Abstract
Although apoptosis (programmed cell death type I) is more frequently reported in the literature in imatinib-treated gastrointestinal stromal tumor (GIST) cell lines,morphological features consistent with autophagic changes aremore often encountered in surgical specimens of treated patients. Autophagy (programmed cell death type II) is highly regulated by a tumor-suppressor mechanism that mainly involves the genes beclin1, PI3KIII, and bcl2. Being our material not suitable for electron microscopy analysis (not paraformaldehyde-glutaraldehyde-fixed), we evaluated the morphological, biochemical, and immunophenotypical profiles expected to be related to autophagy and apoptosis in a series of surgically resected samples taken from 11 imatinib-treated patients with molecularly characterized GISTs. The samples were examined for imatinib-induced morphological changes, the presence/interactions of the autophagic-related proteins (beclin1, PI3KIII, bcl2, and LC3-II) and the presence of apoptosis-related proteins (caspase 3, caspase 7, and lamin A/C) by means ofWestern blot analysis and coimmunoprecipitation, complemented by immunohistochemistry. We also studied samples of two untreated GISTs used as controls. Sampling areas with different residual cellularity scores fromboth the imatinib-treated and untreated patients showed biochemical and immunohistochemical evidence of high levels of proautophagy beclin1/PI3KIII and low levels of antiautophagy beclin1/bcl2 complexes, together with the presence of LC3-II detected by Western blot analysis, thus supporting the presence of autophagy. There was no expression of cleaved/activated caspase 3 or 7 or cleaved lamin A/C. Our descriptive results support the idea that GISTs activate autophagy rather than apoptosis in response to imatinib treatment and that their molecular makeup includes fingerprints of autophagy.
Introduction
There have been frequent descriptions of apoptosis in gastrointestinal stromal tumor (GIST) cell lines treated with imatinib (Glivec, Novartis, Basle, Switzerland) or receptor tyrosine kinase (RTK) inhibitors [1,2], but apoptosis-associated morphological changes are only occasionally encountered in surgical samples of GISTs.
For the last year, we have histologically examined a number of surgical specimens taken after imatinib treatment in which, in addition to previously described highly responding areas of complete cell depletion and poorly responding areas with unchanged tumoral cellularity [3,4], we found areas in which the tumoral cells showed prominent intracytoplasmic vacuoles and relatively intact nuclei in the absence of nuclear and cytoplasmic condensation and nuclear fragmentation. These findings, which have previously been described at microscopic level in untreated GISTs [5], and at microscopic and ultrastructural levels in imatinib-treated GISTs [4], are consistent with autophagic rather than apoptotic changes.
Autophagy is a dynamic process in which portions of the cytoplasm are sequestrated within a double-membrane vesicle (or autophagosome) that fuses to the lysosome to generate what are known as autolysosomes or vacuoles. The content of the autophagosome is released into the autolysosome as a result of the action of lysosomal enzymes and then degraded by the same enzymes [6,7]. This degradation can promote cell survival by recycling the degraded nucleotides, amino acids, and fatty acids that maintain energy production or can promote cell death as a result of self-cannibalization [8].
The main difference between autophagy and apoptosis is that the latter invariably leads to cell death, whereas the former may contribute to cell survival or death depending on the threshold level: high levels of autophagy promote cell death [9].
Autophagy is known as programmed cell death type II (apoptosis is programmed cell death type I) and is highly regulated by means of a tumor-suppressor mechanism [10]. It is molecularly controlled mainly by constitutively expressed autophagy-related genes: beclin1 (the major player) and PI3K class I (PI3KI) and class III (PI3KIII). Beclin1 is a haploinsufficient tumor-suppressor that is not required for apoptosis but is necessary for autophagy [11,12]. PI3KI and PI3KIII have opposite effects on autophagy, which is inhibited by the activation of PI3KI but initiated by PI3KIII forming a complex with beclin1 [13,14]. Beclin1 can also bind bcl2, which, in addition to being an important regulator of apoptosis, may inhibit autophagy by directly interacting with beclin1. Growing evidence suggests that the beclin1/bcl2 complex may act as a rheostat ensuring the threshold for cell homeostasis or cell death depending on the presence of a beclin1 function that is respectively checked or unchecked by bcl2 [15,16]. Another widely accepted molecular autophagic marker is the posttranslational modified microtubule-associated protein light chain 3 (LC3-II), which is derived from cytosolic LC3-I because of lipidization during autophagy. LC3-II closely binds to autophagosomes, and its amount closely correlates with the number of autophagosomes [17]. Interestingly, the regulatory pathway of autophagy shares a number of molecules with the oncogenic pathways activated by RTKs, such as the PI3KI/AKT/mTOR and RAS/RAF/MEK1/2/MAPKs pathways. The first inhibits autophagy and activates cancer growth, whereas the second can promote autophagy and cancer growth [18]. The activated RTKs in GISTs areKITand PDGFRA, together with their downstream effectors (PI3KI/AKT and MAPKs), and imatinib is their main starvation inducer.
Within the framework of this quite new scenario, we evaluated the morphological, biochemical, and immunophenotypical profiles expected to be related to autophagy and apoptosis in a series of molecularly characterized GISTs taken from surgically resected imatinib-treated patients. The results showed no signs of apoptosis but a number of indirectmarkers of autophagy, which is in keeping with clinical observations suggesting prompt tumoral regrowth when imatinib is discontinued.
Materials and Methods
Patients and Materials
Table 1 summarizes the details. The case material consisted of 13 surgical specimens: 6 from a previous study of imatinib-treated advanced GISTs (numbers in bold) [3,19]; 5 from patients undergoing a neoadjuvant GIST protocol (numbers in italics); and 2 (used as controls) from untreated patients. Paraffin-embedded and frozen material was available for each patient. The GIST diagnosis was confirmed on paraffin-embedded material by means of immunohistochemical (IHC) analyses using antibodies against CD117, CD34, PDGFRA, desmin, actin, andMib-1, as previously described [3,20]. Written informed consent was obtained from all of the patients.
