Abstract
Limb-girdle muscular dystrophy type 2A (LGMD2A) is a recessive genetic disorder caused by mutations in calpain 3 (CAPN3). Calpain 3 plays different roles in muscular cells, but little is known about its functions or in vivo substrates. The aim of this study was to identify the genes showing an altered expression in LGMD2A patients and the possible pathways they are implicated in. Ten muscle samples from LGMD2A patients with in which molecular diagnosis was ascertained were investigated using array technology to analyze gene expression profiling as compared to ten normal muscle samples. Upregulated genes were mostly those related to extracellular matrix (different collagens), cell adhesion (fibronectin), muscle development (myosins and melusin) and signal transduction. It is therefore suggested that different proteins located or participating in the costameric region are implicated in processes regulated by calpain 3 during skeletal muscle development. Genes participating in the ubiquitin proteasome degradation pathway were found to be deregulated in LGMD2A patients, suggesting that regulation of this pathway may be under the control of calpain 3 activity. As frizzled-related protein (FRZB) is upregulated in LGMD2A muscle samples, it could be hypothesized that β-catenin regulation is also altered at the Wnt signaling pathway, leading to an incorrect myogenesis. Conversely, expression of most transcription factor genes was downregulated (MYC, FOS and EGR1). Finally, the upregulation of IL-32 and immunoglobulin genes may induce the eosinophil chemoattraction explaining the inflammatory findings observed in presymptomatic stages. The obtained results try to shed some light on identification of novel therapeutic targets for limb-girdle muscular dystrophies.
Introduction
Limb-girdle muscular dystrophy type 2A (LGMD2A) is a recessive genetic disorder caused by mutations in calpain 3 (CAPN3), a muscle-specific, calcium-dependent cystein protease. Calpain 3 structure is similar to that of the ubiquitous calpains 1 and 2, but calpain 3 has specific regions (NS, IS1, and IS2) that confer it special characteristics such as autocatalytic and nuclear translocation capacity. Although calpain 3 was identified in 1989 [1], little is known about its function or its in vivo substrates. It has been reported to play different roles in the cell. Calpain 3 has a certain role in direct and indirect regulation of conventional calpains by proteolytic degradation of calpains and calpastatin respectively [2]. It may be involved in muscle contraction due to its link to titin and to its regulation by calcium [3]–[7].
Calpain 3 was shown to be in complex with dysferlin, suggesting a membrane homeostasis role of calpain 3 [8], and more recent studies demonstrated that AHNAK, a novel component of the dysferlin protein complex, serves as a direct substrate of calpain 3 in cell culture [9].
On the other hand, it has been confirmed that calpain 3 can cleave the C-terminal portion of FLNC in vitro and suggested that FLNC may be an in vivo substrate for calpain 3, functioning to regulate protein-protein interactions with sarcoglycans. Thus, calpain-mediated remodeling of cytoskeletal-membrane interactions, such as those occurring during myoblast fusion and muscle repair, may involve regulation of FLNC-sarcoglycan interactions [10].
Its presence in the nucleus has led to suggest that calpain 3 plays an important role in regulation of transcription factors indirectly controlling apoptotic processes [11], [12]. Recent studies reported that the antiapoptotic factor, cellular FLICE inhibitory protein (c-FLIP), is NF-κB dependent and is only expressed when CAPN3 is present [13]. However, other studies suggest that apoptosis may be secondary to muscle damage and inflammatory response [14].
Based on the observation of the C3 knockout (C3KO) mice, it has been suggested that calpain 3 is necessary for ubiquitination and acts upstream of the ubiquitination machinery [15].
Inflammatory cells have been detected in muscle tissue from patients with mutations in the CAPN3 gene in early stages [16] as happen in other distrophies. The role of inflammation in many dystrophies seems to be unexplained, and it has been related to the presence of signaling factors (cytokines) that withstand inflammatory mechanisms and regulatory phenomena [17]–[19].
In this study, the RNA expression profiling in muscle from biopsies of LGMD2A patients and control subjects were compared in order to determine the potential functions and the pathways in which calpain 3 is implicated.
Materials and Methods
Muscle samples and RNA processing
Muscle biopsies were taken from 10 LGMD2A patients (3 females and 7 males aged 13–48 years, mean age 29,5 years) and 10 controls (2 females and 8 males aged 22–84 years, mean age 50,2 years). Two out of the 10 LGMD2A patients showed an inflammatory pattern with eosinophilic infiltrates in their biopsies.
For diagnostic purposes deltoid, quadriceps, and biceps muscle specimens were collected using institutionally approved protocols and after obtaining informed consent (Table 1). Muscle tissues were snap frozen and stored at −80°C. Most of the 7 symptomatic cases showed similar necrosis and regenerating phenomena (data not available in one case).
Table 1. Distribution of muscle biopsies taken from 10 LGMD2A patients and 10 control subjects.
| Biopsy Number | Status | Gender | Muscle | Age | Myopathological data | Gardner-Medwin-Walton Scale | CAPN3 mutations | |
| Mutation 1 | Mutation 2 | |||||||
| EXP-01 | LGMD2A | M | Quadriceps | 34 | Mild myopathic changes (fiber size alteration, centrally located nuclei and splitting) No necrosis, no regeneration, no lobulated fibers. | 2 | p.(Gly222Arg) | p.(Arg748Gln) |
| EXP-02 | LGMD2A | F | Deltoid | 33 | Necrosis and regenerating phenomena | 3 | c.946-1G>A | p.(Gln660Arg) |
| EXP-03 | LGMD2A | M | Quadriceps | 37 | Necrosis, regenerating phenomena and fibrosis | Unknown | p.(Met248Arg) | p.(Arg769Gln) |
| EXP-04 | LGMD2A | M | Quadriceps | 44 | Necrosis and regenerating phenomena | Unknown | p.(Gln300X) | p.(Gln660Arg) |
| EXP-05 | LGMD2A | M | Deltoid | 13 | Inflammatory reaction around necrotic and non necrotic fibers. Inflammation collects at endomysial site sometimes with perivascular infiltrate without destruction of walls of arterioles and venules. Numerous eosinophilic leucocytes are present. | Asymptomatic | p.(Arg788SerfsX14) | p.(Arg788SerfsX14) |
| EXP-09 | LGMD2A | F | Biceps braquialis | 14 | Myositis with local infiltration of eosinophils. Patchy, focal inflammatory cell infiltrate with minor changes in the architecture of fibers without changes in the distribution of the fiber type. | Asymptomatic | p.(Arg490Trp) | p.(Gly691TrpfsX7) |
| EXP-35 | LGMD2A | M | Deltoid | 48 | Necrosis, fibrosis, lobulated fibres | 7 | p.(Gln142X) | p.(Gln142X) |
| EXP-36 | LGMD2A | M | Quadriceps | 26 | Necrosis and regenerating phenomena | 2 | p.(Lys254Glu) | c.1910delC |
| EXP-40 | LGMD2A | M | Quadriceps | 29 | Mild myopathic changes (centrally located nuclei and fibrosis) | 7 | p.(Ala160Gly) | c.1029+3A>G |
| EXP-41 | LGMD2A | F | Deltoid | 17 | No data available | 2 | c.2185-12_2194del | p.(Arg788SerfsX14) |
| EXP-25 | Control | F | Deltoid | 57 | ||||
| EXP-27 | Control | M | Quadriceps | 50 | ||||
| EXP-28 | Control | M | Quadriceps | 22 | ||||
| EXP -29 | Control | F | Quadriceps | 73 | ||||
| EXP -30 | Control | M | Quadriceps | 84 | ||||
| EXP-31 | Control | M | Quadriceps | 46 | ||||
| EXP-32 | Control | M | Quadriceps | 48 | ||||
| EXP-33 | Control | M | Deltoid | 51 | ||||
| EXP-38 | Control | M | Quadriceps | 31 | ||||
| EXP-39 | Control | M | Quadriceps | 41 | ||||
The quality of all RNAs obtained from muscle biopsies (RNAPlus, QBiogene) was verified using spectrophotometry and the Bioanalyzer system (Agilent). All of them showed acceptable quality and integrity (RIN above 7) to be eligible for the experiment.
All RNAs were reverse-transcribed, and biotinylated cRNA probes were generated by in vitro transcription (Ambion, CA, USA). Fragmented cRNA of each sample was hybridized individually to human HG-U133A (22.283 probe sets) and HG-U133B (22.645 probe sets) GeneChips (Affymetrix, Santa Clara, California) in order to analyze the expression of 44.928 probes, comprising more than 33.000 genes.
Data analysis
In-depth quality controls were performed to analyze the validity of the hybridization processes in accordance with four criteria. First, the correct presence of the signal corresponding to the spike control BioB. Second, the expression ratio between the 3′ and 5′ ends of the housekeeping GAPDH should not exceed a value of three. Third, the full percentage of presences detected by the Affymetrix Detection algorithm for each array must be in the range 40–60. And finally, the percentage of outlier probe sets detected within each microarray should be less than 5%. All hybridized arrays on the study met all four quality criteria, demonstrating the reliability of data generated.
