Abstract
MicroRNAs (miRNAs) are a recently discovered class of non-coding genes that regulate the translation of target mRNA. More than 300 miRNAs have now been discovered in humans, although the function of most is still unknown. A highly sensitive, semi-quantitative RT-PCR method was utilised to reveal the differential expression of a number of miRNAs during the development of both mouse and human lung. Of note was the upregulation in neonatal mouse and fetal human lung of a maternally imprinted miRNA cluster located at human chromosome 14q32.21 (mouse chromosome 12F2), which includes the miR-154 and miR-335 families and is situated within the Gtl2-Dio3 domain. Conversely, several miRNAs were upregulated in adult compared to neonatal/fetal lung including miR-29a and miR-29b. Differences in the spatial expression patterns of miR-154, miR-29a and miR-26a was demonstrated using in situ hybridisation of mouse neonatal and adult tissue using miRNA-specific LNA probes. Interestingly, miR-154 appeared to be localised to the stroma of fetal but not adult lungs. The overall expression profile was similar for mouse and human tissue suggesting evolutionary conservation of miRNA expression during lung development and demonstrating the importance of maternally imprinted miRNAs in the developmental process.
Keywords: microRNA, lung, mouse, human, post-natal, neonatal, fetal, adult
Introduction
MicroRNAs (miRNAs) comprise a large family of small, non-coding genes that have been implicated in regulating the translation of many protein-coding genes (Ambros, 2001). miRNAs are expressed as long primary-miRNAs in the nucleus where they undergo initial processing by the RNase III type enzyme Drosha to form pre-miRNAs approximately 70 nucleotides in length (Pillai, 2005). Further processing in the cytoplasm by the enzyme Dicer results in the formation of mature 21-23 nucleotide length, double stranded miRNAs, which are incorporated into a RNA-induced silencing complex (RISC). Following the loss of one strand of the miRNA, binding of a guide strand to a target mRNA occurs in a non-complimentary manner resulting in repression of protein translation through an unknown mechanism (Pillai, 2005).
The importance of miRNAs during vertebrate development was clearly demonstrated in zebrafish (Giraldez et al., 2005) and mice deficient in the enzyme Dicer (Bernstein et al., 2003; Harfe et al., 2005; Yang et al., 2005). Thus, loss of Dicer resulted in embryonic lethality in both species. Large-scale screening using miRNA-specific microarrays revealed the differential expression of many miRNAs in various animal tissues (Monticelli et al., 2005; Lagos-Quintana et al., 2002; Babak et al., 2004) whilst in situ hybridisation studies have shown that many miRNAs are even cell-type specific within individual organs (Wienholds et al., 2005). Moreover, a single miRNA often dominates the population of all expressed miRNAs in a given tissue, suggesting a role in maintaining tissue and/or cell phenotype (Lagos-Quintana et al., 2002).
Although the function of most miRNAs is unknown, they do play a significant role during mammalian morphogenesis. Thus, limb development was shown to be affected by miR-196 (Hornstein et al., 2005), miR-1 and miR-133 have been implicated in cardiogenesis (Kwon et al., 2005; Zhao et al., 2005) and skeletal muscle development (Chen et al., 2006), while miR-181 expression enhanced the differentiation of myoblasts (Naguibneva et al., 2006). miRNAs have also been shown to regulate the differentiation of neuronal progenitor cells (Krichevsky et al., 2006), inhibit neuronal dendrite growth (Schratt et al., 2006), affect skin morphogenesis (Yi et al., 2006), hair follicle growth (Andl et al., 2006), and angiogenesis (Yang et al., 2005).
Significantly, miRNAs have been strongly implicated in the morphogenesis of embryonic lung tissue. Thus, a transgenic mouse model containing a conditional Dicer knockout exhibited defective development (Harris et al., 2006). In this study, we have attempted to build upon this observation by identifying individual miRNA that might be involved in lung development and morphogenesis. To this end, we have used a novel and highly sensitive real time PCR (RT-PCR) approach to determine the differential miRNA expression profile in adult mouse and human lung compared with that in mouse neonates and human fetal lung. This data revealed remarkable similarities in the overall expression profile in adult mouse and human lung, as well as the differential expression of miRNAs during lung development. Notably, a miRNA cluster located within the maternally imprinted Gtl2-Dio3 region at human chromosome 14q32.21 was upregulated in neonatal/fetal lung. In situ hybridisation revealed miRNA expression predominantly in epithelia, although a member of the miRNA cluster miR-154 was predominantly localised in the stromal tissue in neonate lungs and suggested that differences in expression were partly due to changes in distribution of cell types within the lung.