Table 1.
Clinical Findings in Imatinib-Treated and Untreated Patients, Molecular Characterization, and Histological Response Assessment on Nodules of Treated Cases.
| Clinical Findings | KIT Gene Mutation | Morphology | ||||||||||
| Tumor Site* | Imatinib Dose (mg/day) | Duration on Imatinib (months) | Clinical Response† | Primary‡ | Secondary | No. of Nodules Examined and Residual Cellularity§ | Mitosis | IHC Mib-1 | Histological Response Assessment (Cases) | Residual Cellularity Assessment (Frozen Samples)¶ | ||
| Treated patients# | ||||||||||||
| 1 | Stomach P | 800 → 400 | 11 (8 + 3) | r | Exon 11 | V559D | Not present | 18 (4a; 14b) | 0 | 0 | High | b |
| 2 | Stomach P | 400 | 6 | r | Exon 11 | del 557–558 | Not present | 10 (4a; 6b) | 0 | 0 | High | b |
| 3 | Esophagus P | 400 | 9 | r | Exon 11 | del 554–558 | Not present | 11 (1a; 10b) | 0 | 0 | High | b |
| 4 | Abdominal R | 400 | 14 | r | Exon 11 | K558N + Del 559 | Not present | 6 (1a; 1b; 2c; 2d) | 0 | 0 | Moderate | c |
| 5 | Small intestine P | 400 | 6 | r | Exon 11 | V559D | Not present | 12 (2a; 5b; 5c) | 0 | 0 | Moderate | c |
| 6 | Abdominal R | 800 | 15 | r | Exon 11 | Del 552–553 + E554K | Not present | 4 (2c; 2d) | 0 | 0 to <10% | Moderate | c |
| 7 | Abdominal R | 400 | 16 | r | Exon 9 | Dupl 502–503 | Not present | 5 (1a; 1b; 3d) | >10/50 (47) | 20–30% | Low | a |
| 8 | Stomach P | 400 | 15 | r | Wild type | Not present | 14 (2a; 2b; 10d) | >10/50 (70) | >30% | Low | d | |
| 9 | Abdominal M | 400 | 12 | r | Exon 13 + 17** | K642E + N822K | Not present | 10 (9c; 1d) | >10/50 (15) | 20–30% | Low | d |
| 10 | Abdominal R | 400 | 16 | p | Exon 11 | L576P | Not present | 6 (1b; 5d) | >10/50 (70) | 0–10% + area >30% | Low | d |
| 11 | Abdominal M | 400 → 800 | 33 | p | Exon 11 | Dupl 577–586 | Exon 17 N822T | 9 (2a; 1b; 6d) | >10/50 (21) | 20–30% | Low | d |
| Untreated patients | ||||||||||||
| 1 | Stomach P | / | / | / | Exon 11 | Del 558–563 | / | / | >10/50 (112) | >30% | / | / |
| 2 | Small intestine P | / | / | / | Exon 9 | Dupl 502–503 | / | / | 2/50 | 0 to <10% | / | / |
M indicates metastasis; R, recurrence; P, primary.
p indicates progression; r, response.
PDGFRA was sequenced in all cases and the analyzed exons were wild type.
a <10%, b 10% to 50%, c 50% to 90%, d >90%; the letters in bold indicate the residual cellularity of the samples analyzed by immunohistochemistry.
a <10%, b 10% to 50%, c 50% to 90%, d >90%.
The numbers in bold are patients who participated in a study of advanced GIST (three of which, nos. 4, 6, and 7, were described in a previous article [3] as 14, 19 and 25, respectively); the numbers in italics are those treated using the neoadjuvant protocol.
The same mutations have been reported in Heinrich et al. [23].
Clinical findings
Tumor location and TNMstatus (primary [P], recurrence [R], and metastasis [M]) and the dose and duration of imatinib treatment applied to advanced (number in bold) and neoadjuvant (number in italic) cases are shown in Table 1. The neoadjuvant treatment protocol was generally shorter (approximately 12 months) than that used for surgically treated advanced cases. Imatinib treatment was discontinued 12 to 36 hours before the surgical procedures in all cases. Nine patients were judged to be clinical responders (r) and two clinical progressors (p). The two clinical progressors belonged to advanced cases and showedmicroscopically both high mitotic index andMib1 labeling, and the GIST corresponding to case no. 11, harbored a secondarymutation. Clinically, the responsewas assessed by means of both the Response Evaluation Criteria in Solid Tumors and the Choi criteria [21,22].
Gene mutation analysis
The samples of the patients that had not been previously characterized were analyzed for c-Kit and PDGFRA gene mutations as described by Miselli et al. [3]. Among c-kit mutations, one secondary mutation at exon 17, known to be related to imatinib resistance, was observed. No PDGFRA gene mutations were detected. The molecular findings in all of the patients are shown in Table 1.
Morphology
Histological response assessed on paraffin-embedded surgical specimens
One hundred five samples/nodules from the 11 surgical specimens were assessed for tumor response based on residual cellularity, the mitotic index, and Ki-67 labeling, and the results were averaged for each case. Residual cellularity was scored as follows: a, <10%; b, 10% to 50%; c, 50% to 90%; and d, >90%. Averaging yields the following scores: high responders, 0 to <50% residual viable tumoral cells with no mitosis and no obvious Ki-67 immunostaining; moderate responders, >50% to 90% tumoral cells, no mitosis, and Ki-67 immunostaining in 0 to <10% of cells; low responders, >50% to 90% tumoral cells, mitotic index >10/50 high-power fields, Ki-67 immunostaining in 20% to 30% or >30% of cells.
