Abstract
Members of the baculovirus p35 gene family encode proteins that specifically inhibit caspases, cysteine proteases that are involved in apoptosis. To date, p35 homologs have only been found in baculoviruses. We have identified AMVp33, a gene from Amsacta moorei entomopoxvirus with low but significant homology to baculovirus p35 genes. Expression of AMVp33 blocked apoptosis in several different insect and human cell lines. Purified recombinant P33 protein was an efficient inhibitor of insect and human effector caspases, but not initiator caspases. P33 was cleaved by effector caspases, and the resulting cleavage fragments stably associated with the caspases. Mutation of the predicted caspase cleavage site in P33 eliminated cleavage, caspase inhibition, and anti-apoptotic function. Thus, AMVp33 encodes a caspase inhibitor similar to baculovirus P35 with a preference for effector caspases. This is the first report of a p35 homolog from any viral or cellular genome outside of the baculovirus family.
INTRODUCTION
Apoptosis or programmed cell death is a normal physiological process used to remove unwanted cells including damaged and virus-infected cells. Upon receiving an apoptotic signal a group of cysteine proteases called caspases are activated. The first caspases activated are the upstream initiator caspases which then cleave and activate the downstream effector caspases (Riedl and Shi, 2004). The effector caspases then selectively cleave various cellular substrates leading to the eventual dismantling of the cell (Hengartner, 2000). Caspases have a high degree of substrate specificity, and will only cleave proteins that contain caspase recognition sites. Cleavage occurs immediately after an Asp residue found in the recognition site. In addition, the residues immediately surrounding the cleavage site Asp also play an important role in determining whether a particular caspase will cleave a given protein (Riedl and Shi, 2004).
Many viruses have evolved strategies to combat this process and thereby allow for their multiplication. Baculoviruses encode two families of anti-apoptotic genes, iap(inhibitor of apoptosis) and p35, that can inhibit apoptosis induced by a wide range of stimuli, including virus infection (Clem, 2005). One of the most widely studied viral IAPs, Op-IAP from Orgyia pseudotsugata M Nucleopolyhedrovirus is able to inhibit apoptosis in insects and mammals by interacting with pro-apoptotic proteins such as Hid, Reaper, Grim in Drosophila and Smac/Diablo in mammals (Vucic et al., 1997; Vucic et al., 1998; Wilkinson et al., 2004; Wright and Clem, 2002). Unlike some cellular IAP proteins, Op-IAP does not appear to function by directly inhibiting caspases (Wright et al., 2005). P35, which was originally found in the baculovirus Autographa californica M Nucleopolyhedrovirus (AcMNPV), is also able to block apoptosis in both insects and mammals. However, the mode of action of P35 is quite different from that of baculoviral IAPs. P35 acts as a substrate inhibitor of effector caspases by virtue of a solvent-exposed reactive site loop that contains a caspase cleavage site, 84DQMD87. Upon cleavage after Asp87 by an active caspase, a stable complex is formed between P35 and the caspase (Bump et al., 1995; Fisher et al., 1999).
While IAP homologs are found in nearly all baculoviruses examined to date, homologs of P35 exist in only a limited number of other baculoviruses. The most divergent of these is found in Spodoptera littoralis Nucleopolyhedrovirus (SlNPV) and is called P49 (Du et al., 1999). P49 also acts as a substrate inhibitor of caspases, but unlike P35 has been shown to inhibit the activity of Drosophila initiator caspase DRONC (Jabbour et al., 2002; Pei et al., 2002; Zoog et al., 2002).
Amsacta moorei entomopoxvirus (AmEPV) can infect the gypsy moth, Lymantria dispar and has been recently sequenced and shown to contain a single functional iap gene (Li et al., 2005). In addition to the iap gene, we identified a novel homolog of the baculovirus p35 genes, which we have named AMVp33. In this report, we show that P33 is a potent apoptosis inhibitor, able to inhibit apoptosis in both insect and mammalian cells, and functions to regulate apoptosis by acting as a substrate inhibitor of effector caspases.
RESULTS AND DISCUSSION
P33 Structure
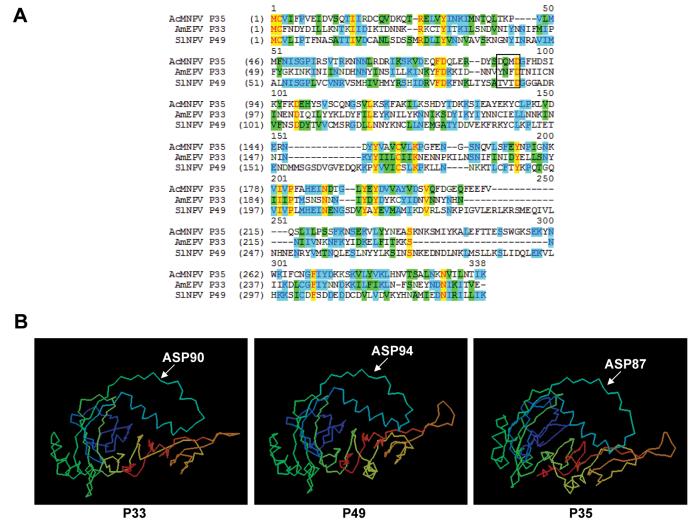
As part of a search for potential anti-apoptotic genes we performed a standard BLAST search using the AcMNPV p35 gene sequence and identified AMV010, an ORF found in AmEPV with low but significant homology to p35 (Fig. 1A). AMV010 encodes a predicted protein of 32.7 KDa which lacks identifiable sequence motifs. To be consistent with the nomenclature of other known p35 homologs, we have renamed this gene AMVp33. At the amino acid level, the identity of the predicted P33 protein to AcMNPV P35 was 25%. To determine if the predicted structure of P33 correlated to its possible function, we performed computer-assisted modeling of P33 based on the known structure of the caspase inhibitor P35. This comparison predicted a similar overall structure to that of both P35 and P49 (Fig. 1B). While computer-based protein structure predictions are of limited value, the most significant aspect of the computer-generated model was the potential for P33 to contain a reactive site loop similar to that found in P35 and P49, with a potential caspase cleavage site, 87YNFD90, in the same position as the cleavage site 84DQMD87 found in AcMNPV P35. Since P35 is a potent caspase inhibitor, we decided to analyze the ability of P33 to inhibit apoptosis, and in particular caspases.
