Abstract
Arabidopsis sol2 mutants showed CLV3 peptide resistance. Twenty-six synthetic CLE peptides were examined in the clv1, clv2 and sol2 mutants. sol2 showed different levels of resistance to the various peptides, and the spectrum of peptide resistance was quite similar to that of clv2. SOL2 encoded a receptor-like kinase protein which is identical to CORYNE (CRN). GeneChip analysis revealed that the expression of several genes was altered in the sol2 root tip. Here, we suggest that SOL2, together with CLV2, plays an important role in the regulation of root meristem development through the CLE signaling pathway.
Keywords: Arabidopsis thaliana, CLE peptides, CLV, Meristem, Receptor kinase, SOL2
In multicellular organisms, intercellular communication is a fundamental mechanism for coordinating growth and differentiation. In the plant shoot apical meristem (SAM), the CLAVATA (CLV) signaling pathway plays an important role in regulating stem cell fate. CLV3 is a peptide ligand of Arabidopsis thaliana that interacts with the receptor complex of CLV1 and CLV2 to restrict the stem cell population in the SAM in a non-cell-autonomous manner (Fletcher et al. 1999). CLV1 is a leucine-rich repeat receptor-like kinase (LRR-RLK), and CLV2 is an LRR protein without a kinase domain (Clark et al. 1997, Jeong et al. 1999). CLV3 belongs to the 32 member CLV3/ESR (CLE) gene family in Arabidopsis. CLE genes encode small proteins with a conserved 14 amino acid motif (CLE motif) at or near the C-terminus (Sharma et al. 2003, Kondo et al. 2006).
It has been suggested that there is functional redundancy among the CLE proteins in the regulation of meristem homeostasis (Fiers et al. 2004, Kinoshita et al. 2007). Similarly, several lines of evidence suggest that multiple receptors for CLE peptides function redundantly (Diévart et al. 2003).
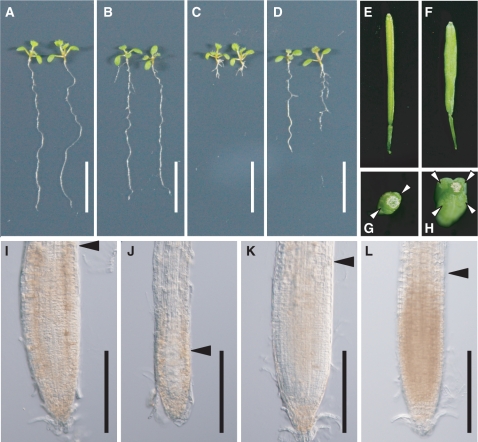
The suppressor of overexpression of LLP1-1 and -2 (sol1, sol2) mutants have been isolated, which suppress a short root phenotype of transgenic plants constitutively overexpressing the CLE19 gene (Casamitjana-Martínez et al. 2003). We examined the CLV3 peptide resistance of the sol2 mutant. In the absence of CLV3 peptide, the sol2 mutant roots were slightly shorter than the wild type (Figs. 1A, B and 2A). We could not detect any difference regarding root apical meristem (RAM) size between the sol2 and wild-type plants grown on agar medium without peptide (Fig. 1I, K). The application of 1 µM CLV3 peptide severely inhibited root growth in the wild type; however, the sol2 mutant was more resistant to the peptide treatment (Fig. 1A–D, I-L). These results suggested that the synthetic CLV3 peptide induces RAM consumption, as has been demonstrated previously (Fiers et al. 2005, Kinoshita et al. 2007), and functions in a SOL2-dependent signaling pathway. Carpel number is used as an indicator of the clv phenotype (Ni and Clark 2006). The carpel number per flower was significantly increased in the sol2 mutant (Fig. 1E–H), as has been reported previously (Casamitjana-Martínez et al. 2003). The presence of extra carpels in the sol2 flowers, which is the typical phenotype of the clv mutants, also indicates that SOL2 functions in the same CLV signaling pathway.
Fig. 1.