Histological response assessed on frozen material
The 11 frozen specimens obtained from the 11 treated patients (one sample for each case) were analyzed to assess the response to imatinib based on the morphological criterion of residual cellularity (scored as described above). For details, see Table 1.
Biochemical Analysis
Positive controls
The NIH3T3 (American Type Culture Collection, Manassas, VA), HeLa (kindly provided by Dr. A. Greco), and HEK293 cell lines (kindly provided by Dr. A. Greco) were used as positive controls for the beclin1, PI3KIII, bcl2, and LC3-II blots; the staurosporine-treated HeLa cell line was used as a positive control for the caspase 3, caspase 7, lamin A/C, and LC3-II blots.
Protein extraction, Western blot analysis, and coimmunoprecipitation
The cytoplasmic and nuclear proteins were extracted as previously described [24,25]. The protein extracts underwent electrophoresis and immunoblot analysis using standard protocols and 30 µg of cytoplasmic or nuclear protein lysates per sample.
The antibodies tested by means of immunoblot analysis included rabbit polyclonal anti-beclin1 (H-300: sc-11427; Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:200; rabbit polyclonal anti-PI3KIII (anti-Vps34, 38–2100; Zymed Laboratories Invitrogen immunodetection, South San Francisco, CA) diluted 1 µg/ml; mouse monoclonal anti-bcl2 (clone 124 code: M0887; DakoCytomation, Glostrup, Denmark) diluted 1:200; rabbit-polyclonal anti-LC3-II (LC3B, #2775; Cell Signaling Technology, Danvers, MA) diluted 1:1000; rabbit polyclonal anti-caspase 3 (#9662; Cell Signaling Technology) diluted 1:500; rabbit polyclonal anti-cleaved-caspase 7 (Asp 198, #9491; Cell Signaling Technology) diluted 1:1000; rabbit polyclonal anti-lamin A/C (#2032; Cell Signaling Technology) diluted 1:1000; and antiactin (A2066; Sigma, St. Louis, MO) diluted 1:5000.
For the coimmunoprecipitation experiments, 1 mg of protein lysate per sample was immunoprecipitated by means of incubation with 5 µl of rabbit polyclonal anti-beclin1 antibody (sc-11427; Santa Cruz Biotechnology) and protein A Sepharose (Sigma). Immunoblot analysis was performed using the same anti-beclin1 antibody, and the filters were then stripped and incubated with anti-PI3KIII and antibcl2 antibodies. The secondary antibodies included anti-rabbit (Sigma), anti-mouse (Sigma), and protein A-HRP (Amersham Biosciences, Piscataway, NJ) used at the recommended dilutions. After hybridization with the secondary antibody, the blots were incubated with ECL Western Blotting Detection Reagents (Amersham Biosciences) and exposed onto ECL Hyperfilm (Amersham Biosciences).
Immunohistochemistry
The IHC analyses were made using 2 µm of the formalin-fixed and paraffin-embedded samples of those surgical specimens whose residual cellularity was comparable with that of the corresponding frozen sample (see letters in bold in Table 1) and the UltraVision LP Large Volume Detection System HRP Polymer (code TL-125-HL; Lab Vision Corporation,Newmarket, UK) with the following primary antibodies (all except anti-PI3KIII previously used in the biochemical analysis): anti-beclin1 diluted 1:100, anti-PI3KIII [PI3KC3 antibody (N-term) AP8014a; Abgent, San Diego, CA] diluted 1:50, and antibcl2 diluted 1:500 to investigate the expression of autophagy-related proteins; and anti-cleaved caspase 3 (Asp175, #9661; Cell Signaling Technology) diluted 1:600, anti-lamin A/C diluted 1:100, and anti-cleaved lamin A/C (Small Subunit, #2035; Cell Signaling Technology) diluted 1:50 to investigate the presence of apoptosis. Peroxidases were blocked in 3% H2O2 (Sigma) for 10 minutes. The antigens were retrieved in an autoclave at 95°C in 10 mM citrate buffer, pH 6, for 15 minutes (PI3KIII, cleaved caspase 3, lamin A/C, and cleaved lamin A/C) or 30 minutes (beclin1), or in 5 mM citrate buffer, pH 6, for 6 minutes (bcl2). The primary antibodies were incubated as follows: anti-beclin1 and anti-bcl2 for 1 hour at 25°C; anti-PI3KIII overnight at 25°C; and anti-cleaved caspase 3, anti-lamin A/C, and anti-cleaved lamin A/C overnight at 4°C. The reactions were developed using 3,3′-diaminobenzidine (DakoCytomation) as chromogen for 10 minutes at room temperature.
Results
Pathology
Regression changes in treated patients
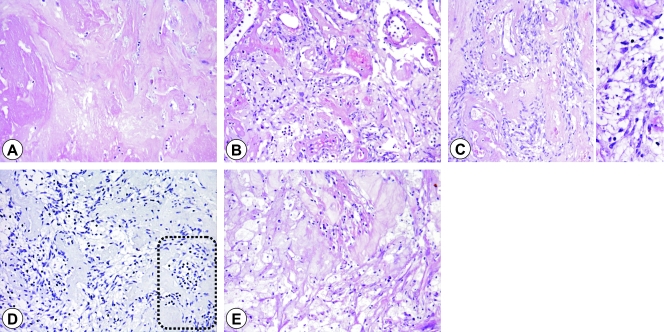
We analyzed b samples (with residual cell component of 10–50%) and c samples (with a residual cell component of 50–90%) in which acellular areas characterized by the presence of an eosinophilic proteinaceous matrix variously defined as myxoid, collagenous, or hyaline [5,26], and often associated with ectatic vessels surrounded by hyaline sclerosis (called a: residual cell component, <10%), were intermixed with clusters of viable tumoral cells with prominent perinuclear vacuolization and relatively intact nuclei in the absence of nuclear or cytoplasmic condensation and fragmentation: i.e., devoid of apoptosis hallmarks. These multivacuolated tumoral cells showed nuclear immunostaining with full-length lamin A/C antibody and a null immunophenotype with cleaved lamin A/C antibody. Scattered lymphocytes negative for lamin A/C and positive for CD3 (data not shown) were also present. Areas of more regressive changes were found in which the tumoral cells had lost their morphology and acquired histiocytic features (corresponding to a samples Figure 1). We also analyzed d samples (residual cell component, >90%) and two samples from untreated patients.