Figure 1.
P33 alignment and predicted structure. (A) Alignment of the predicted amino acid sequences of AmEPV P33, AcMNPV P35, and SlNPV P49. Yellow bars represent identical amino acids found in all 3 sequences, blue bars represent identical amino acids found in 2 of the 3 sequences, and green bars represent similar amino acids. The known caspase cleavage sites in P35 and P49 and the putative cleavage site in P33 are boxed. Sequence alignment was performed using Vector Nti. (B) Predicted structures of AmEPV P33, SlNPV P49, and AcMNPV P35. Computer assisted modeling of P33 and P49 was performed based on the structure of P35 using the 3D-PSSM Web server. Arrows indicate the positions of caspase cleavage sites.
P33 inhibits apoptosis induced by diverse stimuli
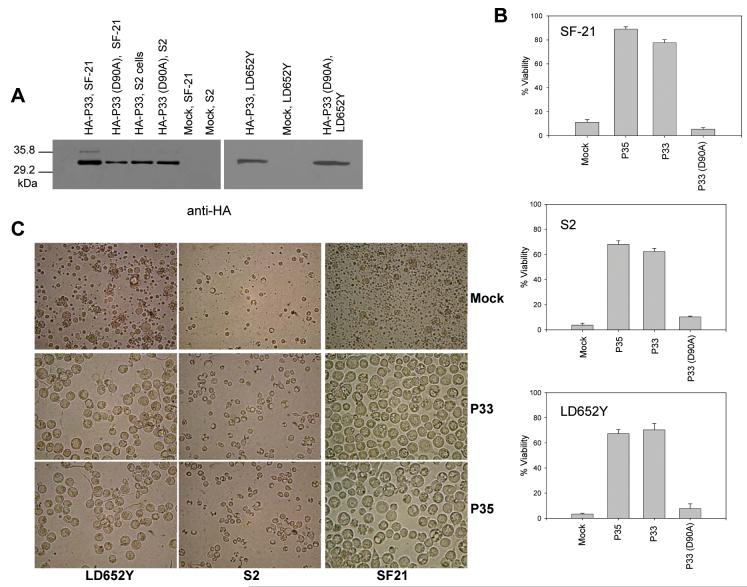
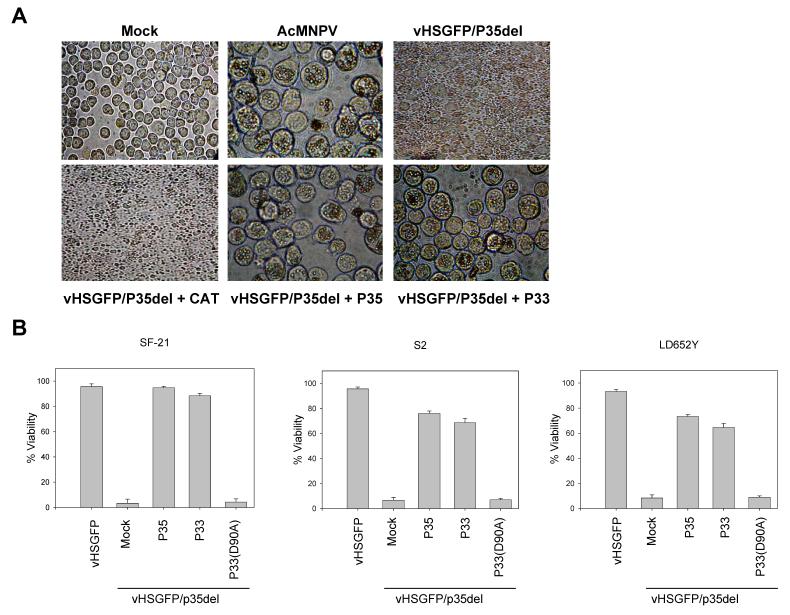
To test the ability of p33 to block caspase-dependent apoptosis, we transiently expressed N-terminally HA-tagged P33 in LD652Y, S2 and SF-21 cells and induced the cells to undergo apoptosis with UV radiation. Immunoblot analysis was performed to confirm expression of P33 in each cell type (Fig. 2A). P33 expression inhibited UV-induced apoptosis in all three cell lines, to a degree comparable to that of P35 (Fig. 2B-C). P33 also blocked apoptosis induced by vHSGFP/P35del, a mutant of AcMNPV that lacks p35 (Fig.3A-B). P33 prevented apoptosis and allowed for virus replication as indicated by the formation of polyhedra (Fig. 3A).
Figure 2.
P33 blocks apoptosis induced by UV irradiation in insect cell lines. (A) Anti-HA immunoblot of lysates from LD652Y, S2, and SF-21 cells collected 24 h after transfection with plasmids expressing HA-tagged P33 or point mutant P33(D90A). Lanes labeled “Mock” represent lysates from mock-transfected cells. (B) SF-21, S2, and LD652Y cells were mock transfected or transfected with plasmids expressing P33, P33(D90A) or P35, and 24 h after transfection the cells were UV-irradiated and cell viability was determined 24 h later. Each data point represents the mean of three experiments +/- SE. (C) Photomicrographs of LD652Y, S2, and SF-21 cells taken 24 h after UV treatment. Magnification, × 400.
Figure 3.
P33 blocks AcMNPV-induced apoptosis. (A) SF-21 cells were mock transfected or transfected with plasmids expressing P33, P35 or negative control chloramphenicol acetyl transferase (CAT) and 24 h later the cells were infected with vHSGFP/P35del at an MOI of 10 PFU/cell. At 96 h post infection the cells were photographed. Magnifications, X400 (i, iii, iv) or X1,000 (ii, v, vi). (B) SF-21, S2, and LD652Y cells were mock transfected or transfected with plasmids expressing P35, P33 or P33(D90A) and 24 h later infected with vHSGFP/P35del at an MOI of 10 PFU/cell. Cell viability was determined 24 h after infection. One well of each was infected with AcMNPV expressing P35 (vHSGFP) and cell viability was determined as above. The data represent the mean of three experiments +/- SE.