Results
Comparison of miRNA expression profile in adult mouse and human lung
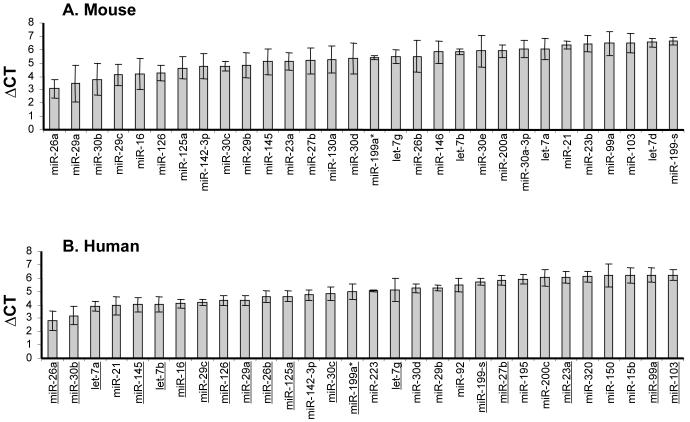
Initial studies were undertaken to determine the relative miRNA quantities in adult mouse and human lungs using a novel RT-PCR based approach (Chen et al., 2005) (Figure 1). The relative expression (ΔCT) of individual miRNA was determined by normalisation to 18S. The absolute CT values for 18S remained constant across samples and time points obtained from each species, therefore the ΔCT values (miRNA assays versus 18S) could be used to compare the relative miRNA expression in mouse and human lung samples (data not show). The relative levels (ΔCT) of all miRNAs expressed in adult mouse and human lungs are shown in Supplemental Table 1 and the 30 most expressed in both species shown in Figure 1 (only those miRNA that were most highly expressed are presented for purposes of clarity and physiological relevance). These data indicate that there is substantial similarity between species. For example, 23 of the 30 most highly expressed miRNAs are shared in adult lungs from mice (Figure 1A) and humans (underlined in Figure 1B) and the relative expression levels (ΔCT) are very similar. Of note, miR-26a is the most highly expressed individual miRNA, whilst the let-7 family, miR-29 family, miR-30 family and miR-199 are also expressed highly in both species.
Figure 1. Most highly expressed miRNAs in adult mouse and human lung.
Total RNA was extracted from the adult lung of mice and human. The relative expression of miRNA was determined by RT-PCR, normalised to 18S and are therefore represented as the mean ΔCT ± SEM (n=3-4). The top 30 most expressed miRNA in mouse (A) and human (B) lung are plotted. Those underlined in the human profile B) show those that are also present in the mouse (A).
Differential expression of miRNAs in the post-natal and adult lung of mice
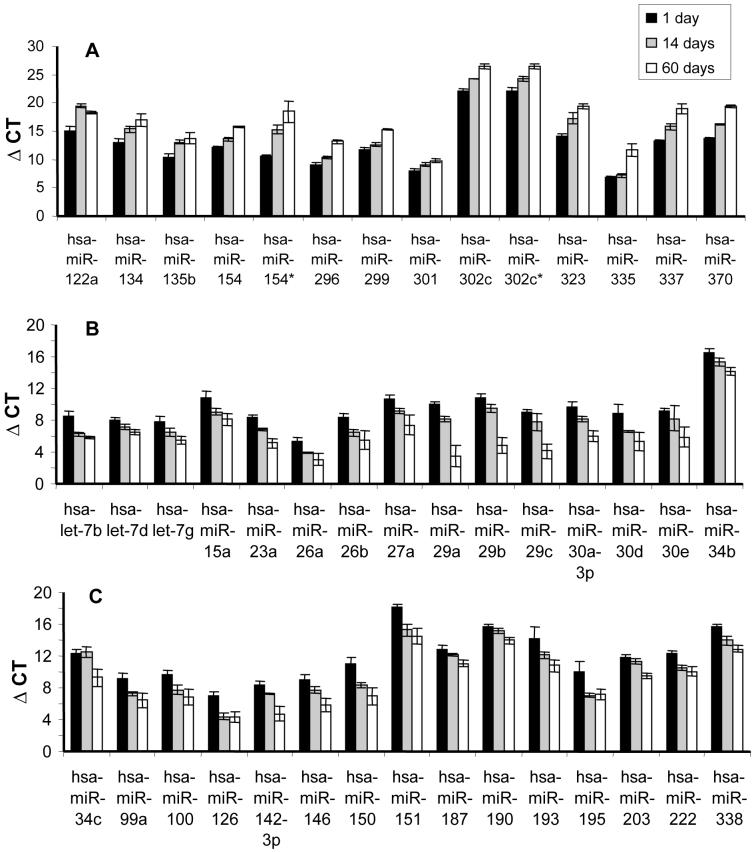
Detailed account of the miRNA expression profile during post-natal lung development was previously unreported. For this reason, the levels of miRNAs were measured (ΔCT) in whole lung tissue from 1 day and 14 day old neonatal mice (Supplemental Table 2) and 60 day old adult mice (Supplemental Table 1). The fold change in miRNA expression between neonatal and adult lung was also calculated (Supplemental Table 3). The differential miRNA expression in neonatal lung compared to adult lung (Figure 2) revealed a group of 14 miRNAs that were more highly expressed in the neonatal lung (Figure 2A). In particular miR-154, miR-323, miR-335, miR-337 and miR-370 were expressed more than 20-fold higher (an increase of > 4 CT units) compared to adult lung. In contrast, 30 miRNAs were upregulated in adult lung compared to neonatal lung (Figure 2B/C). In particular miR-29a, miR-29b, miR-29c, miR-150 and miR-151 were expressed more than 50-fold higher in adult lung compared to neonatal lung. The miRNA levels at 14 days were often intermediate between 1 day and 60 days, which implies a gradual developmental shift in the expression pattern.
