fluorescent signals were pseudo-coloured by the AxioVision software and optimised for contrast.
Confocal microscopy was performed with a Nikon fluorescence microscope and a Bio-Rad confocal imaging system using LaserSharp 2000 (Bio-Rad) for validation of the anti-hSNM1B antibody, VMRC10.
Immunostaining of fixed cells in photoinduction experiments was carried out using the primary antibodies, anti-γH2A.X (Upstate Biotechnology) and anti-hSNM1B (VMRC10). Images of fixed cells were obtained using a 63× 1.4 NA objective mounted onto a Zeiss Axioplan 2 microscope equipped with a Hammamatsu Orca ER camera. 12 bit grey scale images captured using Openlab software (Improvision) were subsequently merged into 8 bit color images with Adobe Photoshop.
For foci quantification, slides were coded and, if not otherwise indicated, 175 (IR induced hSNM1B-foci) or 500 (siRNA knockdown) nuclei assessed for the presence of foci using the DAPI stain to count total nuclei. We used no threshold for foci number per nucleus. Results from at least two independent experiments are shown in the figures (mean counts per nucleus are given together with error bars indicating the standard error of the mean). Statistical analysis was done by Fishers exact test (two-tailed) using the GraphPad QuickCalc internet tools (http://www.graphpad.com/quickcalcs/).
Irradiation of cells was carried out using a Machlett OEG-60 X-ray apparatus (UA = 50kV, I = 10mA, filter 1mm Be, dose rate: 3.63 Gy/min).
Mitotic index determination by flow cytometry
The mitotic index was determined as described before [26]. Briefly, siRNA treated GM00637 cells were non irradiated or irradiated with 3Gy. Cells were fixed in 75% ice-cold ethanol at the indicated timepoints, and permeabilized for 10 min on ice in 0.1% Triton X-100 in phosphate buffered saline (PBS) containing 1% bovine serum albumin (BSA). After washing, the cells were incubated overnight at 4°C in 1% BSA in PBS with a polyclonal rabbit anti-phosphorylated histone H3 antibody (Upstate Biotechnology, USA) at 1 : 100 dilution. The cells were washed and incubated in 1% BSA in PBS with a Cy2- conjugated goat anti-rabbit antiserum (Jackson ImmunoResearch) at 1 : 100 dilution. The cells were washed and stained with propidium iodide at 25 mg/ml in 1% BSA in PBS containing 100 mg/ml RNase A. Cytometry was performed in the FACSCalibur (Becton Dickinson). At least 30,000 cells were counted per sample. Statistical analysis was done by t- test using the GraphPad QuickCalc internet tools (http://www.graphpad.com/quickcalcs/).
Yeast Two Hybrid screening
Yeast two hybrid screens were performed using the LexA-B42 system. The yeast strain EGY48, harbouring pEG202-hSNM1B (full length) and pSH18-34, tested negative for autoactivation and was subsequently transformed by the lithium acetate method with a HeLa cDNA library fused to the activation domain vector, pJG4-5 (Origene). Colonies of the resulting transformants were pooled and replated on selective agar lacking the amino acid leucine (leu). Cells proficient for growth on leu− agar were tested for activation of the LacZ gene - the second reporter of interaction.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (DE842-2-1, DE842-2-2) and the National Cancer Institute (CA57569). The authors wish to thank Lara Eckelt and Uwe Gneveckow for their assistance with irradiation of cells and Karl Sperling for critical comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Demuth I, Digweed M, Concannon P. Human SNM1B is required for normal cellular response to both DNA interstrand crosslink-inducing agents and ionizing radiation. Oncogene. 2004;23:8611–8618. doi: 10.1038/sj.onc.1207895. [DOI] [PubMed] [Google Scholar]
- 2.Poinsignon C, Moshous D, Callebaut I, de Chasseval R, Villey I, de Villartay JP. The metallo-beta-lactamase/beta-CASP domain of Artemis constitutes the catalytic core for V(D)J recombination. J Exp Med. 2004;199:315–321. doi: 10.1084/jem.20031142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 4.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 5.O'Driscoll M, Gennery AR, Seidel J, Concannon P, Jeggo PA. An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair (Amst) 2004;3:1227–1235. doi: 10.1016/j.dnarep.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 6.Demuth I, Digweed M. Genomic organization of a potential human DNA-crosslink repair gene, KIAA0086. Mutat Res. 1998;409:11–16. doi: 10.1016/s0921-8777(98)00037-8. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Richie C, Legerski RJ. Translation of hSNM1 is mediated by an internal ribosome entry site that upregulates expression during mitosis. DNA Repair (Amst) 2002;1:379–390. [PubMed] [Google Scholar]