To assess the importance of the P33 predicted cleavage site (YNFD90) for its anti-apoptotic function, Asp90 was substituted with Ala. Although expressed at similar levels as wild type P33 (Fig. 2A), the point mutant P33(D90A) failed to block apoptosis induced by either UV irradiation or viral infection (Fig. 2B, 3B). In examining the sequence of P33, two other Asp residues (Asp83 and Asp101) were found in the predicted reactive site loop near Asp90. These Asp residues were mutated to Ala and the resulting point mutants were tested for their ability to block apoptosis. P33(D83A) and P33(D101A) were able to prevent apoptosis induced by UV irradiation or viral infection as efficiently as wild type P33 (data not shown), further illustrating the importance of residue Asp90.
P33 expression suppresses caspase activity in cells
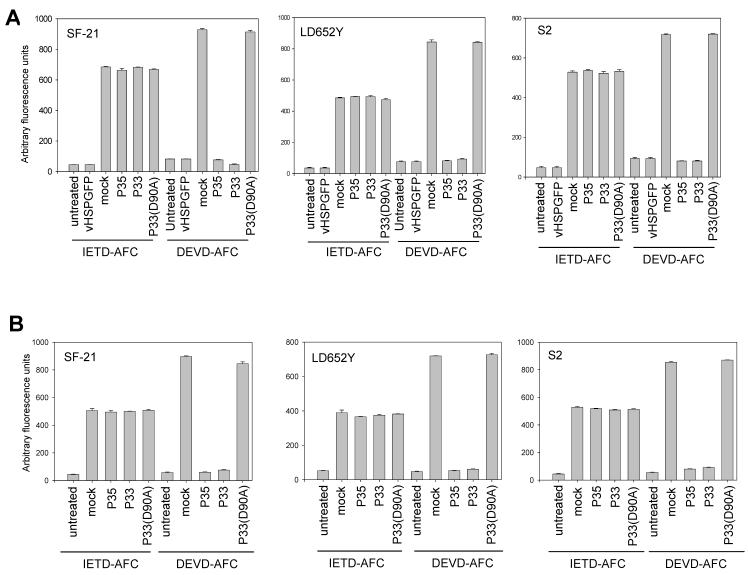
To determine whether inhibition of apoptosis by P33 correlated with reduced caspase activity, whole cell lysates from either mock transfected, P35- or P33-expressing cells were harvested 12 h after UV or viral infection and caspase activity was determined using the fluorogenic caspase substrates Ac-IETD-AFC (to measure initiator caspase activity) or Ac-DEVD-AFC (to measure effector caspase activity). P33-expressing cells showed significantly reduced effector caspase activity as compared to mock transfected cells, comparable to that seen with P35 (Fig. 4). However, initiator caspase (IETD) activity was unaffected by P33 expression, again similar to what was seen in P35-expressing cells. These results suggest that P33, like P35, functions by inhibiting effector caspase activity. The point mutant P33(D90A) was unable to inhibit either type of caspase activity, which correlated with its inability to block apoptosis (Fig. 4).
Figure 4.
P33 expression prevents effector but not initiator caspase activity in cells. (A) SF-21, S2 or LD652Y cells were either left untreated, infected with control virus vHSGFP, or mock transfected or transfected with plasmids encoding P35, P33 or P33(D90A) and 24 h later infected with p35 deletion virus vHSGFP/P35del. Twelve h later caspase activity was determined using Ac-DEVD-AFC or Ac-IETD-AFC as substrate. The lysates used for the DEVD assay were diluted 50-fold before use in the assay to maintain enzyme activity in the linear range. Panel (B) is similar to (A) except the cells were UV irradiated 24 h after transfection instead of being infected with vHSGFP/P35del. Untreated cells were not irradiated or infected. Each data point represents the mean of three experiments +/- SE.
P33 is a direct inhibitor of caspases
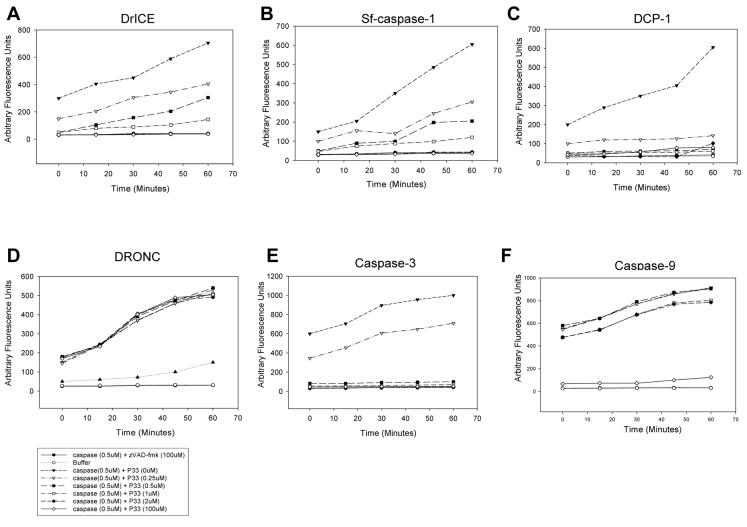
Next, we determined if P33 was a direct inhibitor of caspases by purifying recombinant C-terminally His6-tagged P33 from bacteria and testing its ability to inhibit purified recombinant caspases. We used the fluorogenic caspase substrates Ac-DEVD-AFC (DrICE, DCP-1, Sf-Caspase-1, caspase-3), Ac-IETDAFC (DRONC), or Ac-LEHD-AFC (caspase-9) performed a dose-response assay, with increasing amounts of P33-His6. The results showed that the effector caspases Sf-caspase-1 from Spodoptera frugiperda, human caspase-3, and DrICE and DCP-1 from Drosophila were efficiently inhibited by purified P33-His6 (Fig. 5A-C, E), but the initiator caspases DRONC from Drosophila and human caspase-9 were not efficiently inhibited by P33- His6, with approximately 4-fold excess showing no inhibition. A 200-fold excess of P33-His was able to inhibit DRONC and caspase-9 (Fig. 5D and F), indicating that P33 has a limited ability to inhibit initiator caspases. However, given its lack of inhibition at lower concentrations, P33 is probably not able to inhibit initiator caspases under physiological conditions.
