Abstract
Three short (7 to 9 nucleotides) highly conserved nucleotide sequences were identified in the putative promoter regions (150 bp upstream and 50 bp downstream of the ATG translation start site) of three members of the genus Chlorovirus, family Phycodnaviridae. Most of these sequences occurred in similar locations within the defined promoter regions. The sequence and location of the motifs were often conserved among homologous ORFs within the Chlorovirus family. One of these conserved sequences (AATGACA) is predominately associated with genes expressed early in virus replication.
Keywords: Chlorella viruses, Phycodnaviridae, Chlorovirus, Virus PBCV-1, Virus NY-2A, Virus MT325, Promoters
INTRODUCTION
Chlorella viruses (Family Phycodnaviridae, genus Chlorovirus) are large, icosahedral, plaque-forming, dsDNA-containing viruses that infect and replicate in certain isolates of chlorella-like green algae. The 330-kb genome of the prototype virus, Paramecium bursaria chlorella virus 1 (PBCV-1), was sequenced and annotated more than 10 years ago (Li et al., 1997). The virus contains 366 putative protein-encoding genes and a polycistronic gene that encodes 11 tRNAs (Li et al., 1997; Van Etten, 2003). Approximately 40% of its predicted gene products resemble proteins of known function and many are unexpected for a virus. Currently, three species are included in the genus Chlorovirus: i) Viruses that infect Chlorella NC64A (NC64A viruses), ii) viruses that infect Chlorella Pbi (Pbi viruses) and iii) viruses that infect symbiotic zoochlorella in the coelenterate Hydra viridis (Yamada et al., 2006). Chlorella NC64A and Chlorella Pbi are normally endosymbionts of the protozoan Paramecium bursaria, but they can be cultured independently of the protozoan. The current study involves three chlorella viruses whose genomes have been sequenced: viruses PBCV-1 and NY-2A (Fitzgerald et al., 2007b) are NC64A viruses with 330-kb and 369-kb genomes, respectively, and virus MT325, a Pbi virus with a 314-kb genome (Fitzgerald et al., 2007a).
PBCV-1 infects its host by attaching rapidly to the external surface of the algal cell wall (Meints et al., 1984). Attachment occurs at a unique virus vertex (Onimatsu et al., 2006) and is followed by digestion of the cell wall at the attachment point. Following host wall degradation, the PBCV-1 internal membrane presumably fuses with the host membrane, which leads to the entry of virus DNA and probably associated proteins into the host. An empty capsid remains on the host surface. Circumstantial evidence suggests that the infecting PBCV-1 DNA, and DNA associated proteins, rapidly move to the nucleus to initiate virus transcription (Van Etten, 2003). Support for this hypothesis includes the fact that neither PBCV-1 nor any of the chlorella viruses encode a recognizable RNA polymerase or RNA polymerase subunit. Furthermore, RNA polymerase activity was not detected in PBCV-1 virions (Rohozinski and Van Etten, unpublished results). Consequently, assuming that the infecting viral DNA moves to the nucleus, it must commandeer one of the host’s RNA polymerases (probably RNA polymerase II) to initiate viral transcription (Van Etten, 2003). Therefore, the host polymerase(s), possibly in combination with a virus protein(s), must recognize some virus DNA promoter sequence(s) to initiate transcription. This process occurs rapidly because early PBCV-1 transcripts can be detected within 5 to 10 min post-infection (p.i.) (Schuster et al., 1986). Virus DNA replication starts 60 to 90 min p.i., followed by transcription of late virus genes. Nascent virus capsids begin to assemble in localized regions of the cytoplasm, called virus assembly centers, at 2 to 4 hr p.i. By 5 to 6 hr p.i. the cytoplasm contains many progeny viruses and by 6 to 8 hr p.i. the cell lyses and releases progeny viruses (~1,000 particles/cell).
Thus, PBCV-1 transcription is temporally programmed. Genes defined as “early” are transcribed within 5-60 min p.i.; some of the earliest transcripts form in the absence of de novo protein synthesis (Schuster et al., 1986, Yanai-Balser et al., unpublished results). Transcripts of genes defined as “late” begin to appear 60-90 min p.i.; their appearance probably requires translation of early viral genes. However, some early gene transcripts can also be detected in later stages of infection. The PBCV-1 genes are not spatially clustered on the genome by either temporal or functional class. Therefore, temporal regulation of transcription must occur via cis-and possibly trans-acting regulatory elements.
The purpose of the current study is to identify conserved DNA sequences that might be involved in activation and regulation of viral transcription by using bioinformatic procedures. We identified three conserved nucleotide sequences that appear within 150 nucleotides of the ATG translation start codon of many virus open reading frames (ORFs). One of these motifs is associated predominately with early viral gene transcription and is likely to serve as a promoter for early genes.
RESULTS and DISCUSSION
Viruses analyzed and criteria used to define genes and promoter regions in this study
The three chlorella viruses chosen for this study are PBCV-1, NY-2A, and MT325 that have 366, 404, and 329 putative protein-encoding genes, respectively. Approximately 80% of the genes are present in all three viruses. PBCV-1 and NY-2A infect the same host, Chlorella NC64A, and presumably are more closely related, in terms of evolutionary distance, to each other than to MT325, which infects Chlorella Pbi. However, the two NC64A viruses are among the most diverse of the NC64A viruses. The average amino acid identity between PBCV-1 and NY-2A homologs is ~75% (Fitzgerald et al., 2007b), whereas the average amino acid identity between PBCV-1 and MT325 is ~50% (Fitzgerald et al., 2007a). Most PBCV-1 and NY-2A gene homologs are located co-linearly; in contrast, homologous genes in PBCV-1 and MT325 have almost no co-linearity with each other (Fig. 1). Thus, the promoter elements of the two NC64A viruses might be expected to be more similar to each other than between NC64A and Pbi viruses.