Root and shoot phenotypes of the sol2 mutant. (A–D) Ten-day-old seedlings of the wild type, Utr (A), and the sol2 mutant (B) grown on MS agar plates, and of the wild type (C) and the sol2 mutant (D) grown on MS agar plates containing 1 µM CLV3 peptide. Scale bars: 10 mm. (E, G) Carpels of the wild type. The wild type has two carpels. (F, H) Carpels of the sol2 mutant. This sample has four carpels. The arrowhead shows the point of the carpels. (I–L) Nomarski images showing the root meristem boundary (arrowhead) of a 10-day-old wild-type root (I, J) and a sol2 mutant root (K, L) treated without (I, K) and with (J, L) 1 µM CLV3 peptide. Scale bars: 200 µm.
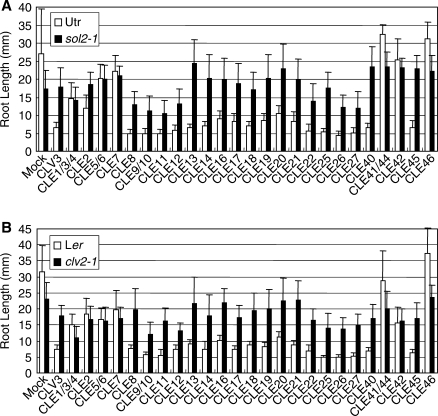
To examine the sensitivity of sol2 against the various peptides, we tested whether 26 synthetic 12 amino acid peptides derived from the CLE motif could trigger RAM consumption. Arabidopsis seeds were germinated on vertical plates with medium containing each peptide (1 µM), and the root length was measured at 10 d after germination. Treatment with CLE41/44 or CLE46 did not affect root elongation, and CLE1/3/4, CLE2, CLE5/6, CLE7 and CLE42 affected root growth only slightly in the wild types, Utr and Ler (Fig. 2A, B). These five peptides did not have obvious effects in the wild type Col (Kinoshita et al. 2006), indicating that peptide sensitivity may be slightly different in each Arabidopsis wild-type accession. The other 19 peptides inhibited root growth and reduced the size of the RAM in wild-type plants (Figs. 1A, C, I, J, and 2A, B). Next, we used two clv1 alleles, clv1-4 and clv1-6, and the clv2-1 and sol2 mutants to compare root elongation. The strong allele of clv1-4 and the weak allele of clv1-6 tended to have shorter roots in the absence of externally added peptides (Supplementary Figs. S1, S2). This result may suggest that CLV1 has a positive role in root elongation in the absence of CLE peptides. The inhibition of root elongation by CLE peptides was not dramatically affected by the clv1 mutations (Supplementary Figs. S1, S2), suggesting that CLV1 is not involved in CLE signaling in roots or that a gene(s) with redundant functions to CLV1 operates in the roots. The sol2 and clv2 roots were also slightly shorter than those of the wild type. These mutant roots were partially resistant to the CLE peptides that inhibit wild-type root elongation (Fig. 2A, B). Interestingly, the sol2 mutants showed various degrees of resistance to different synthetic peptides, and the level of the resistance to each peptide was quite similar to that of the clv2 mutant (Fig. 2A, B). This result indicated the cooperative roles of CLV2 and SOL2 in mediating CLE signaling in roots.
Fig. 2.
CLE peptides affect root elongation in Arabidopsis clv2 and sol2 mutants. (A) Root length of the wild type Utr (parental strain of the sol2) and sol2 mutant was measured 10 d after germination. (B) Root length of the wild type Ler and clv2. Mean values and standard deviations were calculated from at least seven samples grown on agar medium containing various CLE peptides (1 µM).