Figure 5.
P33 directly inhibits effector caspase activity in vitro. Recombinant purified (A) DrICE, (B) Sf-caspase-1, (C) DCP-1, (D) DRONC, (E) caspase-3, or (F) caspase-9 were incubated with increasing concentrations of recombinant P33-His6 and caspase activity was determined using 0.2 μM of the indicated substrates. Caspase inhibitor z-VAD-fmk (100 μM) was used as a control.
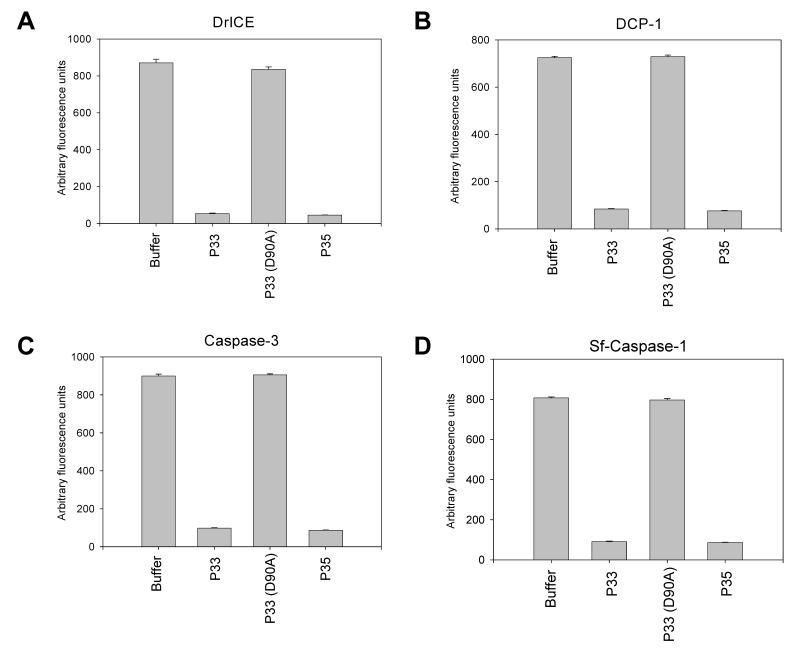
To assess the ability of the P33(D90A) mutant to inhibit caspase activity in vitro, recombinant P33(D90A)-His6 was incubated in increasing amounts with effector caspases. P33(D90A)-His6 was unable to block the caspase activity of DrICE, DCP-1, caspase-3, or Sf-caspase-1 (Fig. 6), indicating that residue Asp90 is important for caspase inhibition.
Figure 6.
P33 (D90A) does not inhibit caspase activity in vitro. P33 (D90A) (100 μM) was tested for its ability to inhibit recombinant effector caspases (A) DrICE, (B) DCP-1, (C) caspase-3, or (D) Sf-caspase-1 (0.5 μM each) using the substrate Ac-DEVD-AFC. P33 and P35 were added at a concentration of 2 μM. The data represent the mean of three experiments +/- SE.
Mechanism of caspase inhibition by P33
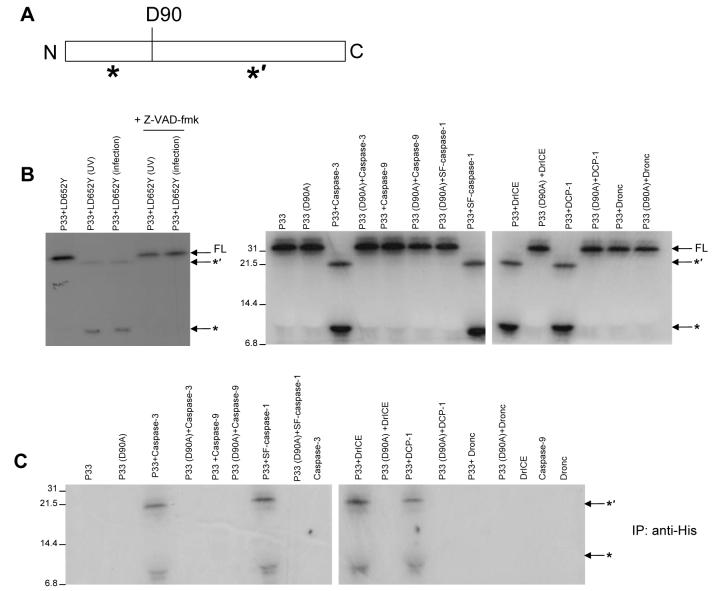
AcMNPV P35 is a substrate inhibitor of caspases. The P35 protein is cleaved at position Asp87 by caspases and then the P35 cleavage products become covalently linked to the caspase by a thioester bond (Xu et al., 2001). To determine whether P33 acts by a similar mechanism, in vitro translated, 35S-labeled P33 was incubated with lysate prepared from LD652Y cells that had been UV irradiated or infected with vHSGFP/P35del. P33 was cleaved by the apoptotic lysates and this cleavage was inhibited by the pan-caspase inhibitor z-VAD-fmk, indicating that caspases were involved in the cleavage (Fig. 7B). To further determine if P33 was a substrate for caspases, recombinant purified DrICE, DCP-1, Sf-caspase-1, caspase-3, caspase-9 or DRONC were incubated with in vitro translated P33. P33 was cleaved by the effector caspases caspase-3, DrICE, DCP-1 and Sf-caspase-1 (Fig. 7C). Cleavage was not observed with DRONC or caspase-9 (Fig. 7C). To determine whether the cleavage fragments associated with the target caspase, we immunoprecipitated the His-tagged caspases using anti-His antibody. 35S-labeled P33 cleavage fragments associated with the effector caspases caspase-3, DrICE, DCP-1, Sf-caspase-1, but not with DRONC or caspase-9 (Fig. 7D). The point mutant P33(D90A) was not cleaved by any of the caspases tested (Fig. 7C) and did not associate with any of the caspases (Fig. 7D), indicating that Asp90 is likely the site of caspase cleavage in P33.