Figure 2. Differential expression of miRNAs during murine lung development.
All values were normalised against 18s CT values and are therefore represented as ΔCT (the lower the value the more highly expressed). A. The miRNAs expressed more highly in 1 day neonatal lung compared to 14 day and 2 month old adult lung. B/C. The miRNAs that are more highly expressed in adult 2 month old lung compared to 1 day old neonatal lung. Only those miRNAs that were statistically significant (p<0.05) are represented and are given as mean ± SEM (n=3).
Differential expression profile in human fetal and adult lung
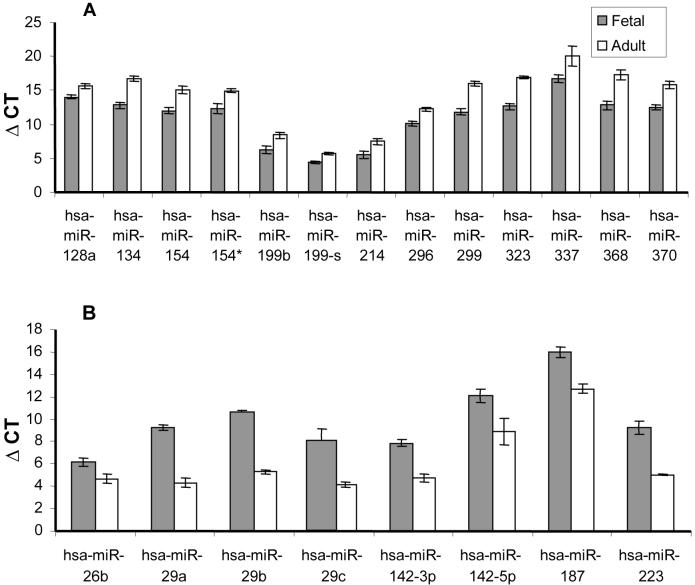
To determine miRNAs that have an evolutionary conserved role in lung development, we examined the differential expression profile between fetal and adult human lung (Supplemental Table 4). In this case 13 miRNAs were expressed more in fetal lung (Figure 3A), while 8 miRNAs were up-regulated in adult lung (Figure 3b). In particular miR-299, miR-323, and miR-368 levels were more than 15-fold higher in fetal than adult lung. Conversely, miR-29a, miR-29b and miR-29c were expressed the highest in adult lung compared to fetal human lung. Significantly, many of these changes in miRNA expression were common to mice. Specifically, the expression of miR-134, miR-154, miR-214, miR-296, miR-299, miR-323, miR-337 and miR-370 were highly expressed in both neonatal mouse and fetal human lung and subsequently down-regulated in adult lung. In contrast, miR-26b, miR-29a/b, miR-146-3p and miR-187 were up-regulated in adult tissue in both mice and humans.
Figure 3. Differential expression of miRNAs during human lung development.
All values were normalised against 18s CT values and are therefore represented as ΔCT (the lower the value the more highly expressed). A. The miRNAs expressed more highly in fetal human lung compared to adult human lung. B. The miRNAs that are more highly expressed in adult human lung compared to fetal human lung. Only those miRNAs that were statistically significant (p<0.05) are represented and are given as mean ± SEM (n=3).
Genomically clustered miRNAs that are differentially expressed during lung development
Genomic analysis of those miRNAs that were highly expressed during development, in both neonatal mouse and fetal human lung, showed that many are clustered at mouse chromosome 12F2 or the syntenic region at human chromosome 14q32.31 (Table 1). These miRNAs, which include miR-134, miR-154, miR-299, miR-323, miR-337, miR-368 and miR-370, might be co-ordinately expressed. Significantly, these regions on mouse chromosome 12F2 and human chromosome 14q32.31 incorporate the imprinted Gtl2-Dio3 domain, which extends over 1.1 Mb. Importantly two miRNA clusters within this domain are only expressed from the maternal chromosome, one cluster is located in a retrotransposon-like gene Rtl1 (miR-337 and miR-370), while the second cluster is located 150 kb upstream in the miRNA-containing gene Mirg (including miR-154). The remainder of the miRNAs upregulated in fetal lung do not appear to be associated with any known genomic clusters (Table 1). Of the miRNAs whose expression is increased in adult lung, few are associated with known clusters (Table 2). In mice, the exceptions are miR-23a and miR-27a that cluster on mouse chromosome 8, and miR-34b and miR-34c that cluster on mouse chromosome 9. miR-29a and miR-29b cluster on mouse chromosome 6/human chromosome 7 and were found to be up-regulated in both adult mouse and adult human lung.