The hybridized arrays were scanned, and raw data were extracted using the Microarray Analysis Suite 5.0 (MAS5; Affymetrix). The raw data were normalized using RMA (Robust Multichip Average) expression summary in Bioconductor [20]. RMA consists of three steps: a background adjustment, quantile normalization, and finally summarization [21]–[23].
The sensitivity of microarray-generated data to noise from experimental variables is well documented [24]. For the analysis, the average values of each tested group (patients and controls) were used in order to obtain the most homogeneous results, trying to avoid variability between individual cases due to different characteristics (genetic background, sex, age, muscles, mutations, etc.). Two statistical methods were applied in order to distinguish significant and substantial differential expression from noise and variation due to either genetic heterogeneity or experimental procedures.
First, in order to identify significantly different genes between LGMD2A patients and normal controls, a geometric fold-change analysis was used [24], [25]. The threshold was set at a two-fold change value. Using the criterion of fold-change implies that larger fold changes are most likely to be real and no hypothesis is assumed. Principal component analysis (PCA) was performed after array normalization. PCA is a technique that summarizes a large set of variables in a smaller set that retains the essential variance of the original data set [26]. PCA derives an equivalent, uncorrelated set of new variables from the original set of correlated variables according to their contribution to a ranked set of principal components [27].
Second, Class Comparison Difference Analyses were performed using BRB-ArrayTools developed by Dr. Richard Simon and BRB-ArrayTools Development Team. In order to identify probe sets with significant intensity differences between disease classes, a two-sample univariate t-test was applied to the unaffected control data set vs. the LGMD2A data set. The use of p-values implies hypothesis testing. It is assumed in the null hypothesis that there is no fold change and then evidence was looked for to reject it using a type-1 error. The threshold was set at p 0.001.
To minimize false positives, only the probe sets commonly yielded by both methodologies were included into the final list of genes differentially expressed in LGMD2A.
Moreover, as an additional supporting process, two machine learning feature selection techniques were run. Symmetrical uncertainty ranking [28] was first applied as an univariate criterion to measure the worth of each probe set alone: this computes the mutual information with respect to the class phenotype and compensates for the bias of the information gain. Correlation-based Feature Subset (CFS) selection [29], a multivariate feature selection that evaluates the merit of a probe set subset by measuring the individual predictive power of each probe set along with the redundancy within that subset, was then used. CFS outputs a subset of features instead of individual relevances.
The same procedure was used to compare samples from patients who were asymptomatic but had eosinophilic infiltrates in their muscle biopsies (2 cases) and samples from healthy controls (10 cases).
Microarray data have been submitted to the GEO (Gene Expression Omnibus) public database (accession GSE11681).
Quantitative Real-Time PCR
To investigate the validity of array data, expression levels of the differentially expressed genes were measured using the TaqMan quantitative RT-PCR assay. Relative expression levels initially determined with the cDNA microarrays were correlated to the expression levels assessed using quantitative RT-PCR for each patient sample.
Whereas microarrays identify target genes of interest among thousands of genes, truly quantitative information relies on quantitative RT-PCR. Some of the significantly regulated changes found on the microarray could be replicated by quantitative RT-PCR. Quantitative RT-PCR was performed using the 7900 HT Fast Real-Time PCR System (Applied Biosystems). Because of the limiting RNA amount isolated from muscle biopsies used for microarray analysis, only a few samples (6 cases) were used for confirmation with quantitative RT-PCR experiments.
The TaqMan Low Density Arrays (TLDA) were purchased from Applied Biosystems, and the protocol recommended by the manufacturer was used. Customer-designed TLDAs were used in order to test a series of 63 genes. In order to select these genes, genes with unknown function, hypothetical proteins, and open reading frame regions were excluded. Gene families were represented including only some of the members, such as collagens, etc. Moreover, genes showing differential expression profiling in the comparison between patients with eosinophilic infiltrates and healthy controls were included in the TLDA, as well as genes with expression variation in other studies.
Expression levels for all transcripts were determined relative to the internal housekeeping control gene GAPDH in the TLDAs which, as expected, did not demonstrate altered expression according to microarray analysis.
In order to identify probe sets with significant intensity differences, the method applied to the unaffected control data set vs. the LGMD2A data set was Benjamini-Hochberg method using Stat Miner program (Integromics).
Results
After having adjusted the background, normalized and summarized the data, the fold change obtained generates a list by magnitude of response. As a result of this method, the fold change analysis identified 156 differentially expressed probe sets in LGMD2A skeletal muscle compared to control skeletal muscle. Of these, 92 were significantly overexpressed and 64 showed a reduced expression in LGMD2A patients compared to the unaffected controls.
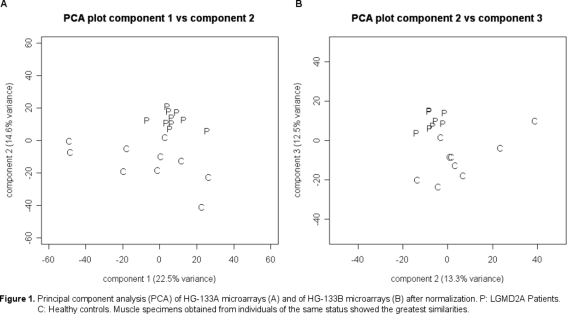
PCA grouped together on the one hand patient samples and on the other hand control samples and a greater variability was seen among controls due to the heterogeneity of this group (Figure 1).
Figure 1. Principal component analysis (PCA) of HG-U133A microarrays (A) and of HG-U133B microarrays (B) after normalization.
P: LGMD2A Patients. C: Healthy controls. Muscle specimens obtained from individuals of the same status showed the greatest similarities.
On the other hand, the additional statistical method used to analyze the data, the Class Comparison Differences method, applied a two-sample univariate t-test to the unaffected control data set vs. the LGMD2A data set. This method identified 627 probe sets with a p value higher than 0.001.
However, the final list of genes comprised 86 probe sets (74 genes) commonly yielded by the two methodologies which were differentially expressed in LGMD2A compared to unaffected muscle biopsies. Of these 74 genes, 53 were overexpressed and 21 had a reduced expression in the LGMD2A patients and all the genes were clustered into functional groups (Table 2). Transcripts were classified according to different biological processes, as obtained from LocusLink (www.ncbi.nlm.nih.gov/LocusLink/): extracellular matrix proteins/phosphate transport, cell adhesion, muscle development, transcription factors, signaling pathways, metabolic process, transport, ubiquitin cycle, and other functions.
Table 2. Significantly differentially regulated transcripts, NV: Not validated by TLDAs.