Figure 7.
P33 is cleaved and associates stably with effector caspases. (A) Diagram of P33 showing the putative cleavage site D90 and the predicted 22 kDa (*’) and 11 kDa (*) cleavage fragments. (B) 35S-labeled P33 was incubated with lysates from untreated LD652Y cells, cells infected with vHSGFP/P35del, or cells treated with UV. The reactions were then analyzed by SDS-PAGE and autoradiography. The caspase inhibitor z-VAD-fmk was used to show that cleavage was due to caspase activity. Migration of the cleavage fragments and full length P33 (FL) is indicated. (C) Purified recombinant His6-tagged caspase-3, caspase-9, DRONC, DCP-1, Sf-caspase-1, or DrICE (0.5 μM each) were incubated with 35S-labeled P33 or 35S-labeled P33 (D90A), followed by SDS-PAGE and autoradiography. (D) 35S-labeled P33 or 35S-labeled P33 (D90A) were incubated with recombinant His6-tagged caspases (0.5 μM). The reactions were then immunoprecitated with anti-His antibody, and immunoprecipitated proteins were analyzed by SDS-PAGE and autoradiography.
Comparison of caspase inhibition by P33 and P35
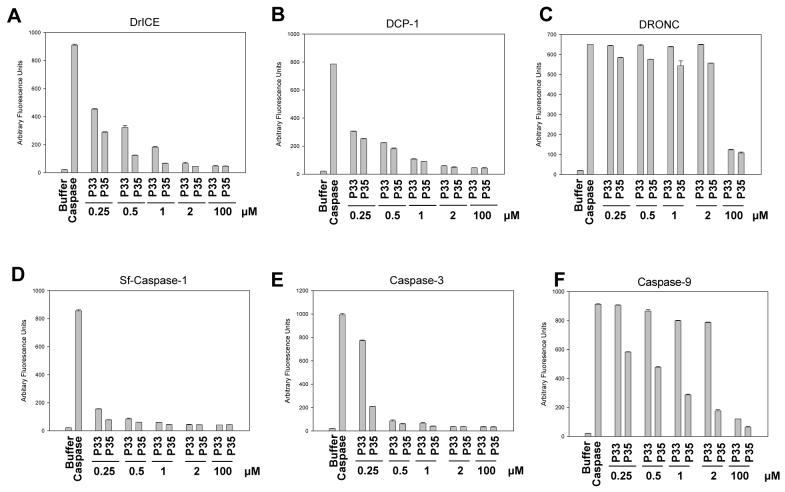
To compare the caspase inhibiting ability of P33 in relation to P35, increasing concentrations of recombinant P33 or P35 were incubated with recombinant effector caspases DrICE, DCP-1, Sf-caspase-1, caspase-3 or initiator caspases DRONC or caspase-9 and caspase activity was determined. Both P33 and P35 inhibited effector caspase activity to a similar extent (Fig. 8A-B, D-E), but were unable to inhibit the initiator caspase DRONC, except at 200-fold excess (Fig. 8C). It has been shown previously that P35 inhibits caspase-9 in vitro, but not in vivo (Ryan et al., 2002). While our results verified inhibition of caspase-9 by P35, we did not observe inhibition of caspase-9 by P33 unless P33 was in high molar excess (Fig. 8F). In addition, P35 was cleaved by caspase-9 in vitro, but P33 was not (data not shown). These results suggest that P33 is more similar to P35 than to P49, since P49 is able to inhibit DRONC in addition to effector caspases, but that P33 differs from P35 in being unable to inhibit human caspase-9 in vitro.
Figure 8.
Comparison of P33 and P35 caspase inhibiting activity. Increasing concentrations of recombinant purified P33-His6 or P35-His6 were incubated with the indicated caspases (0.5 μM) and caspase activity was determined using the indicated substrates. Each data point represents the mean of three experiments +/- SE.
P33 protects mammalian cells against UV-induced apoptosis
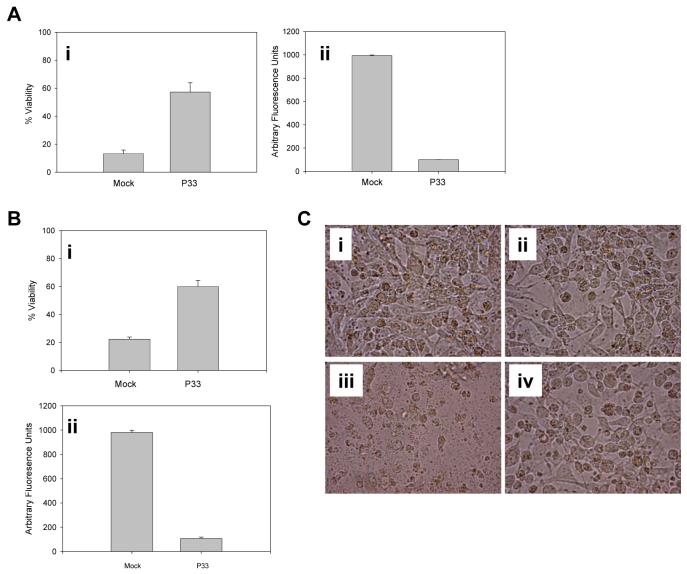
Expression of AcMNPV P35 inhibits apoptosis in a wide variety of organisms, ranging from nematodes to human cells. To determine whether P33 could also protect against apoptosis in a phylogenetically diverse organism, P33 was expressed in the HT-1080 human fibroblastoma cell line and the cells were UV-irradiated to induce apoptosis. P33-expressing cells were protected against apoptosis compared to cells that were mock transfected (Fig. 9Ai, C) and this protection correlated with reduced caspase activity (Fig. 9Aii). P33 was also able to block apoptosis in human embryonic kidney 293 cells induced by UV irradiation (Fig. 9B).