Figure 1.
Post-imatinib morphological regression changes ranging from an acellular area characterized by the presence of eosinophilic proteinaceous matrix (A) sometime intermingled with ectatic vessels surrounded by hyaline sclerosis (B) and clusters of viable tumoral cells with prominent perinuclear vacuolization and relatively intact nuclei (C) in the absence of nuclear or cytoplasmic condensation and fragmentation (D, insert). The multivacuolated tumoral cells showed a null immunophenotype with cleaved lamin A/C antibody, with negative lymphocytes (bottom right, dotted area) as an in-built control. (E) Area of more regressive changes in which the tumoral cells have lost their morphology and acquired histiocytic features.
Biochemical Analyses
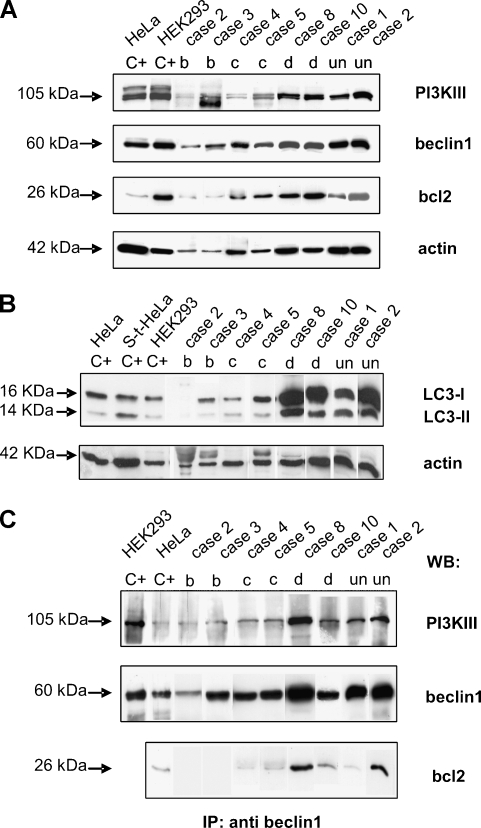
All of the samples were analyzed by means of Western blot (WB) analysis to assess the expression of the beclin1, PI3KIII, bcl2, and LC3-II proteins. The biochemical data are shown in Table 2, which also shows the data regarding clinical and histological responses (on both fixed and frozen material), details of which are reported in Table 1; some representative cases are shown in Figure 2, A and B.
Table 2.
Biochemical and Immunophenotypical Characterization of Samples From Imatinib-Treated and Untreated Patients.
| Clinical Response* | Histological Response Assessment (Cases) | Biochemistry | Immunohistochemistry† | ||||||||||||||||||
| Residual Cellularity Assessment (Frozen Samples)‡ | WB Beclin1 | WB PI3KIII | WB Bcl2 | WB LC3B-II | Co-IP Experiments | WB Caspase 3 | WB Caspase 7 | WB Lamin A | Beclin1 | PI3KIII | Bcl2 | Caspase 3 | Lamin A | ||||||||
| IP Beclin1/WB Beclin1 | IP Beclin1/WB PI3KIII | IP Beclin1/WB Bcl2 | Full Length | Cleaved | Cleaved | Full Length | Cleaved | Cleaved | Full Length | Cleaved | |||||||||||
| Treated patients§ | |||||||||||||||||||||
| 1 | r | High | b | 1+ | +w | +w | - | 1+ | ne | - | +w | - | - | 1+ | - | + | +w | +w | - | + | - |
| 2 | r | High | b | 1+ | +w | +w | - | 1+ | +w | - | 1+ | - | - | 1+ | - | + | + | + | - | + | - |
| 3 | r | High | b | 1+ | +w | +w | +w | 1+ | 1+ | - | 1+ | - | - | 1+ | - | + | +w | +w | - | + | - |
| 4 | r | Moderate | c | 2+ | 1+ | 1+ | +w | 2+ | 1+ | +w | 1+ | - | - | 1+ | - | + | + | + | - | + | - |
| 5 | r | Moderate | c | 2+ | 1+ | 1+ | 1+ | 2+ | 1+ | +w | 1+ | - | - | 1+ | - | + | + | +w | - | + | - |
| 6 | r | Moderate | c | 2+ | +w | +w | +w | 2+ | 1+ | +w | 1+ | - | - | 1+ | - | + | + | +w | - | + | - |
| 7 | r | Low | a | 1+ | - | - | - | 1+ | - | - | 1+ | - | - | 1+ | - | + | + | + | - | + | - |
| 8 | r | Low | d | 2+ | 2+ | 1+ | 2+ | 2+ | 2+ | 1+ | 1+ | - | - | 1+ | - | + | + | +w | - | + | - |
| 9 | r | Low | d | 2+ | 2+ | 2+ | 2+ | 2+ | 1+ | 1+ | 1+ | - | - | 1+ | - | + | + | + | - | + | - |
| 10 | p | Low | d | 2+ | 2+ | 2+ | 2+ | 2+ | 1+ | +w | 1+ | - | - | 1+ | - | + | + | + | - | + | - |
| 11 | p | Low | d | 2+ | 1+ | 1+ | +w | 2+ | 1+ | +w | 1+ | - | - | 1+ | - | + | + | + | - | + | - |
| Untreated patients | |||||||||||||||||||||
| 1 | / | 2+ | 2+ | 1+ | 2+ | 2+ | 1+ | +w | 1+ | - | - | 1+ | - | + | + | + | - | + | - | ||
| 2 | / | 2+ | 2+ | 2+ | 2+ | 2+ | 2+ | 1+ | 1+ | - | - | 1+ | - | + | + | ++ | - | + | - | ||
+w indicates weak positive; Co-IP, coimmunoprecipitation; IP, immunoprecipitation; ne, not evaluable.
p indicates progression; r, response.