| Affymetrix ID | Biological process/Gene title | Gene symbol | Fold change | Parametric p value Class Comparison | Fold change Validated by RT-PCR | Significant p value Stat Miner |
| Extracellular matrix proteins | ||||||
| 202310_s_at | collagen, type I, alpha 1 | COL1A1* | 4.71 | 0.0003482 | 5.86 | 0.012178774 |
| 202404_s_at | collagen, type I, alpha 2 | COL1A2* | 4.72 | 6.61e-05 | NV | – |
| 211161_s_at | collagen, type III, alpha 1 | COL3A1* | 7.72 | 1.1e-06 | 13.71 | – |
| 201852_x_at | 5.12 | 7.4e-06 | ||||
| 215076_s_at | 4.79 | 3.48e-05 | ||||
| 212488_at | collagen, type V, alpha 1 | COL5A1 | 2.07 | 8.18e-05 | 9.24 | 1.13E-04 |
| 221729_at | collagen, type V, alpha 2 | COL5A2 | 3.12 | <1e-07 | NV | – |
| 221730_at | 2.12 | 3e-07 | ||||
| 203477_at | collagen, type XV, alpha 1 | COL15A1* | 2.80 | 2.36e-05 | NV | – |
| 225681_at | collagen triple helix repeat containing 1 | CTHRC1 | 2.96 | 3.74e-05 | NV | – |
| Cell adhesion | ||||||
| 201005_at | CD9 antigen (p24) | CD9 * | 2.38 | 6.36e-05 | 2.16 | 5.71E-02 |
| 212063_at | CD44 antigen (homing function and Indian blood group system) | CD44 * | 2.35 | 0.0002703 | 2.23 | 0.3071496 |
| 211719_x_at | fibronectin 1 | FN1* | 2.33 | 0.0006326 | 3.30 | 1.16E-02 |
| 210495_x_at | 2.30 | 0.0001008 | ||||
| 216442_x_at | 2.21 | 0.0001307 | ||||
| Muscle development | ||||||
| 205940_at | myosin, heavy polypeptide 3, skeletal muscle, embryonic | MYH3* | 11.78 | 1.4e-06 | 40.62 | 2.15E-04 |
| 205145_s_at | myosin, light polypeptide 5, regulatory | MYL5 | 4.28 | 3.49e-05 | 7.21 | 1.83E-02 |
| 204173_at | myosin light chain 1 slow a | MLC1SA ( = MYL6B) | 2.41 | 0.0003024 | 3.28 | 1.77E-02 |
| 219829_at | integrin beta 1 binding protein (melusin) 2 | ITGB1BP2 | 2.23 | 1.3e-06 | 2.50 | 0.013391304 |
| Transcription factors | ||||||
| 202431_s_at | v-myc myelocytomatosis viral oncogene homolog (avian) | MYC* | 0.48 | 0.0006587 | 0.35 | – |
| 201466_s_at | v-jun sarcoma virus 17 oncogene homolog (avian) | JUN* | 0.47 | 4.98e-05 | 0.69 | 0.443749559 |
| 219990_at | E2F transcription factor 8 | E2F8 | 3.22 | 0.0001105 | 4.52 | 4.80E-03 |
| 201473_at | jun B proto-oncogene | JUNB* | 0.41 | 0.0002329 | NV | – |
| 209189_at | v-fos FBJ murine osteosarcoma viral oncogene homolog | FOS* | 0.12 | 7.52e-05 | 0.10 | – |
| 203973_s_at | CCAAT/enhancer binding protein (C/EBP), delta | CEBPD* | 0.39 | 0.0009174 | 1.61 | 0.266597408 |
| 201694_s_at | early growth response 1 | EGR1* | 0.31 | 5.16e-05 | 0.13 | 1.74E-03 |
| 227404_s_at | 0.12 | 8.11e-05 | ||||
| 209357_at | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | CITED2 | 0.39 | 9e-07 | 0.57 | 0.255171653 |
| 210479_s_at | RAR-related orphan receptor A | RORA | 0.46 | 0.0003675 | NV | – |
| 202393_s_at | Kruppel-like factor 10 | KLF10 | 0.45 | 0.0000651 | 0.49 | 0.130862236 |
| 221778_at | KIAA1718 protein | KIAA1718 (JHDM1D) | 0.48 | 0.0009927 | NV | – |
| Signal transduction | ||||||
| 209541_at | insulin-like growth factor 1 (somatomedin C) | IGF1* | 2.81 | 1e-07 | 1.96 | 7.01E-02 |
| 209822_s_at | very low density lipoprotein receptor | VLDLR* | 2.63 | 3e-07 | NV | – |
| 213880_at | leucine-rich repeat-containing G protein-coupled receptor 5 | LGR5 | 0.30 | 0.0004577 | 0.23 | 6.35E-02 |
| 219654_at | protein tyrosine phosphatase-like (proline instead of catalytic arginine), member a | PTPLA | 2.42 | 4.83e-05 | NV | – |
| 222918_at | RAB9B, member RAS oncogene family | RAB9B | 2.04 | 1.09e-05 | NV | – |
| 212099_at | ras homolog gene family, member B | RHOB | 0.41 | 0.0000594 | NV | – |
| 217728_at | S100 calcium binding protein A6 (calcyclin) | S100A6* | 2.26 | 0.0008602 | 2.43 | 5.00E-02 |
| Signal pathways | ||||||
| 200665_s_at | secreted protein, acidic, cysteine-rich (osteonectin) | SPARC* | 2.02 | 0.000903 | 2.55 | 5.82E-02 |
| 218087_s_a | sorbin and SH3 domain containing 1 | SORBS1 | 2.05 | 0.000123 | NV | – |
| 214844_s_at | docking protein 5 | DOK5 | 2.24 | 3.16e-05 | 3.16 | 6.09E-03 |
| 203697_at | frizzled-related protein | FRZB | 5.42 | 0.000466 | 13.16 | 2.41E-04 |
| 203698_s_at | 2.99 | 0.000466 | ||||
| 203789_s_at | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C | SEMA3C | 2.86 | 0.0001054 | NV | – |
| 201309_x_at | chromosome 5 open reading frame 13 | C5orf13 | 2.09 | 0.0005666 | NV | – |
| 236860_at | Neuropeptide Y receptor Y6 (pseudogene) | NPY6R | 4.09 | 0.0007927 | NV | – |
| Metabolic process | ||||||
| 201425_at | aldehyde dehydrogenase 2 family (mitochondrial) | ALDH2 | 0.43 | 8.62e-05 | 0.67 | 0.459523739 |
| 209301_at | carbonic anhydrase II | CA2* | 0.48 | 0.0007153 | 0.79 | 0.522535838 |
| 202464_s_at | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | PFKFB3 | 0.15 | 1.5e-06 | 0.45 | 0.141846652 |
| Transport | ||||||
| 208691_at | transferrin receptor (p90, CD71) | TFRC* | 8.11 | 6.47e-05 | 4.52 | 4.80E-03 |
| 207332_s_at | 5.99 | 6.28e-05 | ||||
| 204430_s_at | solute carrier family 2 (facilitated glucose/fructose transporter), member 5 | SLC2A5 | 2.17 | 0.0003736 | 3.74 | 2.69E-03 |
| 201560_at | chloride intracellular channel 4 | CLIC4 | 2.20 | 0.0001832 | NV | – |
| 202236_s_at | solute carrier family 16 (monocarboxylic acid transporters), member 1 | SLC16A1 | 2.55 | <1e-07 | NV | – |
| 205073_at | Cytochrome P450, family 2, subfamily J, polypeptide 2 | CYP2J2 | 2.04 | 3.4e-05 | NV | – |
| 224579_at | Solute carrier family 38, member 1 | SLC38A1 | 2.58 | 0.0007761 | NV | – |
| 219525_at | hypothetical protein FLJ10847 | FLJ10847 (SLC47A1) | 0.47 | 6.42e-05 | NV | – |
| 217966_s_at | chromosome 1 open reading frame 24 | C1orf24 (FAM129A) | 0.47 | 0.00045 | 0.78 | 0.55888133 |
| Ubiquitin cycle | ||||||
| 218306_s_at | hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 | HERC1 | 2.22 | 0.0000045 | 2.45 | 2.29E-02 |
| 218575_at | Anaphase promoting complex subunit 1 | ANAPC1 | 2.02 | 9e-07 | 1.31 | 0.447268323 |
| 229267_at | 2.07 | 0.0001629 | ||||
| Other functions | ||||||
| 218273_s_at | protein phosphatase 2C, magnesium-dependent, catalytic subunit (mitochondrial) | PPM2C | 2.49 | 1.55e-05 | 3.08 | 2.97E-02 |
| 201609_x_at | isoprenylcysteine carboxyl methyltransferase | ICMT* | 2.25 | 7.2e-06 | NV | – |
| 201611_s_at | 2.00 | 1.88e-05 | ||||
| 202965_s_at | calpain 6 | CAPN6 | 2.05 | 1.71e-05 | 5.31 | 9.12E-04 |
| 212848_s_at | chromosome 9 open reading frame 3 | C9orf3 | 2.01 | 0.0003013 | NV | – |
| 201010_s_at | Thioredoxin interacting protein | TXNIP* | 0.43 | 1e-05 | 0.49 | 0.133790906 |
| 201009_s_at | 0.42 | 0.0009904 | ||||
| 202917_s_at | S100 calcium binding protein A8 (calgranulin A) | S100A8 | 0.34 | 0.0002179 | 0.27 | 1.51E-02 |
| 209398_at | histone 1, H1c | HIST1H1C | 0.45 | 0.0003089 | 0.76 | 0.491015255 |
| 225061_at | DnaJ (Hsp40) homolog, subfamily A, member 4 | DNAJA4* | 2.53 | 0.0000529 | 3.65 | 8.54E-03 |
| 209596_at | matrix-remodelling associated 5 | MXRA5 | 2.90 | 5e-07 | NV | – |
| 219087_at | asporin (LRR class 1) | ASPN | 4.58 | 4e-07 | NV | – |
| 235022_at | chromosome 18 open reading frame 19 | C18orf19 | 2.34 | 1e-07 | NV | – |
| 218820_at | chromosome 14 open reading frame 132 | C14orf132 | 2.14 | 2.87e-05 | NV | – |
| 202016_at | mesoderm specific transcript homolog (mouse) | MEST | 2.11 | 0.000709 | NV | – |
| 218999_at | hypothetical protein FLJ11000 | FLJ11000 (TMEM140) | 0.49 | 218999_at | NV | – |
| 224836_at | tumor protein p53 inducible nuclear protein 2 | TP53INP2* | 2.46 | 0.0003853 | NV | – |
| Unknown function | ||||||
| 238124_at | Myomesin family, member 3 | MYOM3 | 2.30 | 0.0000126 | 6.74 | 1.86E-04 |
| 230284_at | 2.01 | 0.0000003 | ||||
| 202759_s_at | PALM2-AKAP2 protein | PALM2-AKAP2 | 2.16 | 0.0000007 | NV | – |
| 229778_at | Hypothetical protein MGC10946 | MGC10946 (C12orf39) | 2.75 | 0.0000022 | NV | – |
| 211071_s_at | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11 | MLLT11 | 3.92 | 0.0002385 | NV | – |
| 218876_at | brain specific protein | CGI-38 (TPPP3) | 2.86 | 0.0000867 | NV | – |
| 221104_s_at | nipsnap homolog 3B (C, elegans) | NIPSNAP3B | 2.68 | 0.0001963 | NV | – |
| 225242_s_at | steroid sensitive gene 1 | URB (CCDC80) | 3.22 | 0.0004132 | NV | – |
Dysregulated genes in FSHD, DMD, α-sarcoglycan, and congenital myopathies (Campanaro et al 2002, Winokur et al 2003, Haslett et al 2003, Taniguchi et al 2006, Osborne et al 2007).