Figure 9.
P33 blocks UV-induced apoptosis in HT-1080 and HEK293 cells. (A) HT-1080 or (B) 293 cells were mock transfected or transfected with plasmids expressing P33 and 24 h after transfection the cells were UV irradiated. Twenty four h later viability (i) or twelve h later caspase activity (ii) was determined using Ac-DEVD-AFC as substrate. The lysates were diluted 50-fold before use in the caspase assay to maintain enzyme activity in the linear range. Each data point represents the mean of three experiments +/-SE. (C) Photomicrographs of HT-1080 cells that were mock transfected (i, iii) or transfected with a plasmid expressing P33 (ii, iv) and either left untreated (i, ii) or UV irradiated (iii, iv). Pictures were taken twenty four h after UV treatment. Magnification X400.
Conclusions
In this study we have identified and characterized AMVp33, the first homolog of p35 genes found outside of the baculoviruses. Computer-assisted modeling of P33 showed an overall predicted structure similar to that of P35, including a potential reactive site loop and caspase cleavage site. Expression of P33 was found to inhibit apoptosis induced by UV radiation and baculovirus infection in cells from diverse organisms, including insects and human. P33 was able to potently inhibit effector caspases from phylogenetically diverse organisms, but had only limited ability to inhibit initiator caspases. Thus P33 appears to be more similar in action to P35 than to P49. Mutation of the potential cleavage site, Asp90, resulted in complete loss of anti-apoptotic activity and caspase inhibition, and also eliminated caspase cleavage of P33, indicating that Asp90 is likely a site for caspase cleavage similar to that seen in P35 and P49. These results indicate that P33 functions like P35 by acting as a substrate inhibitor of effector caspases.
P35 family members are able to block apoptosis in diverse organisms (Hay et al., 1994; Rabizadeh et al., 1993; Sugimoto et al., 1994), and are among the most widely acting anti-apoptotic proteins known. Despite this, P35 homologs have only been identified to date in baculoviruses and entomopoxviruses, both of which infect only insects. Whether or not P35 homologs exist in cellular genomes or in the genomes of viruses that infect higher organisms is still an unanswered question. The reason for this may be that such homologs have evolutionarily diverged to the point where the sequence identity is too low to be recognized by currently available algorithms. However it is interesting to note that iap genes, which are also found in insect viruses including baculoviruses, entomopoxviruses, and iridoviruses, do have readily recognizable homologs in higher organisms, but have not been found in viruses that infect higher organisms (with the exception of African swine fever virus, which has an obligate stage in ticks). This work significantly expands the P35 family of protease inhibitors and may aid in the eventual identification of cellular P35 homologs.
MATERIALS AND METHODS
Cells and viruses
S. frugiperda (SF-21), L. dispar (LD652Y), and Trichoplusia ni (TN-368) cells were maintained in TC-100 medium (Invitrogen) supplemented with 10% tryptose broth and 10% fetal bovine serum (FBS) (Invitrogen). Drosophila S2 cells were maintained in Schneider’s medium (Invitrogen) supplemented with 10% FBS. Human fibrosarcoma HT-1080 cells and human embryonic kidney 293 cells were maintained in Dulbeccco’s Modification of Eagle’s Medium (DMEM) with 10% FBS. The wild-type AcMNPV L1 strain and the p35 deletion virus vHSGFP/P35del (Clarke and Clem, 2002) were propagated and titered by plaque assay using TN-368 cells.
p33 mutagenesis
The p33(D90A) mutant was generated using the QuikChange Site-Directed Mutagenesis kit (Stratagene) with the following primers: forward primer 5′-aatgtatataattttgccacaaatattatatgt-3′ and reverse primer 5′-acatataatatttgtggcaaaattatatacatt-3′, which resulted in mutation of the Asp residue at position 90 to Ala. The plasmid inserts was sequenced to verify the mutation and to ensure the absence of inadvertent mutations.
Plasmid construction
For expression in insect cells the AMVp33 gene was PCR amplified from AmEPV genomic DNA (provided by Teri Shors, University of Wisconsin-Oshkosh) using primers containing ApaI and EcoRI sites to allow for cloning into pHSEpiOpIAPVI+ (Vucic et al., 1997). The following primers were used: forward primer 5′-gggcccgaatgtgttttaatgattatga-3′ and reverse primer 5′-gaattcttattcgactgtaatttttatattg-3′. The PCR product was gel-purified with the Gel Extraction Kit (Qiaex II), cloned into the pCRII vector (Invitrogen), digested with ApaI and EcoRI, and cloned into the pHSEpiOpIAPVI+ vector after the coding sequence of Op-IAP was removed by digesting with ApaI and EcoRI. This allowed for P33 to be expressed with the HA tag in frame at the N-terminus. For expression in mammalian cells, AMVp33 was cloned into the pcDNA3.1/V5His TOPOTM TA vector (Invitrogen) using primers containing BamHI and SacII sites. For recombinant protein production AMVp33 or AcMNPV p35 genes were PCR amplified using primers containing XhoI and NdeI sites to allow for cloning into the pET23b expression vector (Novagen), which resulted in expression in frame with C-terminal His6 tags. Point mutations were constructed using the Quikchange kit (Stratagene). The nucleotide sequences of all constructs were verified before use.
Recombinant protein expression and purification
His-tagged P33, P35, DRONC, DrICE, DCP-1 and Sf-Caspase-1 were expressed in BL21pLysS(DE)3 Eschericia coli (Stratagene). Cultures were grown at room temperature to OD600=0.4, at which time they were induced with 0.1 M IPTG for 1 hour. The bacteria were sonicated in Lysis Buffer A (200 mM Tris-Cl pH 8.0, 0.4 M ammonium sulfate, 10 mM MgCl2, 10% glycerol, and protease inhibitor cocktail (Roche)) and purified using Talon Metal Affinity Resin (Clontech) according to the manufacturer’s instructions. Active human recombinant caspase-3 and -9 were purchased from Chemicon International.