- 8.Dronkert ML, de Wit J, Boeve M, Vasconcelos ML, van Steeg H, Tan TL, Hoeijmakers JH, Kanaar R. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol Cell Biol. 2000;20:4553–4561. doi: 10.1128/mcb.20.13.4553-4561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richie CT, Peterson C, Lu T, Hittelman WN, Carpenter PB, Legerski RJ. hSnm1 colocalizes and physically associates with 53BP1 before and after DNA damage. Mol Cell Biol. 2002;22:8635–8647. doi: 10.1128/MCB.22.24.8635-8647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahkter S, Richie CT, Zhang N, Behringer RR, Zhu C, Legerski RJ. Snm1-deficient mice exhibit accelerated tumorigenesis and susceptibility to infection. Mol Cell Biol. 2005;25:10071–10078. doi: 10.1128/MCB.25.22.10071-10078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akhter S, Richie CT, Deng JM, Brey E, Zhang X, Patrick C, Jr, Behringer RR, Legerski RJ. Deficiency in SNM1 abolishes an early mitotic checkpoint induced by spindle stress. Mol Cell Biol. 2004;24:10448–10455. doi: 10.1128/MCB.24.23.10448-10455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freibaum BD, Counter CM. hSnm1B is a novel telomere-associated protein. J Biol Chem. 2006;281:15033–15036. doi: 10.1074/jbc.C600038200. [DOI] [PubMed] [Google Scholar]
- 13.van Overbeek M, de Lange T. Apollo, an Artemis-Related Nuclease, Interacts with TRF2 and Protects Human Telomeres in S Phase. Curr Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Lenain C, Bauwens S, Amiard S, Brunori M, Giraud-Panis MJ, Gilson E. The Apollo 5' Exonuclease Functions Together with TRF2 to Protect Telomeres from DNA Repair. Curr Biol. 2006;16:1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Ishiai M, Kimura M, Namikoshi K, Yamazoe M, Yamamoto K, Arakawa H, Agematsu K, Matsushita N, Takeda S, Buerstedde JM, Takata M. DNA cross-link repair protein SNM1A interacts with PIAS1 in nuclear focus formation. Mol Cell Biol. 2004;24:10733–10741. doi: 10.1128/MCB.24.24.10733-10741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 17.Fairall L, Chapman L, Moss H, de Lange T, Rhodes D. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol Cell. 2001;8:351–361. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- 18.Bradshaw PS, Stavropoulos DJ, Meyn MS. Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat Genet. 2005;37:193–197. doi: 10.1038/ng1506. [DOI] [PubMed] [Google Scholar]
- 19.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, deLange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 21.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 22.Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY, Qin J. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16:571–582. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van Laethem F, Yang YP, Petukhova GV, Eckhaus M, Feigenbaum L, Manova K, Kruhlak M, Camerini-Otero RD, Sharan S, Nussenzweig M, Nussenzweig A. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- 25.Shiloh Y. Shiloh ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 26.Demuth I, Frappart PO, Hildebrand G, Melchers A, Lobitz S, Stockl L, Varon R, Herceg Z, Sperling K, Wang ZQ, Digweed M. An inducible null mutant murine model of Nijmegen breakage syndrome proves the essential function of NBS1 in chromosomal stability and cell viability. Hum Mol Genet. 2004;13:2385–2397. doi: 10.1093/hmg/ddh278. [DOI] [PubMed] [Google Scholar]
- 27.Horejsi Z, Falck J, Bakkenist CJ, Kastan MB, Lukas J, Bartek J. Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene. 2004;23:3122–3127. doi: 10.1038/sj.onc.1207447. [DOI] [PubMed] [Google Scholar]
- 28.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 29.Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Yang Q, Zheng YL, Harris CC. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol Cell Biol. 2005;25:1070–1080. doi: 10.1128/MCB.25.3.1070-1080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Limoli CL, Ward JF. DNA damage in bromodeoxyuridine substituted SV40 DNA and minichromosomes following UVA irradiation in the presence of Hoechst dye 33258. Int J Radiat Biol. 1994;66:717–728. [PubMed] [Google Scholar]
- 32.Williams ES, Stap J, Essers J, Ponnaiya B, Luijsterburg MS, Krawczyk PM, Ullrich RL, Aten JA, Bailey SM. DNA double-strand breaks are not sufficient to initiate recruitment of TRF2. Nat Genet. 2007;39:696–698. doi: 10.1038/ng0607-696. author reply 698–699. [DOI] [PubMed] [Google Scholar]
- 33.Cerosaletti KM, Concannon P. Nibrin forkhead-associated domain and breast cancer C-terminal domain are both required for nuclear focus formation and phosphorylation. J Biol Chem. 2003;278:21944–21951. doi: 10.1074/jbc.M211689200. [DOI] [PubMed] [Google Scholar]