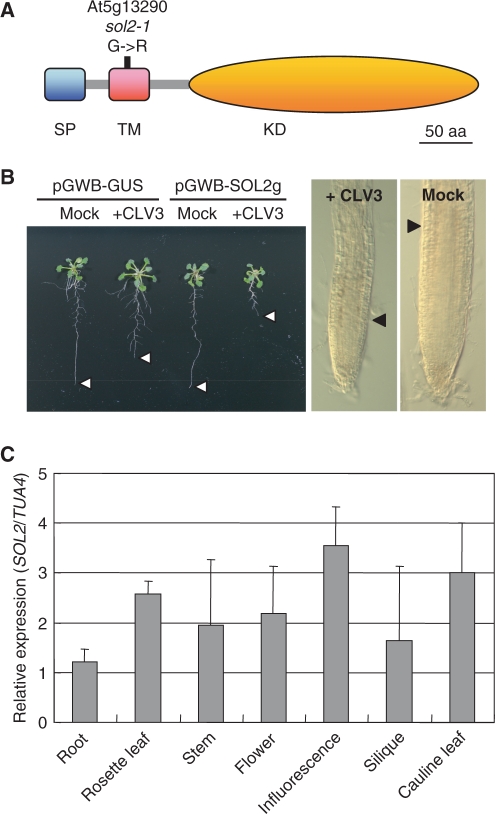
To investigate the molecular basis of the sol2 mutation, we attempted to isolate the affected gene by map-based cloning. For this purpose, the increased carpel number and CLV3 peptide-resistant root phenotypes were used as indicators to map the SOL2 locus to chromosome 5 between markers RCI1B (14/704 recombinant/chromosome tested) and nga151 (1/704). Using the corresponding wild-type sequence, we identified a G→A transition at 220 bp downstream from the ATG start codon of At5g13290 causing a single amino acid substitution (G→R) at codon 74 in a putative RLK (Fig. 3A). At5g13290 encodes a 401 amino acid RLK protein that contains a predicted N-terminal signal peptide, a transmembrane domain and a kinase domain. The sol2 mutation was found in the transmembrane domain (Fig. 3A).
Fig. 3.
Structure and expression of SOL2. (A) Structure of the receptor-like kinase, At5g13290. SP, signal peptide; TM, transmembrane domain; KD, kinase domain. The single amino acid substitution (G→R) is indicated. (B) Complementation of the sol2 mutant. Transgenic hairy roots of the sol2 mutant, harboring the SOL2 (pGWB-SOL2g) genome region, were sensitive to 1 µM CLV3 peptide. As a control, transgenic hairy roots of the sol2 mutant expressing the GUS gene (pGWB-GUS) were resistant to CLV3 peptide. The white arrowhead indicates the root tip region (left). The RAM region of transgenic sol2 hairy roots expressing SOL2 showed RAM consumption (right). The black arrowhead indicates the root meristem boundary. (C) Expression analysis of the SOL2 gene using total RNA from roots, rosette leaves, open flowers, stems, inflorescences, siliques and cauline leaves. SOL2 expression was normalized against that of the TUA4 gene.
To confirm that At5g13290 was the causative gene, we performed a complementation test using the hairy root transformation system. A genomic DNA fragment spanning the At5g13290 locus was cloned into a binary vector and transformed into Agrobacterium rhizogenes. This Agrobacterium strain was then used to generate the sol2 mutant plants with transgenic roots. After the emergence of hairy roots, plants were transferred to MS agar plates with and without the CLV3 peptide, and incubated for 7 d. The transgenic hairy roots of sol2 mutants carrying the wild-type SOL2 genome region showed a short root phenotype and RAM consumption when grown on agar plates with the CLV3 peptide, and we concluded that the At5g13290 gene is the causative gene of the sol2 mutant (Fig. 3B). As a control, we confirmed that the transgenic sol2 hairy roots with the β-glucronidase (GUS) gene showed resistance to the CLV3 peptide (Fig. 3B).
To investigate the expression pattern of SOL2, a quantitative reverse transcription–PCR (RT–PCR) analysis was conducted using total RNA isolated from the roots, rosette leaves, open flowers, stems, inflorescences, siliques and cauline leaves. The expression of SOL2 was observed in all the tissues tested (Fig. 3C), and the highest expression was detected in the inflorescence, suggesting that it probably functions in various developmental processes.