Caspase assays
Purified recombinant caspase (caspase-3-His6, caspase-9-His6, DRONC-His6, DrICE-His6, DCP-1-His6 or Sf-Caspase-1-His6) (0.5μM) was incubated with increasing concentrations of P35-His6 or P33-His6 (0-100μM) in caspase activity buffer A (50 mM Hepes, pH 7.5, 100 mM NaCl, 1mM EDTA, 0.1% CHAPS, 10% sucrose, 5 mM DTT) for 4 h at 30°C. All caspase inhibitors and substrates were purchased from Enzyme Systems Products. Following incubation, caspase substrate was added to the reactions at a concentration of 0.2 μM (Ac-IETD-AFC was used for DRONC, Ac-LEHD-afc was used for caspase-9 and Ac-DEVD-AFC was used for caspase-3, DrICE, Sf-caspase-1 and DCP-1). In control reactions the caspase inhibitor zVAD-fmk was used at a concentration of 100 μM. Caspase activity was determined as an increase in fluorescence detection caused by the enzymatic cleavage of the substrate and the release of AFC (7-amino-(trifluoromethyl)coumarin). The reactions were analyzed fluorometrically (excitation 405nm, emission 535nm) and activity was expressed in relative arbitrary fluorescence units. When necessary, reactions were diluted to maintain enzyme activity in the linear range as indicated.
To determine caspase activity in SF-21, S2, and LD652Y cells, plasmids encoding HA-tagged P33 or P35 were transfected into the cells using a lipid reagent (Means et al., 2003) in TC-100 medium without FBS, and 4 (SF-21, LD652Y) or 5.5 (S2) h later the lipid-DNA mix was replaced with TC-100 medium containing 10% FBS. Twenty-four h after transfection the cells were UV irradiated by placing on a transilluminator for 10 minutes. Twelve h after UV treatment, cells were harvested in caspase activity buffer A and caspase activity determined using Ac-DEVD-afc or Ac-IETD-afc as substrate. For the p35 deletion virus, plasmids expressing P33 or P35 were transfected into SF-21, S2 and LD652Y cells as described and 24 h after transfection cells were infected with p35 deletion virus at an MOI of 10 PFU/cell. Twelve h after virus infection cells were harvested and caspase activity determined as described.
P33 structural modeling
Computer assisted modeling of P33 was based on the structure of AcMNPV P35 using the 3D-PSSM Web server Biomolecular Modeling Laboratory at the Imperial Center Research Fund.
Immunoblot analysis
To examine expression of P33 constructs, LD652Y, S2 or SF-21 cells were transfected as described above and samples were harvested 24 h after transfection and analyzed by immunoblotting using anti-HA.11 mouse monoclonal antibody (Covance Research Products) at a concentration of 1:500, goat anti-mouse IgG horseradish peroxidase-conjugated antibody (1:20,000) and Supersignal chemiluminescent reagent (Pierce).
Viability assays
For UV viability, SF-21, LD652Y, or S2 cells were transfected with plasmids expressing enhanced green fluorescent protein (eGFP) (3 μg) and either P33, P33(D90A), or P35 (3 μg) and 24 h after transfection cells were UV irradiated as described above. Twenty-four h after UV treatment the number of viable eGFP-positive cells remaining was determined by counting three fields of view under 400X magnification and the percent viability was calculated relative to the number of eGFP-positive cells at 1 hour after UV treatment. Cellfectin reagent (Invitrogen) was used for S2 cells and a liposome reagent (Means et al., 2003) was used for SF-21 and LD652Y cells. For viability with the p35 deletion virus, plasmids were introduced into cells as described above. Twenty-four h after transfection cells were infected with vHSGFP/P35del at an MOI of 10 PFU/cell and 24 h after infection viability was determined as above. For viability in HT-1080 and 293 cells, P33-expressing plasmid was co-transfected with a plasmid expressing eGFP into cells using Lipofectamine 2000 (Invitrogen). Twenty-four h after transfection cells were UV irradiated by placing on a transilluminator for 10 minutes and viability was determined twenty-four h later as described above.
Marker rescue assay
Plasmids expressing P33, P35 or CAT were introduced into SF-21 cells and infected with vHSGFP/P35del as described above. Four days after infection, polyhedra formation was examined by microscopy.
Caspase cleavage assay and co-immunoprecipitation
35S-P33 or 35S-P33 (D90A) was synthesized using the TNT T7/SP6 Coupled Reticulocyte Lysate System (Promega) from the pCRII/P33 plasmid. Twenty μl reticulocyte lysate were then incubated with 1μM caspase-3, caspase-9, DRONC, DCP-1, Sf-caspase-1, or DrICE in caspase buffer A for 4 h at 37°C. After incubation, SDS-PAGE was performed followed by autoradiography. To determine if P33 cleavage fragments associated with caspase, the caspase cleavage assay was performed as described, and the reactions were then added to 50 μl of protein G beads (Sigma) that had been preincubated with anti-HIS antibody (His-Probe H-3, Santa Cruz Biotechnology) diluted 1:100 and rocked overnight at 4°C. The beads were washed 3 times with Caspase Buffer A and bound protein was removed from the protein G beads by heating the samples at 100°C in SDS-PAGE sample buffer for 5 minutes. SDS-PAGE and autoradiography was then performed. To examine cleavage of P33 using cell lysate, LD652Y cells were harvested 12 h after UV or p35 deletion virus infection and incubated with 20 μl reticulocyte lysate followed by SDS-PAGE and autoradiography. As a control, caspase inhibitor zVAD-fmk (100 μM) was added 4 h prior to treatment.