Table 1. Genomic location of miRNAs down-regulated during lung development.
| miRNA | Mouse | Human | Species expression |
|---|---|---|---|
| miR-134 | 12F2 | 14q32.31 | Both |
| miR-154 | 12F2 | 14q32.31 | Both |
| miR-299 | 12F2 | 14q32.31 | Both |
| miR-323 | 12F2 | 14q32.31 | Both |
| miR-337 | 12F2 | 14q32.31 | Both |
| miR-368 | Not found (12F) | 14q32.21 | Human |
| miR-370 | 12F2 | 14q32.31 | Both |
| miR-122a | 18E1 | 18q21.31 | Mouse only |
| miR-135b | 1E4 | 1q32.1 | Both |
| miR-199b | 2B | 9q34.11 | Human only |
| miR-214 | 1H1 | 1q24.3 | Human only |
| miR-296 | 2H3 | 20q13.32 | Both |
| miR-301 | 11C | 17q23.2 | Both |
| miR-302c | 3H1 | 4q25 | Mouse only |
| miR-335 | 6A3 | 7q32.2 | Both |
Table 2. Genomic locations of miRNAs up-regulated during lung development.
| miRNA | Mouse | Human | Species expression |
|---|---|---|---|
| miR-23a | 8C3 | 19p13.12 | Mouse only |
| miR-27a | 8C3 | 19p13.12 | Mouse only |
| miR-29a | 6A3 | 7q32.3 | Both |
| miR-29b | 6A3 | 7q32.3 | Both |
| miR-34b | 9B | 11q23.1 | Mouse only |
| miR-34c | 9B | 11q23.1 | Mouse only |
| let-7b | 15E3 | 22q13.31 | Mouse only |
| let-7d | 13B1 | 9q22.32 | Mouse only |
| let-7g | 9F1 | 3p21.2 | Mouse only |
| miR-15a | 14C3 | 13q14.2 | Mouse only |
| miR-26a | 9F4 | 3p22.3 | Mouse only |
| miR-26b | 1C3 | 2q32 | Both |
| miR-29c | 1E4 | 1q32.2 | Both |
| miR-30a-3p | 1A5 | 6q13 | Mouse only |
| miR-30d | 15D3 | 8q 24.22 | Mouse only |
| miR-30e | 4D1 | 1p34.2 | Mouse only |
| miR-99a | 16C3.1 | 21q21.1 | Mouse only |
| miR-100 | 9B | 11q24.1 | Mouse only |
| miR-126 | 2A3 | 9q34.3 | Mouse only |
| miR-142-3p | 11C | 17q23.2 | Both |
| miR-142-5p | 11C | 17q23.2 | Both |
| miR-146 | 11B1.1 | 5q33.3 | Both |
| miR-150 | 7B4 | 19q22.32 | Both |
| miR-151 | 15E1 | 8q24.3 | Mouse only |
| miR-187 | 18B1 | 18q12.2 | Both |
| miR-190 | 9D | 15q22.2 | Mouse only |
| miR-193 | 11B5 | 17q11.2 | Mouse only |
| miR-195 | 11B4 | 17p13.1 | Mouse only |
| miR-203 | 12F3 | 14q 32.33 | Mouse only |
| miR-222 | XA2 | Xp11.1 | Mouse only |
| miR-223 | XC2 | Xq12 | Human only |
| miR-338 | 11E2 | 17q25.3 | Mouse only |
In situ hybridisation on neonatal and adult mouse lung tissue
In order to support the semi-quantitative RT-PCR data, the spatial expression of the most relevant miRNAs was examined by in situ hybridisation using miRNA-specific LNA probes. Of the 156 miRNAs studied, three miRNAs that were either highly expressed or exhibited high levels of differentiated expression were analysed from neonatal and adult mouse lung tissue. These included miR-26a as it is the most highly expressed miRNA in lung tissue irrespective of age or species, miR-29a as this is the most upregulated in adult tissue and miR-154 as this is the most highly expressed miRNA in neonatal tissue compared to adult and is a member of the maternally imprinted miRNA cluster. In addition, a scrambled miRNA probe was used as a non-specific hybridisation control.
The general expression pattern for all three lung-expressed miRNAs was different in neonatal tissue compared to adult tissue (Figure 4). In neonatal lung miR-26a was predominantly expressed at the basal side in epithelia of bronchioles and larger airways, with diffuse expression throughout the stromal tissue. In adult lung tissue miR-26a was highly expressed in the epithelia of large airways, endothelia of arterioles and venules and less intensely within alveolar epithelium (Figure 4A). A similar pattern was observed for miR-29a, which was predominantly expressed in adult airway and alveolar epithelium, while high expression in neonatal tissue was restricted to larger airway epithelium with a less intense, diffuse expression throughout the stroma (Figure 4B). Both miR-26a and miR-29a showed higher staining intensities in adult tissue, although staining was more widely distributed in neonatal tissue. Conversely, miR-154 was highly expressed throughout neonatal lung tissue, including epithelia and stromal cells, while expression was most intense in the airway and alveolar epithelium of adult tissue (Figure 4C). No expression was observed for the scrambled miRNA probe (Figure 4D).