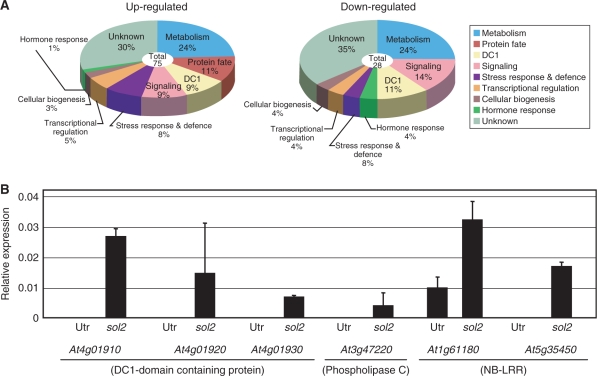
The sol2 mutant did not show RAM consumption in response to the CLV3 peptide treatment, indicating the importance of SOL2 in RAM maintenance. GeneChip analysis was performed to compare the gene expression profiles of the wild-type and the sol2 mutant root apex region, about 2 mm in length, when they were grown on agar plates without CLE peptides. Genes with an expression ratio (sol2/wild type) of <1/4 or >4 were selected. We identified 28 down-regulated genes and 75 up-regulated genes in the RAM region of the sol2 mutant (Fig. 4A; Supplementary Tables S1, S2). Of the up-regulated genes, 24% encoded metabolic enzymes, including oxidoreductase, cytochrome P450 and dioxygenase. Furthermore, 9% of the up-regulated genes encoded various signaling-related proteins and proteins involved in protein fate decision and transcriptional regulation. Expression of peptidase and a set of genes involved in disease resistance/stress response was also up-regulated in the sol2 mutant. Interestingly, 9% of the up-regulated genes and 11% of the down-regulated genes encoded DC1 domain-containing proteins. The DC1 domain is considered to be a type of Zn2+-binding RING finger domain, which probably acts as a transcription factor, although its biological function is largely unknown (Shinya et al. 2007). The Arabidopsis genome encodes 146 putative DC1 domain-containing proteins. Our GeneChip analysis detected 10 of them, suggesting that the DC1 domain-containing transcription factors might have specific roles in SOL2-mediated CLE signaling.
Fig. 4.
(A) Identification of genes down- or up-regulated in the sol2 mutant root tip using GeneChip analysis. Samples were grown on MS agar plates without CLV3 peptide for 4 d and the RAM region of 2 mm from the root tip was collected for this analysis. Gene expression profiles of the wild-type and the sol2 mutant root tips were analyzed with GeneChip. The up- and down-regulated genes, in both the duplicated experiments, were categorized into nine groups by the common functions of their putative gene products. Lists of the genes are given in Supplementary Tables S1 and S2. (B) Quantitative RT–PCR of up-regulated genes in the sol2 mutant root tip. Changes in gene expression of At4g01910, At4g019420, At4g01930, At3g47220, At1g61180 and At5g35450 were calculated relative to the abundance of the TUA4 gene. Bars denote standard deviations (n = 3). For all samples, P <0.01 (t-test, Utr vs. sol2).
To confirm the results of the GeneChip analyses, we conducted a real-time RT–PCR experiment to monitor the transcript levels of six genes, At4g01910 (DC1), At4g01920 (DC1), At4g01930 (DC1), At3g47220 (phospholipase C), At1g61180 (nucleotide-binding site–LRR; NB-LRR) and At5g35450 (NB-LRR), in the wild type and sol2 mutants without CLV3 peptide treatment. As a result, all of the transcripts accumulated to higher levels in the sol2 mutant (Fig. 4B), indicating that these six genes may be involved in CLE signaling under the control of the SOL2 RLK in the RAM.
Here, we suggest that SOL2 functions in the CLE signaling pathway for plant meristem maintenance. From our root elongation assay, we found the sol2 and clv2 mutants showed similar sensitivities to each CLE peptide, and the levels of sensitivities to various CLE peptides differed (Fig. 2A, B). This result may suggest the existence of a complex signaling network for meristem maintenance. However, we cannot exclude the possibility that the different sensitivity simply reflects the different binding affinities of CLE peptides for their receptors.
On the other hand, the spectrum of CLE sensitivity seems very similar between the sol2 and clv2 mutants, suggesting functional similarities between the SOL2 and CLV2 genes in meristem maintenance. Recently, Müller et al. found that the coryne (crn) mutant also showed a clv-like phenotype and resistance to the defect caused by CLV3 overexpression. crn has a mutation in the At5g13290 gene (Müller et al. 2008), and the results support our idea that the At5g13290 gene is involved in the CLE signaling pathway. The RLK has a cytoplasmic kinase domain and a short extracellular domain. In contrast, CLV2 has an extracellular LRR domain and a short intracellular domain. Together with our results showing that both sol2 and clv2 showed similar resistance to various CLE peptides, SOL2/CRN might be responsible for meristem maintenance synergistically working with CLV2, for example in the same receptor complex. The kinase domain of SOL2/CRN might complement the lack of an intracellular domain in CLV2.