Immunohistochemistry of paraffin-embedded sections of samples showing comparable residual cellularity to that of the corresponding frozen samples.
a <10%, b 10% to 50%, c 50% to 90%, d >90%.
The numbers in bold are patients who participated in a study of advanced GIST (three of which, nos. 4, 6, and 7, were described in a previous article [3] as 14, 19, and 25, respectively); the numbers in italics are those treated using the neoadjuvant protocol.
Figure 2.
Autophagy-related protein expression in imatinib-treated and untreated GISTs (A and B) and their interactions in the autophagic process (C). (A) Western blot analyses using anti-PI3KIII, antibeclin1 and anti-bcl2 antibodies: the arrows indicate the bands corresponding to the three proteins and their molecular weights. The anti-actin antibody was used as a control for protein loading and amount, corresponding to the residual cellularity of each analyzed sample. Lanes C+: positive controls (HeLa and HEK293 cell lines). The numbers correspond to the cases listed in Table 1, and letters correspond to the histological responses of the samples (residual tumoral cells: b, 10–50%; c, 50–90%; d, >90%); un: untreated patient. (B) Western blot analyses using anti-LC3B antibody: the arrows indicate the bands corresponding to LC3-I and LC3-II and their molecular weights. The anti-actin antibody was used as control for protein loading and amount, corresponding to the residual cellularity of each sample. Lanes C+: positive controls [HeLa, staurosporine-treated HeLa (S-t HeLa), and HEK293 cell lines]. The numbers correspond to the cases listed in Table 1, and letters correspond to the histological responses of the samples (residual tumoral cells: b, 10–50%; c, 50–90%; d, >90%); un: untreated patient. (C) Coimmunoprecipitation experiments. Total protein extract (1 mg) from each sample was immunoprecipitated using anti-beclin1 antibody, run on gel, and blotted with the indicated antibodies: the arrows indicate the bands corresponding to the three investigated proteins, and their molecular weights. Lanes C+: positive controls (HEK293 and HeLa cell lines for WB PI3KIII and beclin1; HeLa cell line alone for WB bcl2). The numbers correspond to the cases listed in Table 1, and the letters correspond to the histological responses of the samples (residual tumoral cells: b, 10–50%; c, 50–90%; d, >90%); un: untreated patient.
Beclin1 and PI3KIII expression
Similar 2+ expression of beclin1 and PI3KIII was observed in the untreated cases and the treated low responders. In the moderate and high responders, beclin1 expression corresponded to respectively 2+ and 1+, whereas, although retaining the same trend throughout the groups, PI3KIII was less expressed, ranging from weakly positive (+w) to 1+. In no. 7, a sample in which residual cellularity was very low, beclin1 was positive but PI3KIII-negative.
Bcl2 expression
The expression of bcl2 in the untreated cases and low responders was more heterogeneous than the two autophagyrelated markers described above, and this tendency was confirmed by IHC analysis (data shown later in the Table 2). This finding is in keeping with the observed variability in bcl2 expression in primary tumors [27] and that found in the two unselected control GISTs, in which bcl2 expression was 1+ and 2+ (comparing the expressions of tumoral cells and in-built lymphocytes). On the contrary, in the moderate and high responders, the expression profile of bcl2 was the same as that of PI3KIII.
LC3-II expression
Excluding the two negative cases among the high responders, the results were similar to those relating to PI3KIII in 9 of the 11 treated and untreated cases, in keeping with the role of PI3KIII in initiating autophagy and that of LC3-II in a later phase. Furthermore, as the amount of LC3-II correlated well with the number of autophagosomes, the absence of LC3-II in the presence of PI3KIII is consistent with their degradation in the two most regressing samples (b).
Coimmunoprecipitation experiments to confirm the presence of autophagy
The interactions between beclin1/PI3KIII and beclin1/bcl2 were assessed by means of coimmunoprecipitation experiments. The results are shown in Table 2 and some cases in Figure 2C.
Beclin1
All of the protein extracts from each sample immunoprecipitated using an antibody specific for beclin1 protein showed a corresponding 60-kDa band after membrane incubation. The intensity of the band (1+, 2+) varied from sample to sample in accordance with the expression revealed by the WB experiments and was in line with residual cellularity.
Beclin1/PI3KIII interaction
With the exception of one case (no. 7), all of the evaluable samples coexpressed beclin1 and PI3KIII. Given the differences in residual cellularity between the high/moderate and low responders/untreated cases, coexpression was definitely greater in the former.
Beclin1/bcl2 interaction
The range of beclin1 and bcl2 coexpression (from +w to 1+) was smaller than that of beclin1 and PI3KIII. Given also that the expression levels of PI3KIII and bcl2 were similar in the high and moderate responders, these findings support the view that a small amount of bcl2 protein directly interacts with beclin1.
Taken together, the high levels of proautophagy beclin1/PI3KIII complexes and low levels of antiautophagy beclin1/bcl2 complexes are consistent with the presence of autophagy.