Figure 4. In situ hybridisation on mouse lung tissue.
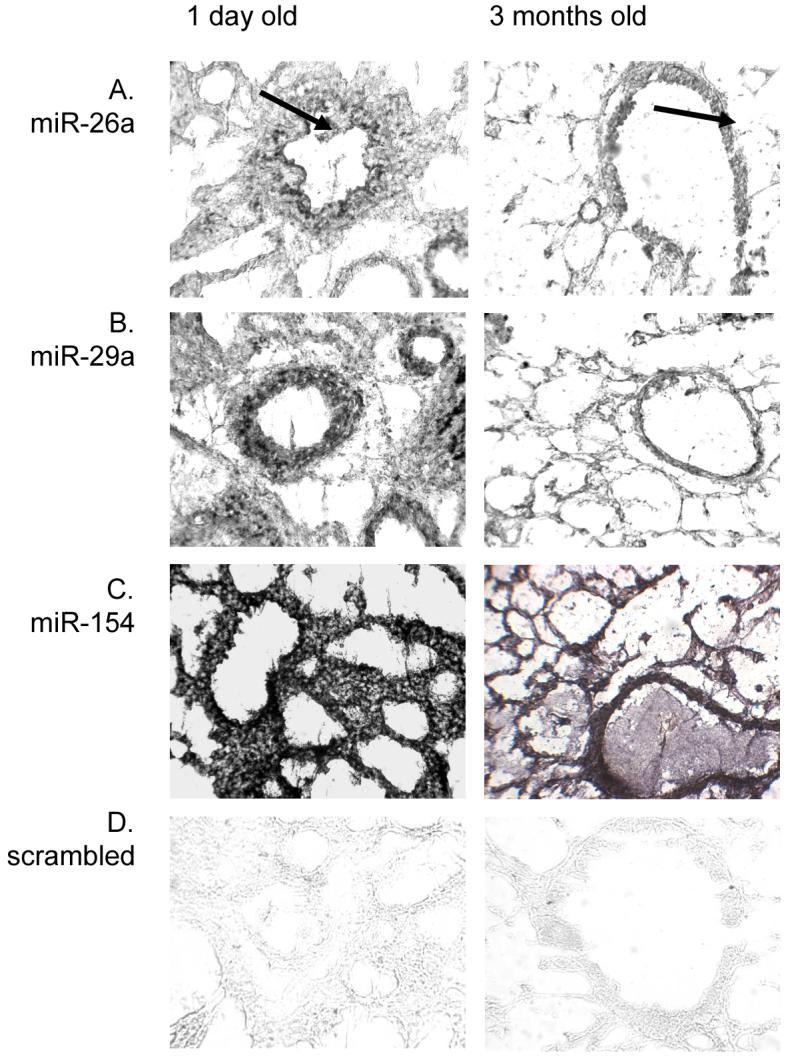
MiRNA expression was compared between the lungs of neonatal (1 day) and adult (3 months) BALB/c mice. Locked nucleic acid (LNA) probes, specific for miR-26a, miR-29a and miR-154 were labelled with DIG and expression visualised using alkaline phosphatase substrate. All slides were taken at magnification x400. Arrows indicate miR-26a expression at the basal surface of airway epithelial cells.
Discussion
At present, little is known of the role of miRNAs in lung physiology and development. For this reason, we used a RT-PCR based approach to compare the expression of miRNAs in adult lungs obtained from mice and humans and investigated their role during lung development. This approach has a number of advantages compared to existing techniques. Principally, unlike DNA based microarrays, it permits relative quantification of mature miRNAs over seven orders of magnitude. Furthermore, compared with the 5-50 μg of total RNA required to detect a single miRNA by northern blotting; only 5 ng is required when using RT-PCR (Chen et al., 2005).
Using this approach, examination of the profile of miRNA expression in adult lung showed similarities between mouse and human. In both species, the most highly expressed miRNAs included miR-26a and members of the let-7, miR-29, miR-30 families. Significantly, this is similar to reports using microarrays (Babak et al., 2004) and northern blotting (Sempere et al., 2004) that determined miRNA expression At present, there have been no reports of the functional roles of these miRNAs although decreased let-7 expression has been associated with poor survival from lung cancer (Yanaihara et al., 2006; Takamizawa et al., 2004). Indeed, over-expression of let-7 in the human lung epithelial A549 cell line was shown to inhibit cell growth (Takamizawa et al., 2004).