The clv1 and wild-type plants showed similar CLE peptide sensitivities, and CLV1 does not seem to function in RAM maintenance. However, we cannot deny the possibility that some other functionally redundant receptors complement the effects of the clv1 mutation. It remains to be investigated whether CLV1 and SOL2/CRN have redundant functions in the RAM maintenance system.
Our GeneChip analysis of the sol2 mutant showed that the expression levels of a number of genes are affected by the loss of the SOL2/CRN gene, giving rise to several remarkable observations. First, the gene expression of two peptidases is significantly up-regulated in the sol2 plants compared with that in the wild type. Because meristem homeostasis is mediated by the CLV system, it is plausible that the loss of SOL2/CRN-mediated signaling up-regulates peptidase gene expression, which is possibly involved in the CLE protein maturation processes. Secondly, a set of genes involved in the disease resistance/stress response, including several genes encoding putative plant immune receptor types of NB-LRR proteins, is significantly up-regulated in sol2 plants. SOL2/CRN may not only be responsible for the meristem maintenance, but may also play a role in disease resistance/stress responses.
In conclusion, our data support a role for SOL2/CRN in RAM homeostasis, possibly in conjunction with CLV2, to perceive CLE signaling molecules leading to the regulation of DC1 and NB-LRR transcript levels. A further analysis of SOL2/CRN downstream gene candidates provides new opportunities to broaden our understanding of the signaling components involved in plant meristem maintenance.
Materials and Methods
Plants (A. thaliana) were grown on soil, as described previously (Kinoshita et al. 2007). Wild-type A. thaliana (Utr and Ler) and several mutants related to CLV signaling, clv1-4 (Ler), clv1-6 (Ler), clv2-1 (Ler) and sol2-1 (Utr), were used in this study. Arabidopsis seeds germinated on MS plates were sterilized, as described previously (Kinoshita et al. 2007). CLE peptides were obtained and stored as described previously (Ito et al. 2006).
The SOL2 locus was mapped by crossing the sol2-1 mutant with Ler. DNA was prepared from 352 sol2 mutant F2 generation plants (96 plants showed increased carpel number; 256 plants showed CLV3 peptide resistance). Using simple sequence length polymorphic markers and cleaved amplified polymorphic sequence markers, recombination events between sol2 and the surrounding regions were identified.
Total RNA was extracted from about 2,000 main root tips (2 mm in length) of 4-day-old wild-type and sol2 plants with TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and purified with the RNeasy Micro Kit (Qiagen, Valencia, CA, USA), according to the manufacturers’ instructions. GeneChip analyses were independently performed twice with the Arabidopsis ATH1 Genome Array (Affymetrix, Santa Clara, California, USA) as described in the GeneChip Expression Analysis Technical Manual. Comparative analysis of the wild type (as the baseline sample) and sol2 (as the experimental sample) was performed with the GeneChip Operating Software (Affymetrix) with the standard parameters. After the elimination of ‘Increased’ genes with ‘Absent’ flags in the experimental sample and ‘Decreased’ genes with ‘Absent’ flags in the baseline sample, genes with a signal log ratio of >2 or <–2, in two experiments, were selected.
For a complementation test, pGWB-SOL2g vector carrying a 5 kb genomic DNA fragment of the SOL2 region amplified using a primer set (5′-cgttaacactttccagagtaatattagt-3′ and 5′-acaatgatccggctcgcaaacatagctaag-3′), and a pGWB-GUS control vector carrying the GUS gene, were transformed into A. rhizogenes LBA1334. The sol2 mutant plants were transformed by the hairy root system with several modifications (Kereszt et al. 2007). Plants were germinated and grown in MS medium in Petri dishes. At 7 d after germination, roots were cut and the aerial parts of plants were inoculated with the bacterial culture. After incubation for 7 d, seedlings were transferred to MS medium with and without 1 µM CLV3 peptide, which contained 200 µg ml−1 cefotaxime to eliminate the bacteria. At 7 d after transfer to MS medium, the transgenic hairy roots obtained were examined for sensitivity to the CLV3 peptide. Twenty plants were transformed, and 80% of plants were phenotypically rescued (Fig. 3B). The insertions of transgene in the longest roots were confirmed by PCR.