The additional supporting process, the Correlation-based Feature Subset selection (CFS) [29] highlighted a set of 21 genes. Of these 21 genes, 7 corresponded to the previously determined group of 74 genes. In turn, Symmetrical Uncertainty Ranking returned correlation coefficients higher than 0.5 for 24 genes within the list. Note that the highest correlation was 0.816 when the coefficient was constrained between 1 (maximum) and 0 (minimum). The average coefficient for the whole gene list was 0.36, with a standard deviation of 0.177.
Overview of expression profiling in LGMD2A muscles
Some transcript classes were of particular interest in this analysis (Table 2). Most genes found to be dysregulated in LGMD2A were genes grouped in the transcription factor category, and some of them showed the lowest fold-change values obtained in the study (FOS, EGR1). By contrast, genes showing the highest fold-change values included extracellular matrix proteins, genes involved in muscle development, and three additional genes with different functions (FRZB, TFRC, and CAPN6).
As a whole, in most of the biologically classified processes, the same trend to up- or downregulation was seen for all genes involved in the same process. Genes associated with extracellular matrix (collagen types I, III, V, and XV, and SPARC), cell adhesion, muscle development (MYH3, MYL5 and ITGB1BP2), signaling pathways, and ubiquitin cycle predominated among upregulated genes. However, all genes involved in metabolic processes and transcription factors (except for the E2F8 gene) were downregulated (Table 2).
On the other hand, upregulation of IGF1, which is a regulator of somatic growth and cell proliferation, was seen in this study. IGFa is an inducer of different pathways such as the phosphatidylinositol 3-kinase survival (through activation of AKT1, AKT2), the calcineurin.mediated signaling pathways, and of GATA2 activation.
HERC1 and ANAPC1 are genes implicated in the ubiquitin cycle and showed upregulation in LGMD2A muscle samples. HERC1, ubiquitously expressed, is located in the cytosol in the Golgi apparatus, stimulating guanine nucleotide, forming a cytosolic ternary complex with clathrin and Hsp70, and is involved in protein trafficking. ANAPC1 is a component of the anaphase promoting complex/cyclosome (APC/C), a cell cycle-regulated E3 ubiquitin ligase that controls progression through mitosis and the G1 phase of the cell cycle.
There are two deregulated genes according to our results whose cell location is the mitochondrion matrix, one of which is involved in the metabolic process, ALDH2 (aldehyde dehydrogenase 2 family) (downregulated), while the other, the PPM2C gene (protein phosphatase 2C, magnesium-dependent, catalytic subunit) (upregulated) is implicated in protein amino acid dephosphorylation.
Expression changes in common with other muscular dystrophies
Twenty four out of the 74 deregulated genes with altered expression in LGMD2A were also deregulated in other muscular dystrophies (DMD, α-SGD, FSHD, dysferlinopathies, Fukuyama-type congenital muscular dystrophy, and laminin-α2 deficient congenital muscular dystrophy) [24], [30]–[35] (Table 2).
LGMD2A and eosinophil infiltration
A comparison was made between biopsies of control muscles (10 cases) and biopsies from two cases showing eosinophil infiltrates. Results of this comparison are summarized in Table 3.
Table 3. Significantly differentially regulated transcripts comparing patients with eosinophilic infiltrates to healthy controls.
| Probe set | Biological process/Gene title | Gene symbol | Fold change | Parametric p-value |
| Phosphate transport | ||||
| 212489_at | collagen, type V, alpha 1 | COL5A1 | 2.39 | 0.0000009 |
| 221729_at | collagen, type V, alpha 2 | COL5A2 | 3.20 | 0.0001189 |
| 225681_at | collagen triple helix repeat containing 1 | CTHRC1 | 3.25 | 0.0002197 |
| 230867_at | hypothetical protein LOC131873 | LOC131873 | 2.28 | 0.0000307 |
| Signaling pathway | ||||
| 203697_at | frizzled-related protein | FRZB | 7.67 | 0.0004106 |
| 203698_s_at | 3.02 | 0.0004541 | ||
| Signal transduction | ||||
| 209541_at | insulin-like growth factor 1 (somatomedin C) | IGF1 | 3.06 | 0.0000059 |
| 209542_x_at | 2.51 | 0.000116 | ||
| 211577_s_at | 2.14 | 0.0001273 | ||
| 209822_s_at | very low density lipoprotein receptor | VLDLR | 3.35 | 0.0002441 |
| Immune response | ||||
| 203828_s_at | interleukin 32///interleukin 32 | IL32 | 5.45 | 0.0005892 |
| 211430_s_at | immunoglobulin heavy locus///immunoglobulin heavy constant gamma 1 (G1m marker)///immunoglobulin heavy constant gamma 2 (G2m marker)///immunoglobulin heavy constant gamma 3 (G3m marker)///immunoglobulin heavy constant mu | IGHIGHG1IGHG2IGHG3IGHM | 4.37 | 0.0001606 |
| 221651_x_at | immunoglobulin kappa constant///immunoglobulin kappa variable 1-5 | IGKC///IGKV1-5 | 2.21 | 0.0001881 |
| Transcription | ||||
| 214608_s_at | eyes absent homolog 1 (Drosophila) | EYA1 | 2.30 | 0.0001005 |
| 209357_at | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | CITED2 | 0.34 | 0.0009738 |
| 227705_at | transcription elongation factor A (SII)-like 7 | TCEAL7 | 2.79 | 0.000246 |
| Metabolic process | ||||
| 43427_at | acetyl-Coenzyme A carboxylase beta | ACACB | 0.44 | 0.0002438 |
| 49452_at | 0.39 | 0.0001128 | ||
| 224918_x_at | microsomal glutathione S-transferase 1 | MGST1 | 0.27 | 0.0003928 |
| Transport | ||||
| 202236_s_at | solute carrier family 16 (monocarboxylic acid transporters), member 1 | SLC16A1 | 2.74 | 0.0000816 |
| 239984_at | sodium channel, voltage-gated, type VII, alpha | SCN7A | 2.64 | 0.0005832 |
| Other functions | ||||
| 205206_at | Kallmann syndrome 1 sequence | KAL1 | 0.25 | 0.0004374 |
| 226312_at | TORC2-specific protein AVO3 | AVO3 | 0.47 | 0.0003421 |
| 227013_at | LATS, large tumor suppressor, homolog 2 (Drosophila) | LATS2 | 0.41 | 0.0005208 |
| 224842_at | PI-3-kinase-related kinase SMG-1 | SMG1 | 0.39 | 0.0002757 |
| 202965_s_at | calpain 6 | CAPN6 | 4.00 | <1e-07 |
| 201609_x_at | isoprenylcysteine carboxyl methyltransferase | ICMT | 2.35 | 0.0007576 |
| 202998_s_at | lysyl oxidase-like 2 | LOXL2 | 2.03 | 0.0004611 |
| 209596_at | matrix-remodelling associated 5 | MXRA5 | 4.66 | 0.0001074 |
| 209398_at | histone 1, H1c | HIST1H1C | 0.26 | 0.0007007 |
| 226322_at | ARG99 protein ( Gene name: TMTC1) | ARG99 | 0.45 | 0.0002993 |
| 205381_at | leucine rich repeat containing 17 | LRRC17 | 2.60 | 0.0001683 |
| 219087_at | asporin (LRR class 1) | ASPN | 5.76 | 0.000238 |
| Unknown | ||||
| 202760_s_at | A kinase (PRKA) anchor protein 2///PALM2-AKAP2 protein | AKAP2///PALM2-AKAP2 | 2.71 | 0.0000528 |
| 202759_s_at | PALM2-AKAP2 protein | PALM2-AKAP2 | 3.29 | 0.0000042 |
| 226155_at | KIAA1600 | KIAA1600 | 0.46 | 0.0004366 |
| 235022_at | chromosome 18 open reading frame 19 | C18orf19 | 2.46 | 0.0002359 |
| 229778_at | hypothetical protein MGC10946 | MGC10946 | 2.92 | 0.0007268 |
| 219525_at | hypothetical protein FLJ10847 | FLJ10847 | 0.42 | 0.0004347 |
| 211724_x_at | hypothetical protein FLJ20323 | FLJ20323 | 0.45 | 0.0003093 |
| 225893_at | clone TESTIS-724 mRNA sequence | 0.41 | 0.0001744 | |
Genes involved in immune response such as IL-32, IGHG1, and IGKC were upregulated, as well as CAPN6, while those involved in chemotaxis or regulation of the protein kinase B signaling cascade were downregulated in asymptomatic cases comparing with controls.