The importance of miRNAs during lung development has been demonstrated using conditional Dicer knockout mice (Harris et al., 2006). Mice lacking the Dicer enzyme exhibit broad developmental abnormalities including defective epithelial cell branching and disregulated programmed cell death (Harris et al., 2006). However, although several miRNAs have been shown to regulate the development of the heart (Zhao et al., 2005; Naguibneva et al., 2006), brain and neuronal system (Krichevsky et al., 2003; Giraldez et al., 2005; Schratt et al., 2006) and be involved in embryonic stem cell differentiation (Houbaviy et al., 2003; Krichevsky et al., 2006), no information is available on the role that specific miRNAs play during lung development. To address this situation, we therefore compared the differential expression profile between mouse adult (60 days) and neonatal (1 and 14 days) lungs. The mouse is an ideal animal to study the latter stages of lung development as their lungs do not fully mature until approximately 30 days after birth, which coincides with the final processes of branching morphogenesis, vascularisation and alveolarisation (Massaro & Massaro, 1996). Since it is was envisaged that those miRNA that are central to lung development might be evolutionarily conserved, we also examined samples obtained from adult and fetal human lungs. A direct comparison can be made with human development as the neonatal period of murine lung growth coincides with late human fetal and postnatal lung development (Thurlbeck, 1975; Thurlbeck, 1975).
Multiple miRNAs were shown to be differentially expressed during lung development. Interestingly, the increased expression of 8 miRNAs in neonatal and fetal lung (miR-134, -154, -214, -296, -299, -323, -337 and -370) and the upregulation of 5 miRNAs in adult mouse and human lung (miR-26b, -29a, -29b, -142-3p and -187) were found to be conserved during development in both species. Although the functional role of these changes in miRNA expression is unknown, examination of their genomic localisation shows that 6 of the miRNAs highly expressed in the developing lung of both species (miR-134, -154, -299, -323, -337 and -370), as well as miR-368 in humans, map to the Gtl2-Dio3 domain at human chromosome 14q32.21 (12F2 in mice), a region that is highly conserved between the two species. These miRNAs are contained within two separate clusters. miR-337 and miR-370 map to an intragenic region within the Rtl1 gene, while miR-134, miR-154, miR-299, miR-323 and miR-368 form part of a separate cluster that is located in the intergenic region known as Mirg (miRNA-containing gene) (Seitz et al., 2004). Importantly, imprinting means that these miRNAs are only expressed from the maternally inherited chromosome and their expression is regulated by an intergenic germline-derived differentially methylated region (IG-DMG) located ∼200 kb upstream of the miRNA cluster (Seitz et al., 2004; Tierling et al., 2006). Imprinting also results in the Gtl-2 and Rian genes being expressed from the maternal chromosome, whist Dio-3 and the retrotransposon-like gene Rtl-1 are expressed only from the paternal chromosome (Lin et al., 2003). This region also contains miR-127 and miR-136 and although their expression did not change during development, they are expressed in an antisense fashion within the Rtl1 gene and actually inhibit its translation via an RNAi mechanism (Seitz et al., 2003; Davis et al., 2005). Overall, although members of this miRNA cluster were previously shown to be expressed in the trunk/head of the mouse embryo and in the adult brain (Seitz et al., 2004; Tierling et al., 2006), this is the first study to demonstrate their specific expression profile in the lung. Furthermore, aberrant regulation of imprinted genes has been associated with development of disease, for example Beckwith-Wiedemann syndrome is related to loss of proper control of the IGF2-H19 domain (Jouvenot et al., 2003). Differential expression and hence transcriptional control of the imprinted miRNAs at 14q32.21 suggests that manipulation of miRNA expression may influence disease outcome.
In a parallel situation, several miRNAs are upregulated in the adult. For example, miR-29a and miR-29b are upregulated in adult tissue, being expressed at a lower level in developing lungs in both mouse and human. These miRNA genes map to human chromosome 7q32.3 and the syntenic region at mouse chromosome 6A3. This region maps to a fragile site that has been associated with a number of cancers (Caldas & Brenton, 2005). A deletion in such fragile sites, including that at chromosome 13q14.2 containing miR-15a and miR-16, often involves a deletion in one or more tumour suppressor genes. However, the fragile site FRA7H that includes miR-29a and miR-29b does not encode any tumour suppressor genes (Calin et al., 2004). This suggests that if a deletion of these miRNAs were to occur it may influence the progression of cancers by preventing the inhibition of genes involved in cell proliferation. A similar mechanism may exist during lung development, in that a low expression of these miRNAs may allow cell proliferation to take place in the neonate/fetus, while upregulation in the mature adult lung may prevent further proliferation and maintain cellular homeostasis. Analysis of potential targets for these miRNAs and over- or under-expression studies will provide a clearer insight into the function of such genes.