Expression of apoptosis-related proteins
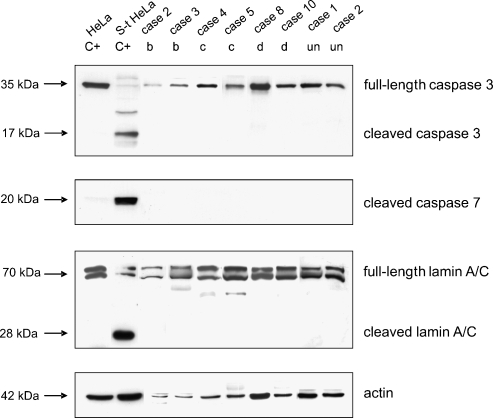
All of the samples were analyzed by means of WB experiments to assess the expression of full-length and cleaved caspase 3, cleaved caspase 7, and full-length and cleaved lamin A/C proteins. The results are shown in Table 2.
Caspase 3 and lamin A/C were only present as full-length proteins in all of the treated patients (regardless of their residual cellularity or clinical/histological group) and the untreated patients. The caspase 7 antibody, which only recognizes the large fragment of the enzyme resulting from apoptosis-induced cleavage, was not found in any of the treated or untreated cases (Figure 3, which shows some representative cases).
Figure 3.
Apoptosis-related protein expression in imatinib-treated and untreated GISTs. The WB analyses were performed using anti-caspase 3, anti-cleaved caspase 7, and anti-lamin A/C antibodies: the arrows indicate the bands corresponding to the expression of the three proteins and their molecular weights. In the case of caspase 3 and lamin A/C, the bands corresponding to both the fulllength and cleaved/activated proteins are indicated. The anti-actin antibody was used as a control for protein loading. Lanes C+: positive controls [HeLa and staurosporine-treated HeLa (S-t HeLa) cell lines]. The numbers correspond to the cases listed in Table 1, and the letters correspond to the histological response of the samples (residual tumoral cells: b, 10–50%; c, 50–90%; d, >90%); un: untreated patient.
Taken together, the results show the absence of apoptotic hallmarks in all of the patients.
Confirmatory IHC Analyses
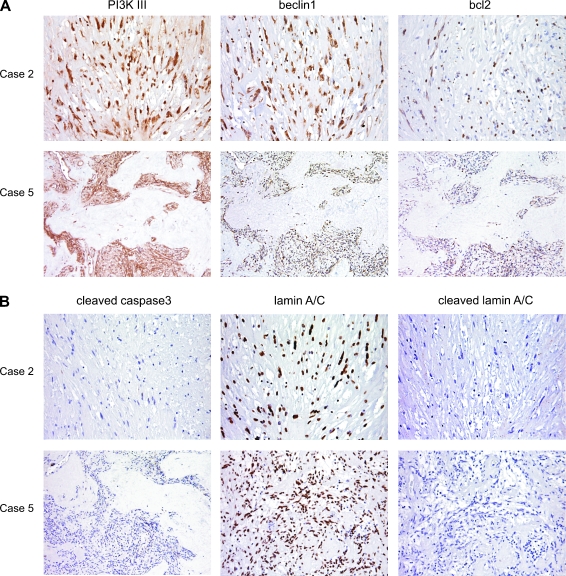
Expression of autophagy-related proteins
To confirm the expression of autophagy-related proteins, sections obtained from paraffinembedded samples whose residual cellularity was comparable with that of the corresponding frozen samples were screened for beclin1, PI3KIII, and bcl2.
The imatinib-treated and untreated cases all showed cytoplasmic reactivity for beclin1, PI3KIII, and bcl2. The results of the IHC analysis are shown in Table 2 and Figure 4A.
Figure 4.
Immunohistochemical analysis of two imatinib-treated GISTs, showing residual cellularity b (case 2) and c (case 5). (A) Expression of autophagy-related proteins. Both cases show cytoplasm decoration for PI3KIII, beclin1, and bcl2. (B) Expression of apoptosisrelated proteins. Both cases show nuclear immunoreactivity for full-length lamin A/C and no immunostaining for cleaved/activated caspase 3 or cleaved lamin A/C.
Expression of apoptosis-related proteins
To investigate the presence of apoptosis, further sections from the same samples were incubated with cleaved caspase 3 and full-length and cleaved lamin A/C antibodies. All of the imatinib-treated and untreated cases were positive for full-length lamin A/C with nuclear reactivity, whereas no immunostaining was observed for cleaved/activated caspase 3 or cleaved lamin A/C (Figure 4B). The results of the IHC analysis are shown in Table 2.
The finding of autophagy-related protein expression in all of the treated and untreated samples in the absence of apoptosis-related protein expression supports the idea that autophagy is part of the molecular profile of GISTs.
Comparison of Biochemical and IHC Results
Given that immunoreactivity relates to individual cells and the biochemical data to residual cellularity (i.e., the actin value), the biochemical and IHC results concerning the expression of autophagyand apoptosis-related proteins correlated in both the treated and untreated patients.
Discussion
Our morphological, biochemical, and immunophenotypical results provide a strong indirect evidence that imatinib-induced tumoral regression in patients with posttreatment surgically resected GISTs is sustained by autophagy rather than apoptosis.
It has recently been reported that, in addition to inhibiting RTK phosphorylation and inducing cell cycle arrest, imatinib activates autophagy [28]. Autophagy is a dynamic rearrangement of the subcellular membranes that leads to autophagic bodies (corresponding to vacuoles at histological level), which may contribute to tumoral death by means of self-cannibalization, although, unlike apoptosis (which invariably leads to cell death), it may also contribute to cell survival [9]. Mainly investigated on cell lines and by means of electron microscopy [29], the only assay able to demonstrate autophagosomes, autophagy is regulated by a tumor-suppressor mechanism in which the major players are the activating beclin1/PI3KIII complex, the suppressing beclin1/bcl2 complex [8], and the presence of LC3-II strictly bound to autophagosomes. In line with this, we found that most of the samples in our high, moderate, and low histological response groups expressed beclin1/PI3KIII, beclin1/bcl2, and LC3-II in the absence of any morphological, IHC, or biochemical marker of apoptosis.