Validation of microarray data by quantitative RT-PCR
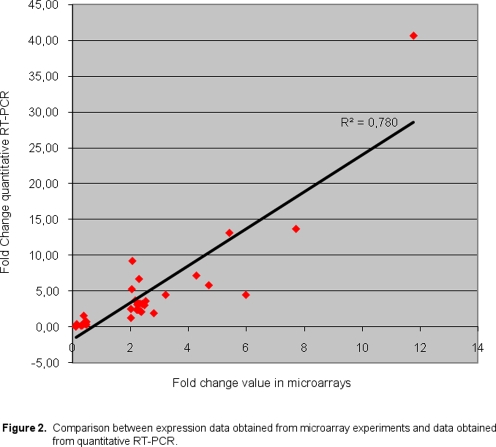
The main correlation between the two assays is showed in Figure 2. Real-time PCR was not only used to confirm the abnormal gene expression profiles detected by microarray analysis, but also to better define fold-change variations using a more sensitive approach. This approach showed mean fold changes in expression levels directionally similar to those determined by microarray analysis. Overall, fold change was lower when the microarray approach was used compared to real-time PCR (Table 2).
Figure 2. Comparison between expression data obtained from microarray experiments and data obtained from quantitative RT-PCR.
Discussion
The main correlation between the two assays, microarrays and quantitative RT-PCR, was high for all genes, indicating a good agreement between both assays for identification of deregulated genes.
On the other hand, by means of the additional supporting process, a high correlation degree among results was shown and it provided more reliability to the final list of genes. Therefore, results of the two machine learning approaches support the degree of relevance of the 74 genes identified.
Calpain 3 was not abnormally regulated in the microarray study (data not shown). While protein analysis usually shows an absence of protein in patients, the microarray data did not reveal a reduction of calpain 3 mRNA indicating that this primary genetic defect cannot be identified by expression profiling. It is worthwhile mentioning that the presence of different missense mutations in most of the patients may explain this observation as well as the variability found in the Western Blot including normal patterns [36].
In LGMD2A muscles, genes associated with ECM/membrane-related, cell adhesion genes, muscle development genes, signaling pathway genes, and ubiquitin cycle genes were upregulated (Table 2).
Extracellular matrix
The general trend for structural genes to be expressed at higher levels in patients could reflect a general upregulation of structural genes in the mutant muscle, as previously reported for other types of muscular dystrophies [37].
It is interesting to note that a large proportion of genes associated to the extracellular matrix were probably upregulated as a result of fibrotic infiltration. These genes include extracellular matrix proteins such as collagen types I and III (the two major collagens in the ECM), cell adhesion proteins such as CD9, CD44, and fibronectin.
SPARC, overexpressed in the fibroblasts of skin biopsy specimens obtained from patients with systemic sclerosis [38], could be the factor involved in the interstitial fibrosis seen in muscles of LGMD2A patients. It is a matricellular glycoprotein that may modulate cell interaction within the ECM by binding to both ECM structural components and growth factors.
Muscle development
Our results showed that MYH3 (myosin, heavy chain 3, skeletal muscle, embryonic) is highly upregulated in samples from LGMD2A patients. Expression of embryonic myosin heavy chain is a hallmark of muscle regeneration after birth and a characteristic marker of human muscular dystrophies. During normal human development, expression is restricted to the embryonic period of development [39]. This could indicate a failed muscular regeneration attempt to compensate a downstream injury.
Another upregulated gene is myosin light chain 1 slow A (MLC1SA), a transcriptional regulator promoting muscle cell proliferation expressed in both slow-twitch skeletal muscle and non-muscle tissue. This gene showed a high individual correlation with class phenotype (0.442), and was one of the seven genes included in the CFS output. This fact flawlessly demonstrates its importance not only from an individual point of view, but also because of its potential interactions. MLC1SA is one of the two phosphorylable regulatory light chains forming the myosin complex. Cohen et al [40] found that MLC1SB (Accession N° P09542 in mice) was a substrate for calpain 3. To date, no contractile proteins have been identified as in vivo substrates for CAPN3. Identification of MLC1 as a potential substrate for CAPN3 was of interest because, in a previous study, Kramerova et al [15] demonstrated that CAPN3 regulates sarcomere remodeling by acting upstream of the ubiquitin–proteasome system.
LGMD2A patients showed upregulation of the IGF-1 gene as previously observed in other muscular dystrophies. Normal skeletal muscle is able to efficiently repair itself in response to injury. IGF-1 has been implicated as a factor that may affect many steps in gene expression control, including cell proliferation, differentiation, and degradation processes. IGF-1 is a peptide that has been shown to have anabolic effects on muscle cells. This action can be explained based on the molecular signaling events initiated by its receptor, a tyrosine kinase activated on IGF-1 binding, and transmitted through a cascade of intracellular events, leading to a general increase in protein synthesis [41], [42].
Integrin β1 binding protein (ITGB1BP2 = melusin), also upregulated in LGMD2A muscles, is present in a costamere-like pattern consisting of two rows flanking a-actinin at Z line. Melusin expression is upregulated during in vitro differentiation of the C2C12 murine myogenic cell line, and is regulated during in vivo skeletal muscle development [43]. Upregulation of the melusin gene may alter a process that is tightly controlled in muscle development, leading to inadequate muscle differentiation and maturation. The generalized inhibition of terminal stages of myogenic differentiation in C3KO myotubes affects at least two events: sarcomere formation and integrin isoform replacement [44]. During myogenesis, two isoforms of β1 integrin are expressed: β1A is expressed in myoblasts and is downregulated during myogenesis, while β1D appears after fusion and eventually displaces β1A in mature myotubes [45]. Neither β1A nor β1D were cleaved by CAPN3, suggesting that changes in the level of integrin isoforms are not a direct result of calpain 3 absence [44].
Ubiquitin cycle and protein degradation
It is still unclear whether CAPN3 directly cleaves proteins to make them available for ubiquitination or whether the effect of CAPN3 is indirect (i.e. through regulation of other proteins involved in ubiquitination) [15]. In LGMD2A muscle samples, the HERC1 and ANAPC1 genes involved in the ubiquitin cycle are upregulated, suggesting that their regulation may be under the control of calpain 3.
Moreover, Ono et al [46] found proteolysis of proteasome regulatory subunit RPS6A by calpain 3, which may indicate that the ubiquitin-proteasome system is subject to regulation by calpain.
As ubiquitination tags proteins for degradation, decreased ubiquitination may lead to excessive accumulation of the proteins that should otherwise be degraded. This in turn could trigger a cell stress response, one manifestation of which is upregulation of heat shock proteins [15]. According to the reported data, the DnaJ (Hsp40) homolog, subfamily A member 4 (DNAJA4), that showed upregulation, may regulate the chaperone function of Hsp70 proteins [47].
Signaling pathways
The protein coded by HERC1, upregulated in LGMD2A patients, has a C-terminal HECT (homologous to E6-AP C-terminus) domain, which suggests that it has an ubiquitin ligase activity.
β-catenin plays a critical role in many cellular and morphogenic processes by performing two distinct functions: in the nucleus, it acts as a mandatory coactivator of TCF/LEF transcription factors in response to Wnt signaling during both embryonic development and adult muscle regeneration, while at the cell membrane, β-catenin associates with the cadherin complex that links adhesion molecules to the cytoskeleton. In both cases, the concentration of β-catenin has been shown to be tightly regulated through ubiquitin-mediated degradation [44].
Two distinct ubiquitin ligase complexes control β-catenin levels in cytoplasm and at the membrane [48]. Ubiquitination and degradation of the cytosolic pool of β-catenin are under the control of Wnt signaling. Degradation of the membrane pool of β-catenin in skeletal muscle is mediated by the Ozz-E3 ubiquitin ligase complex [49]. Thus, it may be suggested that membrane β-catenin is indirectly regulated by CAPN3. It should also be noted that Trim32, found mutated in limb-girdle muscular dystrophy type 2H, is another putative E3-ubiquitin-ligase [50].
On the other hand, frizzled-related protein (FRZB) is upregulated in LGMD2A muscle samples. It could therefore be hypothesized that β-catenin regulation is also altered at the Wnt signaling pathway, leading to an abnormal myotube fusion or incorrect myogenesis.
Deregulation of mitochondrial genes
In our results, the mitochondrial genes found to be deregulated were ALDH2 and PPM2C. ALDH2 was downregulated in patient samples and is implicated in the glycolysis/gluconeogenesis pathway. On the contrary, expression of the PPM2C mitochondrial gene was upregulated in our study. Protein phosphatase 1J (PPM1J_mouse, PP2C family) was found to be an in vivo substrate for calpain 3 [40].