ACKNOWLEDGEMENTS
We thank Dr. Teri Shors for providing AmEPV. We also thank Dr. A. Lorena Passarelli for helpful discussion. This work was supported by NIH grants R29 CA78602 from the National Cancer Institute, P20 RR107686 from the National Center for Research Resources (NCRR), P20 RR16475 from the BRIN Program of the NCRR, and by the Kansas Agricultural Experiment Station. This is contribution number 07-15-J from the Kansas Agricultural Experiment Station.
REFERENCES
- Bump NJ, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, Licari P, Mankovich J, Shi L, Greenberg AH, Miller LK, Wong WW. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- Clarke TE, Clem RJ. Lack of involvement of haemocytes in the establishment and spread of infection in Spodoptera frugiperda larvae infected with the baculovirus Autographa californica M nucleopolyhedrovirus by intrahemocoelic injection. J. Gen. Virol. 2002;83:1565–1572. doi: 10.1099/0022-1317-83-7-1565. [DOI] [PubMed] [Google Scholar]
- Clem RJ. The role of apoptosis in defense against baculovirus infection in insects. Curr. Top. Microbiol. Immunol. 2005;289:113–130. doi: 10.1007/3-540-27320-4_5. [DOI] [PubMed] [Google Scholar]
- Du Q, Lehavi D, Faktor O, Qi Y, Chejanovsky N. Isolation of an apoptosis suppressor gene of the Spodoptera littoralis nucleopolyhedrovirus. J. Virol. 1999;73:1278–1285. doi: 10.1128/jvi.73.2.1278-1285.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AJ, dela Cruz W, Zoog SJ, Schneider CL, Friesen PD. Crystal structure of baculovirus P35: role of a novel reactive site loop in apoptotic caspase inhibition. EMBO J. 1999;18:2031–2039. doi: 10.1093/emboj/18.8.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- Jabbour AM, Ekert PG, Coulson EJ, Knight MJ, Ashley DM, Hawkins CJ. The p35 relative, p49, inhibits mammalian and Drosophila caspases including DRONC and protects against apoptosis. Cell Death Differ. 2002;9:1311–1320. doi: 10.1038/sj.cdd.4401135. [DOI] [PubMed] [Google Scholar]
- Li Q, Liston P, Moyer RW. Functional analysis of the inhibitor of apoptosis (iap) gene carried by the entomopoxvirus of Amsacta moorei. J. Virol. 2005;79:2335–2345. doi: 10.1128/JVI.79.4.2335-2345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means JC, Muro I, Clem RJ. Silencing of the baculovirus Op-iap3 gene by RNA interference reveals that it is required for prevention of apoptosis during Orgyia pseudotsugata M nucleopolyhedrovirus infection of Ld652Y cells. J. Virol. 2003;77:4481–4488. doi: 10.1128/JVI.77.8.4481-4488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z, Reske G, Huang Q, Hammock BD, Qi Y, Chejanovsky N. Characterization of the apoptosis suppressor protein P49 from the Spodoptera littoralis nucleopolyhedrovirus. J. Biol. Chem. 2002;277:48677–48684. doi: 10.1074/jbc.M208810200. [DOI] [PubMed] [Google Scholar]
- Rabizadeh S, LaCount DJ, Friesen PD, Bredesen DE. Expression of the baculovirus p35 gene inhibits mammalian neural cell death. J. Neurochem. 1993;61:2318–2321. doi: 10.1111/j.1471-4159.1993.tb07477.x. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Molec. Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Ryan CA, Stennicke HR, Nava VE, Burch JB, Hardwick JM, Salvesen GS. Inhibitor specificity of recombinant and endogenous caspase-9. Biochem. J. 2002;366:595–601. doi: 10.1042/BJ20020863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto A, Friesen PD, Rothman JH. Baculovirus p35 prevents developmentally programmed cell death and rescues a ced-9 mutant in the nematode Caenorhabditis elegans. EMBO J. 1994;13:2023–2028. doi: 10.1002/j.1460-2075.1994.tb06475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic D, Kaiser WJ, Harvey AJ, Miller LK. Inhibition of Reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs) Proc. Natl. Acad. Sci. U.S.A. 1997;94:10183–10188. doi: 10.1073/pnas.94.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic D, Kaiser WJ, Miller LK. A mutational analysis of the baculovirus inhibitor of apoptosis Op-IAP. J. Biol. Chem. 1998;273:33915–33921. doi: 10.1074/jbc.273.51.33915. [DOI] [PubMed] [Google Scholar]
- Wilkinson JC, Wilkinson AS, Scott FL, Csomos RA, Salvesen GS, Duckett CS. Neutralization of Smac/Diablo by inhibitors of apoptosis (IAPs): A caspase-independent mechanism for apoptotic inhibition. J. Biol. Chem. 2004;279:51082–51090. doi: 10.1074/jbc.M408655200. [DOI] [PubMed] [Google Scholar]
- Wright CW, Clem RJ. Sequence requirements for Hid binding and apoptosis regulation in the baculovirus inhibitor of apoptosis Op-IAP: Hid binds Op-IAP in a manner similar to Smac binding of XIAP. J. Biol. Chem. 2002;277:2454–2462. doi: 10.1074/jbc.M110500200. [DOI] [PubMed] [Google Scholar]
- Wright CW, Means JC, Penabaz T, Clem RJ. The baculovirus anti-apoptotic protein Op-IAP does not inhibit Drosophila caspases or apoptosis in Drosophila S2 cells and instead sensitizes S2 cells to virus-induced apoptosis. Virology. 2005;335:61–71. doi: 10.1016/j.virol.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Xu G, Cirilli M, Huang Y, Rich RL, Myszka DG, Wu H. Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex. Nature. 2001;410:494–497. doi: 10.1038/35068604. [DOI] [PubMed] [Google Scholar]
- Zoog SJ, Schiller JJ, Wetter JA, Chejanovsky N, Friesen PD. Baculovirus apoptotic suppressor P49 is a substrate inhibitor of initiator caspases resistant to P35 in vitro. EMBO J. 2002;21:5130–5140. doi: 10.1093/emboj/cdf520. [DOI] [PMC free article] [PubMed] [Google Scholar]