There was a close correlation between the expression of autophagic gene products and the residual tumoral cellularity, a finding that is underlined by the immunophenotypical profile (Figure 1) and in keeping with the assumption that autophagy represents a single cell's adaptation to starvation [30]. There was in fact a trend toward a threshold decrease in the autophagy pathway in the a and b samples, although b samples showed a definite relative excess of autophagic markers, such as excess beclin1 in beclin1/bcl2 complexes in the presence of beclin1/PI3KIII complexes. This is in line with the idea that our morphological and biochemical findings may represent the end of a phenomenon in which the rapid nonphysiological up-regulation of autophagy triggered by imatinib-induced receptor starvation is in part extinguished.
As PI3KIII is responsible for initiating autophagy by forming complexes with beclin1 and LC3-II expression corresponds to autophagosome formation, the absence of LC3-II protein in two b samples is not unexpected: it is conceivable that most of autophagosomes in high responders (b) have already been digested by lysosomal enzymes and that, in moderate responders (c), autophagy is still at such an early stage that tumoral cells can retain their vitality.
The low bcl2 expression in beclin1/bcl2 complexes in the posttreatment regressing tumors is in keeping with the idea that the down-regulation of bcl2 induces autophagic cell death [31] and does not conflict with the view that GISTs with high bcl2 levels aremore susceptible to a KIT inhibitor such as imatinib [27]. However, as approximately 70% of GISTs express bcl2, and our series did not include any matched pretreatment and posttreatment samples, we can only say that bcl2 contributes to regulating the process of autophagy and that the bcl2 threshold in b and c samples is consistent with cell death.
The fact that the autophagy gene profile in the low responding group did not substantially differ from that in the untreated cases strongly suggests that autophagy is not acquired, but part of the molecular profile of GISTs, and that imatinib-treated GIST cells trigger the same machinery as that used to control cell growth. This assumption, which is also suggested by the marked vacuolization reported to be a notable histological feature in GIST [32,33], is in line with the recently underlined close relationship between the type of cell survival/death used by each tumor type, and its response to drug treatment [34].
Remarkably, we did not observe any morphological changes corresponding to apoptosis, a finding that was corroborated by the negative results of the biochemical and IHC experiments using caspase 3, caspase 7, and lamin A/C antibodies. This absence of apoptosis only apparently conflicts with in vitro experiments because the cell model does not work under nutrient starvation conditions [1,2] and cell lines are treated with imatinib for only a short time. Furthermore, fingerprints of autophagy (but not apoptosis) were also found in the GIST specimens taken from two untreated patients. In this context, it is worth noting that although it inhibits KIT activation, imatinib may fail to induce apoptosis in GIST882 cell lines [35,36], thus suggesting that its antiproliferative effect may be mediated by cell cycle arrest rather than apoptosis. This is in keeping with clinical observations in which the rapid switching off of tumoral TK activity is not paralleled by a decrease in tumor size: also known as “tumor dormancy,” this fits well with the metabolic characteristics of autophagy.
Given the pleiotropic role of autophagy, and the evidence that it and GISTs share a number of RTK pathways, our findings suggest various considerations. First, as autophagy seems to exist in both untreated and treated GISTs, it is conceivable that it may contribute to controlling tumoral cell growth in untreated cases, while being the result of a specific stress response to the imatinib-induced starvation of RTK activation in treated GISTs.
Second, the pleiotropism of autophagy may explain the wide spectrum of posttreatment changes which, depending on the threshold of nutrient starvation and the recognized clonal composition of GISTs, may range from areas showing a complete loss of cellularity or very few scattered and regressing residual cells, to clusters of still viable multi-vacuolated tumoral cells and clusters of tumoral cells lacking posttreatment features in nonresponsive areas.
Third, the pleiotropic role of autophagy may explain the prompt tumoral regrowth that occurs when the treatment is discontinued: removing the starvation block (imatinib) may allow the still viable multivacuolated tumoral cells to resume functioning.
Finally, in relation to the activation of RTK pathways, the evidence that rapamycin may overcome resistance to imatinib once again strongly supports the involvement of autophagy in GISTs: mTOR is a downstream intermediate of the PI3KI/AKT pathway, and rapamycin can eliminate its inhibitory effects and thus favor autophagy [37]. In this light, it is interesting to note that we have recently investigated RTK downstream signaling in 15 treated and 5 untreated GISTs (partially overlapping the present series; manuscript submitted) and found MAPK cascade activation, which is known to promote autophagy [38,39].
In conclusion, although the unavailability of paraformaldehydeglutaraldehyde- fixed material made it impossible to use electron microscopic assay and a direct demonstration of autophagy could not be provided, through the analysis of surgical specimens, we provided a substantial number of descriptive observational data which, together with clinical and imaging evidence, strongly suggest that autophagy rather than apoptosis seems to be activated in GISTs in response to imatinib treatment. Further investigations, including the analysis of imatinib-treated normal tissue, and focusing on mechanistic insights into the role of autophagy in imatinib response through in vitro assays are needed to confirm these preliminary descriptive data.
Acknowledgments
We have no conflict of interest to declare.
Footnotes
Supported by grants from the AIRC (Associazione Italiana per la Ricerca sul Cancro) to S.P. and E.T.