In later stages of the disease, the muscle pathology is characterized mainly by the presence of lobulated fibers (LF), which are composed of misaligned myofibrils that form a lobular pattern, in addition to fiber size variation and interstitial fibrosis. Lobulated muscle fibers reflect an abnormal spatial distribution of the intermyofibrillar mitochondria network [51]. In C3KO mice, abnormal A-bands were seen, suggesting a role for calpain 3 in correct formation of sarcomeres or maintenance of sarcomere alignment [14].
mRNA expression profiles were specifically altered in LGMD2A muscles with lobulated fibers Keira et al [52]. Genes encoding for extracellular matrix (ECM)/membrane-related, cytoskeletal, or sarcomeric proteins were also upregulated in LF muscles.
According to these results, identification of these mitochondrial proteins suggests that CAPN3 may be involved in mitochondrial protein turnover.
Common genes with altered expression in different muscular dystrophies
According to Table 2, LGMD2A can be characterized as an active fibrotic disease with suppressed muscle regeneration, since LGMD2A cases share upregulation of the extracellular matrix (ECM) components with congenital muscular dystrophy cases and share downregulation of the transcription factors with Duchenne muscular dystrophies.
In muscles from patients with Duchenne muscular dystrophy, upregulated genes were mostly those related to immune response, sarcomeric, ECM, and cell growth, whereas downregulated genes were associated to energy metabolism, transcription/translation, signaling, and proteasomes [32].
c-fos and c-jun proteins have been described as showing strong cytoplasmic expression related to the degeneration process occurring in Duchenne and Becker muscular dystrophies [53]. However our results contradict the previously published results and they showed a strong downregulation of c-fos and c-jun in our samples.
Recently Gan et al [54] reported that Dishevelled (Dvl) and c-Jun form a complex with β-catenin-T-cell factor 4 (TCF-4) on the promoter of Wnt target genes and regulate gene transcription. c-Jun mediates Dvl association with the functional TCF-β-catenin complex and functions as a key component of Wnt signaling in vivo. Since genes coding for proteins in this pathway are dysregulated in LGMD2A patients, it may be suggested that the downregulation of c-jun and other transcription factors observed in LGMD2A patients are regulated in an indirect way by calpain 3.
TFRC (transferrin receptor) and VLDLR (very low density lipoprotein receptor) are upregulated in LGMD2A patients as occurred in FSHD muscles [33]. Transferrin is a key myoblast trophic factor, initially promoting myoblast proliferation and subsequently supporting myogenic differentiation. However, TXNIP (thioredoxin-interacting protein) is downregulated in LGMD2A muscles and was also downregulated in FSHD samples [33]. Since TXNIP acts as an oxidative stress mediator, this finding is consistent with the enhanced vulnerability to oxidative stress seen in LGMD2A, as observed in FSHD myoblasts [33]. Many of the genes deregulated in facioscapulohumeral muscular dystrophy (FSHD) are involved in myogenesis, cell differentiation, and cell-cycle control.
According to the available information, it could be suggested that FSHD shared the greatest quantity of differentially expressed genes and the deregulation tendencies (up/downregulation) are the same and in a similar range of variation. However it would be difficult to establish any correlation given that in the FSHD, even in patients with the same deletion fragment, high variability of impairment and of muscle affectation grade is observed. Therefore, the data depend enormously on the place and on the moment that biopsy has been taken.
S100A6 (calcyclin) and S100A8 (calgranulin A), dysregulated in LGMD2A muscles, are involved in various intracellular and extracellular regulatory activities [55]. Upregulation of S100A6 expression was seen also in LGMD2B and as in other muscular dystrophies, the structural defect causes a general membrane instability that leads to an altered uptake of calcium ions into the muscle fibers [31]. Since calpain 3 interacts with dysferlin and AHNAK, a role of calpain 3 in membrane homeostasis has been suggested [8], [9]. The increased Ca2+ concentration probably influences expression of various signaling molecules whose transcription is sensitive to calcium concentration.
Dysferlin was not abnormally regulated in LGMD2A patients in the microarray study. The value obtained for the expression of the DYSF gene did not fulfil the established criteria to be considered as differently expressed (data not shown).
In this study mRNA levels are analysed, not protein quantities. Even if the protein is reduced in the Western Blot, this reduction may not be regulated at a transcriptional level, it may happen at a post-translation level. Since calpain 3 was shown to be in complex with dysferlin and it has been demonstrated that AHNAK, a novel component of the dysferlin protein complex, serves as a direct substrate of calpain 3 in cell culture, the lack of one of these proteins may justify the reduction of the other.
As described in previous works [31] while western blotting tests showed a reduction or the absence of dysferlin protein in most LGMD2B patients, the microarray data showed a reduction of dysferlin mRNA for some of the patients analysed. This could be due to the different types of mutations of the gene that affects the translation efficiency of the mRNA or the stability of the protein. Additionally, while protein analysis usually shows an absence of protein in the C57BL/10.SJL-Dysf mice, the microarray data did not reveal a reduction of dysferlin mRNA indicating that this primary genetic defect cannot be identified by expression profiling [37]. Moreover, it has been observed that neither calpain-3 nor caveolin was consistently reduced in dysferlinopathies.
The vast majority of upregulated genes in Fukuyama-type congenital muscular dystrophy (FCMD) and laminin-a2 deficient congenital muscular dystrophy (MDC1A) encode extracellular matrix components, presumably related to fibrotic change. However, mature muscle components were extremely downregulated in congenital muscular dystrophies [34].
Muscle regeneration is also a process that depends on the skeletal muscle basement membrane. Basement membrane is thought to not only maintain cell integrity but also to mediate signal transmission in cell differentiation, growth, attachment, survival, polarity, proliferation, and apoptosis [56], [57]. It is hypothesized that upregulation of ECM genes might arise from signal transduction defects due to basement membrane dysfunction. It is possible that muscle fibers keep high transcription levels of ECM to create basement membrane components [34].
Costameric proteins can interact with many components of both the sarcolemma and cytoskeleton. Different publications support a role for the costamere/Z-disk axis in mechanotransduction, the dynamic process through which mechanical stimuli are sensed by muscle cells and converted into biochemical responses [57].
Based on our results and since collagens, melusin and fibronectin, were deregulated, we may hypothesize that upregulation of ECM genes found in LGMD2A patients may result from signal transduction defects due to basement membrane dysfunction. Calpain 3 recognizes a wide range of substrates, including cytoskeletal proteins and myofibrillar proteins [40], [46]. These cytoskeletal proteins and matrix proteins contribute to cell shape, mechanical resistance, and morphological integrity of muscle cells, and are part of a complex network of filaments and tubules that transmit mechanical and chemical stimuli between cells. The cytoskeleton is not only involved in cell stability and integrity, but also plays a significant role in signal transmission from the cell membrane to the nucleus.
LGMD2A and eosinophilic infiltrations
Presence of eosinophilic cells has recently been detected in muscle tissue from patients with mutations in the CAPN3 gene [16].
In our study, IL-32 was upregulated in LGMD2A patients with eosinophilic infiltrates (Table 3). Although IL-32 does not share sequence homology with known cytokine families, IL-32 induces various cytokines, human TNFα, and IL-8 in THP-1 monocytic cells. IL-32 activates typical cytokine signal pathways of nuclear factor-kappa B (NF-κB) and p38 mitogen-activated protein kinase [58]. The neutrophil-derived proteinase 3 (PR3) was identified as a putative IL-32 receptor [59], supporting the possibility that IL-32 upregulation in muscle may be chemoattractant for eosinophilic cells.
Eosinophilia has also been reported as a prominent feature of the necrotic phase in dystrophin-deficient mdx mice. This study suggested that eosinophilia was promoted by at least perforin-dependent cytotoxicity of effector T cells and T-cell production of interleukin-5 (IL-5) [60]. However these authors concluded that some eosinophilia of mdx muscle is independent from perforin-mediated processes and that it may be suggested that a similar mechanism of calpain 3 could act in this process.
Inflammatory features may be seen in some muscular dystrophies, such as facioscapulohumeral muscular dystrophy [17] and dysferlinopathies [19]. Moreover, a Becker muscular dystrophy presenting eosinophilic inflammatory myopathy was described by Weinstock et al [18].
Although the comparison between gene expression profiles between LGMD2A with and without eosinophilia would be interesting, it was not possible to perform. The methods used needed a higher quantity of samples to obtain significant results. Moreover, when a PCA plot was performed for LGMD2A patients only (including cases with and without eosinophilia), no different groups were created and this may be due to the low sample number too.
It seems that the comparison between asymptomatic patients with eosinophilia with control samples is more indicated to shed some light onto the initial mechanism that triggers the eosinophilic cell attraction to muscle. The comparison between asymptomatic patients and controls allows a clearer view due to a lower interfering expression variation.
In a first approach asymptomatic cases were considered as affected and were included into the patient group in the general analysis. These cases were included in the affected group due to their abnormal muscle biopsy pattern. Additionally reinforcing this decision, the PCA plots clustered together the LGMD2A case with or without eosinophilia.
Finally in an additional analysis, however, it was decided to consider them as a different group compared to the controls in order to obtain information about eosinofilic attraction in the early stage of the disease.