In situ hybridisation of neonatal and adult mouse lung tissue revealed a differential expression pattern dependent on age. The overall expression patterns for miR-26a, miR-29a and miR-154 corresponded to the data obtained from the semi-quantitative RT-PCR. Therefore, staining intensities for miR-26a and miR-29a were higher in adult tissue, while that for miR-154 was higher in neonatal tissue. Interestingly, the expression of miR-26a appears to be localised to airway and alveolar epithelia in both adult and neonate and within the endothelium of adult lungs. Similarly, miR-29a is expressed in the airway epithelium of adult and neonate lung and dominantly expressed in the alveolar epithelium of adult lung. In contrast, although miR-154 is expressed in epithelium of adult and neonate, the expression is diminished in adult lung. This reflects the varied cell types found within the developing lung tissue, which differs to the epithelial-dominated adult lung. As the lung matures it may be that expression levels alter as a result of loss, or gain, of specific cell types rather than changes within resident cells. For example miR-154 was highly expressed within stromal cells of the neonatal lung, cell types that are largely absent from the mature lung. Therefore, miRNA expression may remain constant in epithelial cells throughout development as a consequence of retaining tissue homeostasis or cell phenotype. Maintenance of lung phenotype during development may require the subtle expression of tissue-specific miRNAs by migratory or transient cells, as well as resident epithelial cells.
In summary, a semi-quantitative RT-PCR based expression profile of mouse and human lung has been used to identify evolutionary conserved changes in miRNA expression during lung development. Importantly, we have shown for the first time that miRNAs expressed from a highly imprinted region at human chromosome 14q32.21 (12F2 in mice) are differentially down-regulated during lung development in both humans and mice. The identification of these miRNAs associated with lung development provides important information on the function of these non-coding genes. Furthermore, manipulation of these miRNAs may provide a novel therapeutic approach for the treatment of diseases that result in disregulation of normal development or homeostasis. For example, miRNAs implicated in epithelial cell proliferation may be manipulated in order to prevent the progression of lung cancer or, alternatively, to promote the regeneration of lung tissue following lung injury.
Experimental Procedures
Animals and Experimental Design
Litters of 1 day old BALB/c mice (Harlan, UK) were housed in specified pathogen-free conditions according to institutional and Home Office (UK) guidelines. Whole lungs were removed from pups and placed immediately in RNAse Later (Sigma), stored overnight at 4 °C and then frozen. Lungs from 1 day old, 14 day old and 2 month old mice were sampled. Total RNA was extracted using MirVana kits (Ambion) according to the manufacturer’s guidelines and stored at −20 °C. Human adult lung and fetal lung RNA, extracted using a modified guanidine thiocyanate technique (Chomczynski & Sacchi, 1987), was supplied by two commercial sources. Adult lung total RNA was supplied by Stratagene (single donor 49 years old), AMS Biotechnology (5 donor pool from 23, 23, 26, 29 and 36 years old), or obtained from adult lung (26 years old), whilst fetal lung total RNA was supplied by Stratagene (2 pooled from 18 and 20 week old lung) and AMS Biotechnology (separate 26 and 29 week old lung).
Semi-quantitative RT-PCR
The miRNA expression profile for each lung sampled was analysed using Applied Biosystems miRNA TaqMan Early Access panel, which consists of 156 individual miRNAs. The panel consists of reverse transcription primers and separate PCR primers for each miRNA and therefore utilises a two step reaction. A total of 2 ng of starting total RNA was used for each miRNA assay.
The RT reaction per specific miRNA contained 0.0775 μl dNTPs, 0.5 μl Multiscribe, 0.75 μl RT buffer, 0.094 μl RNAse inhibitor, 2.081 μl nuclease free water (Promega) and 2.5 μl total RNA (2 ng/μl) from TaqMan MicroRNA RT Kit (Applied Biosystems) plus 1.5 μl RT primer (Applied Biosystems) or 1.5 μl random hexamers (Applied Biosystems) and the reaction conditions were 30 min at 16 °C, 30 min at 42 °C and 5 min at 85 °C. The PCR reaction comprised 5 μl TaqMan 2x Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems), 3.335 μl RNAse free water (Promega), 1 μl TaqMan probe (Appled Biosystems) or 1 μl 18s probe (Applied Biosystems) plus 0.67 μl RT product and the reaction conditions were 30 min at 16 °C, 20 min at 2 °C and 5 min at 85 °C. The amount of RNA from each sample was calibrated to the expression of the ribosomal 18s house-keeping gene. This then gave a delta CT (ΔCT) value for each miRNA (miRNA CT value – 18s CT value). As 18s expression was consistently higher than any miRNA, a higher miRNA expression level corresponded to a smaller ΔCT value. The ΔCT values for all miRNAs on the panel are shown in supplementary data. Those achieving significance <0.05, according to Student’s t-test, after comparing mouse 1 day versus 2 month or human fetal versus adult samples are represented. Fold differences in miRNA expression were also calculated as 2-(ΔCT 1 day - ΔCT 2month) or 2-(ΔCT fetal - ΔCT adult) as an increase in 1 CT value was equivalent to a 2-fold increase in amount of starting cDNA (shown in supplementary data).
Genomic analysis of miRNAs
Mouse and human miRNA sequences were analysed using miRBase:Sequence Release 8.1 (Sanger Institute, Wellcome Trust, UK; http://microrna.sanger.ac.uk/sequences/). The genomic coordinates were determined for each miRNAs and mapped to a specific chromosomal region in both mouse and human genomes. Patterns of clustering of miRNA genes were determined. The synteny between the mouse and human genome was also established.