References
- 1.Duensing A. Mechanisms of oncogenic KIT signal transduction in primary gastrointestinal stromal tumors (GISTs) Oncogene. 2004;23:3999–4006. doi: 10.1038/sj.onc.1207525. [DOI] [PubMed] [Google Scholar]
- 2.Bauer S. KIT oncogenic signaling mechanisms in imatinib-resistant gastrointestinal stromal tumor: PI3-kinase/AKT is a crucial survival pathway. Oncogene. 2007;26:7560–7568. doi: 10.1038/sj.onc.1210558. [DOI] [PubMed] [Google Scholar]
- 3.Miselli FC, et al. c-Kit/PDGFRA gene status alterations possibly related to primary imatinib resistance in gastrointestinal stromal tumors. Clin Cancer Res. 2007;13:2369–2377. doi: 10.1158/1078-0432.CCR-06-1745. [DOI] [PubMed] [Google Scholar]
- 4.Agaram NP, et al. Pathologic and molecular heterogeneity in imatinib-stable or imatinib-responsive gastrointestinal stromal tumors. Clin Cancer Res. 2007;13:170–181. doi: 10.1158/1078-0432.CCR-06-1508. [DOI] [PubMed] [Google Scholar]
- 5.Miettinen M. Gastrointestinal stromal tumors of the stomach: a clinicopathologic, immunohistochemical, and molecular genetic study of 1765 cases with long-term follow-up. Am J Surg Pathol. 2005;29:52–68. doi: 10.1097/01.pas.0000146010.92933.de. [DOI] [PubMed] [Google Scholar]
- 6.Klionsky DJ. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gozuacik D. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 8.Maiuri MC. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 9.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 10.Tsujimoto Y. Another way to die: autophagic programmed cell death. Cell Death Differ. 2005;12:1528–1534. doi: 10.1038/sj.cdd.4401777. [DOI] [PubMed] [Google Scholar]
- 11.Liang XH. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 12.Yue Z. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kihara A. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng X. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 15.Pattingre S. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Pattingre S. Bcl-2 inhibition of autophagy: a new route to cancer? Cancer Res. 2006;66:2885–2888. doi: 10.1158/0008-5472.CAN-05-4412. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 18.Kondo Y. Autophagy and cancer therapy. Autophagy. 2006;2:85–90. doi: 10.4161/auto.2.2.2463. [DOI] [PubMed] [Google Scholar]
- 19.Gronchi A. Surgery of residual disease following molecular-targeted therapy with imatinib mesylate in advanced/metastatic GIST. Ann Surg. 2007;245:341–346. doi: 10.1097/01.sla.0000242710.36384.1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrone F, et al. 9p21 locus analysis in high-risk gastrointestinal stromal tumors characterized for c-kit and platelet-derived growth factor receptor alpha gene alterations. Cancer. 2005;104:159–169. doi: 10.1002/cncr.21113. [DOI] [PubMed] [Google Scholar]
- 21.Benjamin RS. We should desist using RECIST, at least in GIST. J Clin Oncol. 2007;25:1760–1764. doi: 10.1200/JCO.2006.07.3411. [DOI] [PubMed] [Google Scholar]
- 22.Therasse P, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Nat Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 23.Heinrich MC, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006;24:4764–4774. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 24.Lagonigro MS, et al. PDGFRalpha, PDGFRbeta and KIT expression/activation in conventional chondrosarcoma. J Pathol. 2006;208:615–623. doi: 10.1002/path.1945. [DOI] [PubMed] [Google Scholar]
- 25.Lassar AB. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 26.Yantiss RK. Gastrointestinal stromal tumor versus intra-abdominal fibromatosis of the bowel wall: a clinically important differential diagnosis. Am J Surg Pathol. 2000;24:947–957. doi: 10.1097/00000478-200007000-00006. [DOI] [PubMed] [Google Scholar]
- 27.Steinert DM. Expression of Bcl-2 in gastrointestinal stromal tumors: correlation with progression-free survival in 81 patients treated with imatinib mesylate. Cancer. 2006;106:1617–1623. doi: 10.1002/cncr.21781. [DOI] [PubMed] [Google Scholar]
- 28.Ertmer A. The anticancer drug imatinib induces cellular autophagy. Leukemia. 2007;21:936–942. doi: 10.1038/sj.leu.2404606. [DOI] [PubMed] [Google Scholar]
- 29.Lockshin RA. Apoptosis, autophagy, and more. Int J Biochem Cell Biol. 2004;36:2405–2419. doi: 10.1016/j.biocel.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Mizushima N. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akar U. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy. 2008;4(5) doi: 10.4161/auto.6083. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 32.Rubin BP. Gastrointestinal stromal tumours: an update. Histopathology. 2006;48:83–96. doi: 10.1111/j.1365-2559.2005.02291.x. [DOI] [PubMed] [Google Scholar]
- 33.Weiss SW. Enzinger and Weiss's Soft Tissue Tumours. 5th ed. Mosby Elsevier; Extragastrointestinal stromal tumors; pp. 565–581. [Google Scholar]
- 34.Bergmann A. Autophagy and cell death: no longer at odds. Cell. 2007;13:1032–1033. doi: 10.1016/j.cell.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambol EB. Flavopiridol targets c-KIT transcription and induces apoptosis in gastrointestinal stromal tumor cells. Cancer Res. 2006;66:5858–5866. doi: 10.1158/0008-5472.CAN-05-2933. [DOI] [PubMed] [Google Scholar]
- 36.Frolov A. Response markers and the molecular mechanisms of action of Gleevec in gastrointestinal stromal tumors. Mol Cancer Ther. 2003;2:699–709. [PubMed] [Google Scholar]
- 37.Takeuchi H. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 38.Pattingre S. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J Biol Chem. 2003;278:16667–16674. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- 39.Aoki H. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol. 2007;72:29–33. doi: 10.1124/mol.106.033167. [DOI] [PubMed] [Google Scholar]