Conclusions
In conclusion, upregulated genes were mostly those related to extracellular matrix, cell adhesion, sarcomeric proteins, and signal transduction. It is therefore suggested that different proteins located at or participating in the costameric region are involved in processes regulated by calpain 3 during skeletal muscle development. Upregulation of these proteins may indicate a compensatory attempt of the muscle, and since most of these genes are also upregulated in other dystrophic processes, upregulation might be relatively nonspecific.
It was also found that genes participating in the ubiquitin proteasome degradation pathway are deregulated in LGMD2A patients, which suggests that regulation of this pathway may be under the control of calpain 3 activity.
Finally, the upregulation of IL-32 and immunoglobulin genes may cause the eosinophil chemoattraction observed in the inflammatory findings in presymptomatic stages. This upregulation seems to disappear when the disease progresses. However, they might be quite specific markers for the disease.
Though samples taken from different muscles could add variability to the results of the expression array analysis, correlation of the results with the quantitative RT-PCR results gave strength to the findings. Gene expression profiling is presented as a useful approach to mine new data and hopefully open new perspectives for muscular disorders, shedding some light on identification of novel therapeutic targets for limb-girdle muscular dystrophies.
Looking ahead, each of these methods should be individually analyzed in the animal model and in cell models.
This analysis gives a total of 24 genes that may be considered as potential diagnostic or evolutionary biomarkers of the disease. However, this question will not be solved until the predictive value of these markers is proved in a series of patients with different evolutive status and secondly until the consistency of the results in different muscles and different laboratories is proved.
Acknowledgments
We would like to thank Nathalie Deburgrave and Caroline Beugnet for their technical assistance, Harry Zuzan for discussions on data analysis and Daisy E. Hilton (Celer Soluciones) for manuscript editing.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Health Research Fund of the Spanish Ministry of Health (FIS CP06/00099); Gipuzkoako Foru Aldundia (Berrikuntzarako eta Jakintzaren Gizarterako Departamentua, 76/06); European Union (FEDER Funds and CIBERNED); Instituto de Salud Carlos III. AS: Spanish Ministry of Health (FIS) and Basque Foundation for Health Innovation and Research (BIOEF) promoted by the Basque Government Department of Health. RA: Basque Government (AE-BFI-05/430).
References
- 1.Sorimachi H, Imajoh-Ohmi S, Emori Y, Kawasaki H, Ohno S, et al. Molecular cloning of a novel mammalian calcium-dependent protease distinct from both m- and mu-types. Specific expression of the mRNA in skeletal muscle. J Biol Chem. 1989;264:20106–20111. [PubMed] [Google Scholar]
- 2.Ono Y, Kakinuma K, Torii F, Irie A, Nakagawa K, et al. Possible regulation of the conventional calpain system by skeletal muscle-specific calpain, p94/calpain 3. J Biol Chem. 2004;279:2761–2771. doi: 10.1074/jbc.M308789200. [DOI] [PubMed] [Google Scholar]
- 3.Kinbara K, Sorimachi H, Ishiura S, Suzuki K. Skeletal muscle-specific calpain, p94: structure and physiological function. Biochem Pharmacol. 1998;56:415–420. doi: 10.1016/s0006-2952(98)00095-1. [DOI] [PubMed] [Google Scholar]
- 4.García Díaz BE, Gauthier S, Davies PL. Ca2+ dependency of calpain 3 (p94) activation. Biochemistry. 2006;45:3714–3722. doi: 10.1021/bi051917j. [DOI] [PubMed] [Google Scholar]
- 5.Sorimachi H, Kinbara K, Kimura S, Takahashi M, Ishiura S, et al. Muscle-specific calpain, p94, responsible for limb girdle muscular dystrophy type 2A, associates with connectin through IS2, a p94-specific sequence. J Biol Chem. 1995;270:31158–31162. doi: 10.1074/jbc.270.52.31158. [DOI] [PubMed] [Google Scholar]
- 6.Ono Y, Torii F, Ojima K, Doi N, Yoshioka K, et al. Suppressed disassembly of autolyzing p94/CAPN3 by N2A connectin/titin in a genetic reporter system. J Biol Chem. 2006;281:18519–18531. doi: 10.1074/jbc.M601029200. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi C, Ono Y, Doi N, Kitamura F, Tagami M, et al. Multiple molecular interactions implicate connectin/titin N2A region as a modulating scaffold for p94/calpain 3 activity in skeletal muscle. J Biol Chem. 2008;283:14801–14814. doi: 10.1074/jbc.M708262200. [DOI] [PubMed] [Google Scholar]
- 8.Huang Y, Verheesen P, Roussis A, Frankhuizen W, Ginjaar I, et al. Protein studies in dysferlinopathy patients using llama-derived antibody fragments selected by phage display. Eur J Hum Genet. 2005;13:721–730. doi: 10.1038/sj.ejhg.5201414. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, de Morrée A, van Remoortere A, Bushby K, Frants RR, et al. Calpain 3 is a modulator of the dysferlin protein complex in skeletal muscle. Hum Mol Genet. 2008;17:1855–1866. doi: 10.1093/hmg/ddn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guyon JR, Kudryashova E, Potts A, Dalkilic I, Brosius MA, et al. Calpain 3 cleaves filamin C and regulates its ability to interact with gamma- and delta-sarcoglycans. Muscle Nerve. 2003;28:472–483. doi: 10.1002/mus.10465. [DOI] [PubMed] [Google Scholar]
- 11.Baghdiguian S, Martin M, Richard I, Pons F, Astier C, et al. Calpain 3 deficiency is associated with myonuclear apoptosis and profound perturbation of the IkappaB alpha/NF-kappaB pathway in limb-girdle muscular dystrophy type 2A. Nat Med. 1999;5:503–511. doi: 10.1038/8385. [DOI] [PubMed] [Google Scholar]
- 12.Baghdiguian S, Richard I, Martin M, Coopman P, Beckmann JS, et al. Pathophysiology of limb girdle muscular dystrophy type 2A: hypothesis and new insights into the IkappaBalpha/NF-kappaB survival pathway in skeletal muscle. J Mol Med. 2001;79:254–261. doi: 10.1007/s001090100225. [DOI] [PubMed] [Google Scholar]
- 13.Benayoun B, Baghdiguian S, Lajmanovich A, Bartoli M, Daniele N, et al. NF-kappaB-dependent expression of the antiapoptotic factor c-FLIP is regulated by calpain 3, the protein involved in limb-girdle muscular dystrophy type 2A. FASEB J. 2008;22:1521–1529. doi: 10.1096/fj.07-8701com. [DOI] [PubMed] [Google Scholar]
- 14.Kramerova I, Kudryashova E, Tidball JG, Spencer MJ. Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum Mol Genet. 2004;13:1373–1388. doi: 10.1093/hmg/ddh153. [DOI] [PubMed] [Google Scholar]
- 15.Kramerova I, Kudryashova E, Venkatraman G, Spencer MJ. Calpain 3 participates in sarcomere remodeling by acting upstream of the ubiquitin-proteasome pathway. Hum Mol Genet. 2005;14:2125–2134. doi: 10.1093/hmg/ddi217. [DOI] [PubMed] [Google Scholar]
- 16.Krahn M, Lopez de Munain A, Streichenberger N, Bernard R, Pécheux C, et al. CAPN3 mutations in patients with idiopathic eosinophilic myositis. Ann Neurol. 2006;59:905–911. doi: 10.1002/ana.20833. [DOI] [PubMed] [Google Scholar]
- 17.Arahata K, Ishihara T, Fukunaga H, Orimo S, Lee JH, et al. Inflammatory response in facioscapulohumeral muscular dystrophy (FSHD): immunocytochemical and genetic analyses. Muscle Nerve. 1995;2:S56–66. [PubMed] [Google Scholar]
- 18.Weinstock A, Green C, Cohen BH, Prayson RA. Becker muscular dystrophy presenting as eosinophilic inflammatory myopathy in an infant. J Child Neurol. 1997;12:146–147. doi: 10.1177/088307389701200214. [DOI] [PubMed] [Google Scholar]
- 19.Gallardo E, Rojas-García R, de Luna N, Pou A, Brown RH, Jr, et al. Inflammation in dysferlin myopathy: immunohistochemical characterization of 13 patients. Neurology. 2001;57:2136–2138. doi: 10.1212/wnl.57.11.2136. [DOI] [PubMed] [Google Scholar]
- 20.Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S. Springer; 2005. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolstad BM, Irizarry R A, Astrand M, Speed TP. A Comparison of Normalization Methods for High Density Oligonucleotide Array Data Based on Bias and Variance. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 24.Haslett JN, Sanoudou D, Kho AT, Han M, Bennett RR, et al. Gene expression profiling of Duchenne muscular dystrophy skeletal muscle. Neurogenetics. 2003;4:163–171. doi: 10.1007/s10048-003-0148-x. [DOI] [PubMed] [Google Scholar]
- 25.Bakay M, Chen YW, Borup R, Zhao P, Nagaraju K, et al. Sources of variability and effect of experimental approach on expression profiling data interpretation. BMC Bioinformatics. 2002;3:4. doi: 10.1186/1471-2105-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duda RO, Hart PE, Stork DG. Pattern classification. New York: Wiley-Interscience; 2000. [Google Scholar]
- 27.Kang PB, Kho AT, Sanoudou D, Haslett JN, Dow CP, et al. Variations in gene expression among different types of human skeletal muscle. Muscle Nerve. 2005;32:483–491. doi: 10.1002/mus.20356. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Tetko IV, Hall MA, Frank E, Facius A, et al. Gene selection from microarray data for cancer classification – a machine learning approach. Comput Biol Chem. 2005;29:37–46. doi: 10.1016/j.compbiolchem.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Hall MA, Smith LA. Feature Subset Selection: A Correlation Based Filter Approach. 1997. Proceedings of the Fourth International Conference on Neural Information Processing and Intelligent Information Systems, 855–858.