In situ hybridisation
Lungs from 1 day or 3 month old BALB/c mice were inflated and coated with O.C.T compound (Tissue-Tek) and immediately frozen in liquid nitrogen. Cryosections were cut 12 μm thick and the sections fixed in 95% ethanol for 10 min and then dried in a desiccator for 20 min. Sections were fixed in 4% paraformaldehyde (Sigma) in PBS for 10 min and then washed three times in PBS for 2 min each. Sections were incubated with 10 μg/ml proteinase K (Sigma) for 10 min at room temperature and immediately fixed again in 4% paraformaldehyde. After washing three times in PBS sections were pre-hybridised in hybridisation buffer (50% formamide (Sigma), 5x SSC buffer (Sigma), 250 μg/ml yeast RNA (Ambion), 1x Denhardt’s solution (Fluka) in DEPC (Fluka) treated water) for 1 h at room temperature in a humidifying chamber. LNA probes were pre-incubated at 65 °C for 5 min and immediately placed on ice. The pre-hybridisation buffer was removed and replaced with 2.5 μM of miRNA-specific LNA probe or scrambled probe (Exiqon) labelling with digoxigenin (DIG) in hybridisation buffer in a humidifying chamber at 50 °C for 18 h. Sections were washed in stringency buffer (50% formamide, 5x SSC buffer in DEPC-treated water) that was warmed to 50 °C twice for 45 min and once in PBS at room temperature. Sections were incubated in blocking buffer (10% sheep serum (Sigma) in PBS) for 1 h at room temperature and then with sheep anti-DIG FAb fragments (Roche Diagnostics) labelled with alkaline phosphotase (AP) diluted 1:500 for 2 h at room temperature. Sections were washed three times in PBS for 2 min and then incubated with AP-substrate BCIP/NBT substrate kit (Vector Laboratories) for 18 h at room temperature in a humidifying chamber. Sections were washed twice in PBS and once in water and then mounted with aqueous mounting media (DAKO) before microscopic examination.
Supplementary Material
Acknowledgements
Sterghios Moschos is supported by BBSRC (BB/C508234/1), Mark M. Perry is supported by the Royal Brompton Clinical Research Committee, Mark A Lindsay and Andrew E Williams are supported by the Wellcome Trust (076111).
References
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat.Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc.Natl.Acad.Sci.U.S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J.Biol.Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, Rao A, Kosik KS. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr.Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc.Natl.Acad.Sci.U.S A. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat.Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat.Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Yi R, O’Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat.Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE. The miRNA-Processing Enzyme Dicer Is Essential for the Morphogenesis and Maintenance of Hair Follicles. Curr.Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc.Natl.Acad.Sci.U.S A. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev.Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Massaro GD, Massaro D. Formation of pulmonary alveoli and gas-exchange surface area: quantitation and regulation. Annu.Rev.Physiol. 1996;58:73–92. doi: 10.1146/annurev.ph.58.030196.000445. [DOI] [PubMed] [Google Scholar]
- Thurlbeck WM. Postnatal growth and development of the lung. Am.Rev.Respir.Dis. 1975;111:803–844. doi: 10.1164/arrd.1975.111.6.803. [DOI] [PubMed] [Google Scholar]
- Thurlbeck WM. Lung growth and alveolar multiplication. Pathobiol.Annu. 1975;5:1–34. [PubMed] [Google Scholar]
- Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierling S, Dalbert S, Schoppenhorst S, Tsai CE, Oliger S, Ferguson-Smith AC, Paulsen M, Walter J. High-resolution map and imprinting analysis of the Gtl2-Dnchc1 domain on mouse chromosome 12. Genomics. 2006;87:225–235. doi: 10.1016/j.ygeno.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat.Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, Ferguson-Smith AC, Cavaille J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat.Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- Davis E, Caiment F, Tordoir X, Cavaille J, Ferguson-Smith A, Cockett N, Georges M, Charlier C. RNAi-mediated allelic transinteraction at the imprinted Rtl1/Peg11 locus. Curr.Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- Tierling S, Dalbert S, Schoppenhorst S, Tsai CE, Oliger S, Ferguson-Smith AC, Paulsen M, Walter J. High-resolution map and imprinting analysis of the Gtl2-Dnchc1 domain on mouse chromosome 12. Genomics. 2006;87:225–235. doi: 10.1016/j.ygeno.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Jouvenot Y, Ginjala V, Zhang L, Liu PQ, Oshimura M, Feinberg AP, Wolffe AP, Ohlsson R, Gregory PD. Targeted regulation of imprinted genes by synthetic zinc-finger transcription factors. Gene Ther. 2003;10:513–522. doi: 10.1038/sj.gt.3301930. [DOI] [PubMed] [Google Scholar]
- Caldas C, Brenton JD. Sizing up miRNAs as cancer genes. Nat.Med. 2005;11:712–714. doi: 10.1038/nm0705-712. [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell’Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal.Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.