For the quantitative RT–PCR analysis, PCR and detection with a Taqman probe for SOL2 and alpha tubulin A4 (TUA4; At1g04820) were performed on the LightCycler (Roche Diagnostics, Indianapolis, IN, USA). To quantify the transcripts, the copy number of each gene was determined with a standard curve constructed using a diluted series of samples. Each reaction was repeated three times. For the quantitative RT–PCR analysis in Fig. 4, PCR was conducted three times by using the 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Gene-specific primers were described in Supplementary Table S3. The copy numbers of the transcripts were normalized against the copy numbers of the TUA4 transcripts, which are often used as internal controls (Sawa et al. 2002). RNA used for RT–PCR was biologically different from that for GeneChip analysis.
Supplementary data
Supplementary data are available at PCP Online.
Funding
Sumitomo Foundation; Fuji Foundation; Grant-in Aid for Creative Scientific Research; Japan Society of the Promotion of Science Grant-in-Aid for Young Scientists (No. 19677001); the Ministry of Education, Culture, Sports, Science Grant-in-Aid for Scientific Research for Priority Areas (No. 19060009 to H.F., Nos. 20061004 and 19060016); Bio-oriented Technology Research Advancement Institution Program of Basic Research Activities for Innovative Biosciences.
Supplementary Material
Acknowledgments
We would like to thank Dr. Ben Scheres for providing us the sol2 seeds. We also thank Yuki Nakashima for help with CLE peptide sensitivity tests.
Glossary
Abbreviations:
- CLE
CLAVATA3/ESR-related
- CLV
CLAVATA
- GUS
β-glucuronidase
- LRR
leucine-rich repeat
- NB
nucleotide-binding site
- RAM
root apical meristem
- RLK
receptor-like kinase
- RT–PCR
reverse transcription–PCR
- SAM
shoot apical meristem.
References
- Casamitjana-Martínez E, Hofhuis HF, Xu J, Liu CM, Heidstra R, Scheres B. Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 2003;13:1435–1441. doi: 10.1016/s0960-9822(03)00533-5. [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Diévart A, Dalal M, Tax FE, Lacey AD, Huttly A, Li J, et al. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell. 2003;15:1198–1211. doi: 10.1105/tpc.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, et al. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell. 2005;17:2542–2553. doi: 10.1105/tpc.105.034009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Hause G, Boutilier K, Casamitjana-Martinez E, Weijers D, Offringa D, et al. Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene. 2004;327:37–49. doi: 10.1016/j.gene.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell. 1999;11:1925–1934. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszt A, Li D, Indrasumunar A, Nguyen CD, Nontachaiyapoom S, Kinkema M, et al. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nature Protocols. 2007;2:948–952. doi: 10.1038/nprot.2007.141. [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S. Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007;48:1821–1825. doi: 10.1093/pcp/pcm154. [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20:934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Clark SE. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 2006;140:726–733. doi: 10.1104/pp.105.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, et al. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 2002;32:1011–1022. doi: 10.1046/j.1365-313x.2002.01488.x. [DOI] [PubMed] [Google Scholar]
- Sharma VK, Ramirez J, Fletcher JC. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol. Biol. 2003;51:415–25. doi: 10.1023/a:1022038932376. [DOI] [PubMed] [Google Scholar]
- Shinya T, Gális I, Narisawa T, Sasaki M, Fukuda H, Matsuoka H, et al. Comprehensive analysis of glucan elicitor-regulated gene expression in tobacco BY-2 cells reveals a novel MYB transcription factor involved in the regulation of phenylpropanoid metabolism. Plant Cell Physiol. 2007;48:1404–13. doi: 10.1093/pcp/pcm115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.