- 30.Chen YW, Zhao P, Borup R, Hoffman EP. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J Cell Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campanaro S, Romualdi C, Fanin M, Celegato B, Pacchioni B, et al. Gene expression profiling in dysferlinopathies using a dedicated muscle microarray. Hum Mol Genet. 2002;11:3283–3298. doi: 10.1093/hmg/11.26.3283. [DOI] [PubMed] [Google Scholar]
- 32.Noguchi S, Tsukahara T, Fujita M, Kurokawa R, Tachikawa M, et al. cDNA microarray analysis of individual Duchenne muscular dystrophy patients. Hum Mol Genet. 2003;12:595–600. [PubMed] [Google Scholar]
- 33.Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, et al. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 34.Taniguchi M, Kurahashi H, Noguchi S, Sese J, Okinaga T, et al. Expression profiling of muscles from Fukuyama-type congenital muscular dystrophy and laminin-alpha 2 deficient congenital muscular dystrophy; is congenital muscular dystrophy a primary fibrotic disease? Biochem Biophys Res Commun. 2006;342:489–502. doi: 10.1016/j.bbrc.2005.12.224. [DOI] [PubMed] [Google Scholar]
- 35.Osborne RJ, Welle S, Venance SL, Thornton CA, Tawil R. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology. 2007;68:569–577. doi: 10.1212/01.wnl.0000251269.31442.d9. [DOI] [PubMed] [Google Scholar]
- 36.Sáenz A, Leturcq F, Cobo AM, Poza JJ, Ferrer X, et al. LGMD2A: genotype-phenotype correlations based on a large mutational survey on the calpain 3 gene. Brain. 2005;128:732–742. doi: 10.1093/brain/awh408. [DOI] [PubMed] [Google Scholar]
- 37.von der Hagen M, Laval SH, Cree LM, Haldane F, Pocock M, et al. The differential gene expression profiles of proximal and distal muscle groups are altered in pre-pathological dysferlin-deficient mice. Neuromuscul Disord. 2005;15:863–877. doi: 10.1016/j.nmd.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Zhou X, Tan FK, Guo X, Arnett FC. Attenuation of collagen production with small interfering RNA of SPARC in cultured fibroblasts from the skin of patients with scleroderma. Arthritis Rheum. 2006;54:2626–2631. doi: 10.1002/art.21973. [DOI] [PubMed] [Google Scholar]
- 39.Karsch-Mizrachi I, Travis M, Blau H, Leinwand AL. Expression and DNA sequence analysis of a human embryonic skeletal muscle myosin heavy chain gene. Nucleic Acids Res. 1989;17:6167–6179. doi: 10.1093/nar/17.15.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen N, Kudryashova E, Kramerova I, Anderson LV, Beckmann JS, et al. Identification of putative in vivo substrates of calpain 3 by comparative proteomics of overexpressing transgenic and nontransgenic mice. Proteomics. 2006;6:6075–6084. doi: 10.1002/pmic.200600199. [DOI] [PubMed] [Google Scholar]
- 41.Marotta M, Sarria Y, Ruiz-Roig C, Munell F, Roig-Quilis M. Laser microdissection-based expression analysis of key genes involved in muscle regeneration in mdx mice. Neuromuscul Disord. 2007;17:707–718. doi: 10.1016/j.nmd.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Baldwin KM, Haddad F. Skeletal muscle plasticity: cellular and molecular responses to altered physical activity paradigms. Am J Phys Med Rehabil. 2002;81:S40–51. doi: 10.1097/01.PHM.0000029723.36419.0D. [DOI] [PubMed] [Google Scholar]
- 43.Brancaccio M, Guazzone S, Menini N, Sibona E, Hirsch E, et al. Melusin is a new muscle-specific interactor for beta(1) integrin cytoplasmic domain. J Biol Chem. 1999;274:29282–29288. doi: 10.1074/jbc.274.41.29282. [DOI] [PubMed] [Google Scholar]
- 44.Kramerova I, Kudryashova E, Wu B, Spencer MJ. Regulation of the M-cadherin-beta-catenin complex by calpain 3 during terminal stages of myogenic differentiation. Mol Cell Biol. 2006;26:8437–8447. doi: 10.1128/MCB.01296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belkin AM, Zhidkova NI, Balzac F, Altruda F, Tomatis D, et al. Beta 1D integrin displaces the beta 1A isoform in striated muscles: localization at junctional structures and signaling potential in nonmuscle cells. J Cell Biol. 1996;132:211–226. doi: 10.1083/jcb.132.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ono Y, Hayashi C, Doi N, Kitamura F, Shindo M, et al. Comprehensive survey of p94/calpain 3 substrates by comparative proteomics--possible regulation of protein synthesis by p94. Biotechnol J. 2007;2:565–576. doi: 10.1002/biot.200700018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cyr DM, Langer T, Douglas MG. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 48.Wu G, Liu C, He X. Ozz: a new name on the long list of beta-catenin's nemeses. Mol Cell. 2004;13:451–453. doi: 10.1016/s1097-2765(04)00090-5. [DOI] [PubMed] [Google Scholar]
- 49.Nastasi T, Bongiovanni A, Campos Y, Mann L, Toy JN, et al. Ozz-E3, a muscle-specific ubiquitin ligase, regulates beta-catenin degradation during myogenesis. Dev Cell. 2004;6:269–282. doi: 10.1016/s1534-5807(04)00020-6. [DOI] [PubMed] [Google Scholar]
- 50.Frosk P, Weiler T, Nylen E, Sudha T, Greenberg CR, et al. Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am J Hum Genet. 2002;70:663–672. doi: 10.1086/339083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Figarella-Branger D, El-Dassouki M, Saenz A, Cobo AM, Malzac P, et al. Myopathy with lobulated muscle fibers: evidence for heterogeneous etiology and clinical presentation. Neuromuscul Disord. 2002;12:4–12. doi: 10.1016/s0960-8966(01)00245-0. [DOI] [PubMed] [Google Scholar]
- 52.Keira Y, Noguchi S, Kurokawa R, Fujita M, Minami N, et al. Characterization of lobulated fibers in limb girdle muscular dystrophy type 2A by gene expression profiling. Neurosci Res. 2007;57:513–521. doi: 10.1016/j.neures.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 53.Olivé M, Martinez-Matos JA, Pirretas P, Povedano M, Navarro C, et al. Expression of myogenic regulatory factors (MRFs) in human neuromuscular disorders. Neuropathol Appl Neurobiol. 1997;23:475–482. doi: 10.1111/j.1365-2990.1997.tb01324.x. [DOI] [PubMed] [Google Scholar]
- 54.Gan XQ, Wang JY, Xi Y, Wu ZL, Li YP, et al. Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading to stabilization of beta-catenin-TCF interaction. J Cell Biol. 2008;180:1087–1100. doi: 10.1083/jcb.200710050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 56.Campbell KP, Stull JT. Skeletal muscle basement membrane-sarcolemma-cytoskeleton interaction minireview series. J Biol Chem. 2003;278:12599–12600. doi: 10.1074/jbc.R300005200. [DOI] [PubMed] [Google Scholar]
- 57.Ervasti JM. Costameres: the Achilles' heel of Herculean muscle. J Biol Chem. 2003;278:13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- 58.Kim SH, Han SY, Azam T, Yoon DY, Dinarello CA. Interleukin-32: a cytokine and inducer of TNFalpha. Immunity. 2005;22:131–142. doi: 10.1016/j.immuni.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Dinarello CA, Kim SH. IL-32, a novel cytokine with a possible role in disease. Ann Rheum Dis. 2006;65:iii61–64. doi: 10.1136/ard.2006.058511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai B, Spencer MJ, Nakamura G, Tseng-Ong L, Tidball JG. Eosinophilia of dystrophin-deficient muscle is promoted by perforin-mediated cytotoxicity by T cell effectors. Am J Pathol. 2000;156:1789–1796. doi: 10.1016/S0002-9440(10)65050-X. [DOI] [PMC free article] [PubMed] [Google Scholar]