Fig. 1.
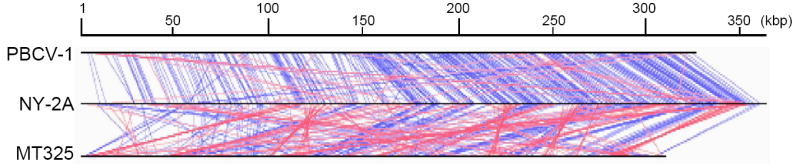
Genomic locations of homologous genes between NY-2A and either PBCV-1 or MT325. When a homologous gene is detected between NY-2A and another genome a line is drawn. If the gene is transcribed in the same direction the line is blue. If the gene is transcribed in the opposite direction the line is red.
The following criteria were originally used to define genes in the three viruses: i) a minimal size of 65 codons initiated by an ATG codon, ii) when genes overlapped, the largest gene was chosen and iii) genes typically contain A+T-rich (>70%) regions in the 50 nucleotides upstream of the ATG translation start codon (Li et al., 1997). For this study, promoter regions were defined as encompassing a 200 bp region (150 bp upstream and 50 bp downstream of the ATG translation start site) of each viral encoded gene. However, the intergenic regions between PBCV-1 genes have an average size of 81 nucleotides with a standard deviation of 83 nucleotides (excluding the two-tailed 5% most extreme data points). In fact, 260 of the 366 PBCV-1 genes have less than 100 nucleotides between them. Using this definition, many of the putative viral promoter regions are located in an adjacent gene.
Three conserved sequences occur in the chlorella virus promoter regions
Using AlignAce software, three highly conserved nucleotide sequences were identified in the PBCV-1 promoter regions (Fig. 2). These sequences were optimized as described in the Materials and Methods section to generate three sequences that range in size from 7 to 9 nucleotides (Table 1); one or more degenerate positions occur in two of the three sequences. Some promoter regions contain more than one copy of either the same or different conserved sequences. As reported in Fig. 3, most of the sequences occurred in the -150 to 0 nucleotide region.
Fig. 2.
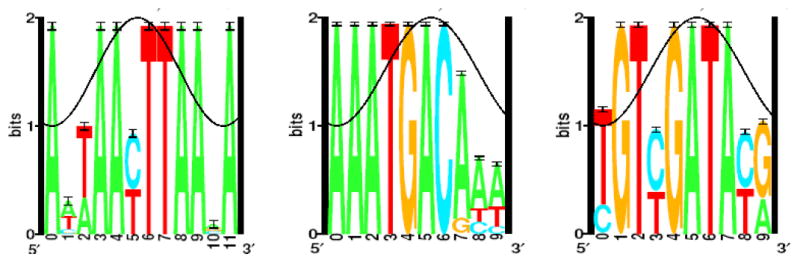
AlignAce results for the three conserved nucleotide sequences that frequently occur in the virus PBCV-1 promoter regions. The black line indicates a potential groove of DNA.
Table 1.
Promoter motif optimization based on generating the motif that has the most occurrences in the promoter region relative to the total number of occurrences in the PBCV-1 genome. *Shown is the total number of times the motif occurred in the promoter regions. Note the same motif can occur multiple times within the same promoter region. For example, there are 91 unique genes that contain the promoter sequence ARNTTAANA; 84 of these genes contain the sequence one time and 7 genes contain the sequence twice.
| ARNTTAANA | |||
|---|---|---|---|
| Sequence | # Promoter Region | Total # in genome | # Promoter Region/ # Genome |
| ANNAANYYAANA | 78 | 208 | 0.38 |
| ANNRRNYYAANA | 99 | 344 | 0.29 |
| NNRRNTTAANA | 108 | 305 | 0.35 |
| ARNTTAANA | 98 | 222 | 0.44 |
| RNTTAANA | 165 | 563 | 0.29 |
| NTTAANA | 323 | 1278 | 0.25 |
| AATGACA | |||
| Sequence | # Promoter Region | Total # in genome | # Promoter Region/ # Genome |
| AAATGACRHH | 89 | 182 | 0.49 |
| ATGACRHH | 107 | 240 | 0.45 |
| AATGACRH | 112 | 251 | 0.45 |
| AATGACR | 116 | 269 | 0.43 |
| AATGACG | 12 | 78 | 0.15 |
| AATGACA | 104 | 191 | 0.54 |
| AATGAC | 128 | 337 | 0.38 |
| ATGACA | 127 | 403 | 0.32 |
| GTNGATAYR | |||
| Sequence | # Promoter Region | Total # in genome | # Promoter Region/ # Genome |
| NGTNGATANN | 69 | 329 | 0.21 |
| GTNGATAYR | 51 | 182 | 0.28 |
| TYGATAYR | 59 | 262 | 0.23 |
| TNGATAYR | 82 | 418 | 0.20 |
| YGATAYR | 103 | 604 | 0.17 |
| YGTYGATAY | 42 | 154 | 0.27 |
| YGTNGATAY | 48 | 194 | 0.25 |
| YGTYGATA | 45 | 189 | 0.24 |
| YGTNGAT | 84 | 513 | 0.16 |
| TNGATAY | 111 | 706 | 0.16 |
Fig. 3.
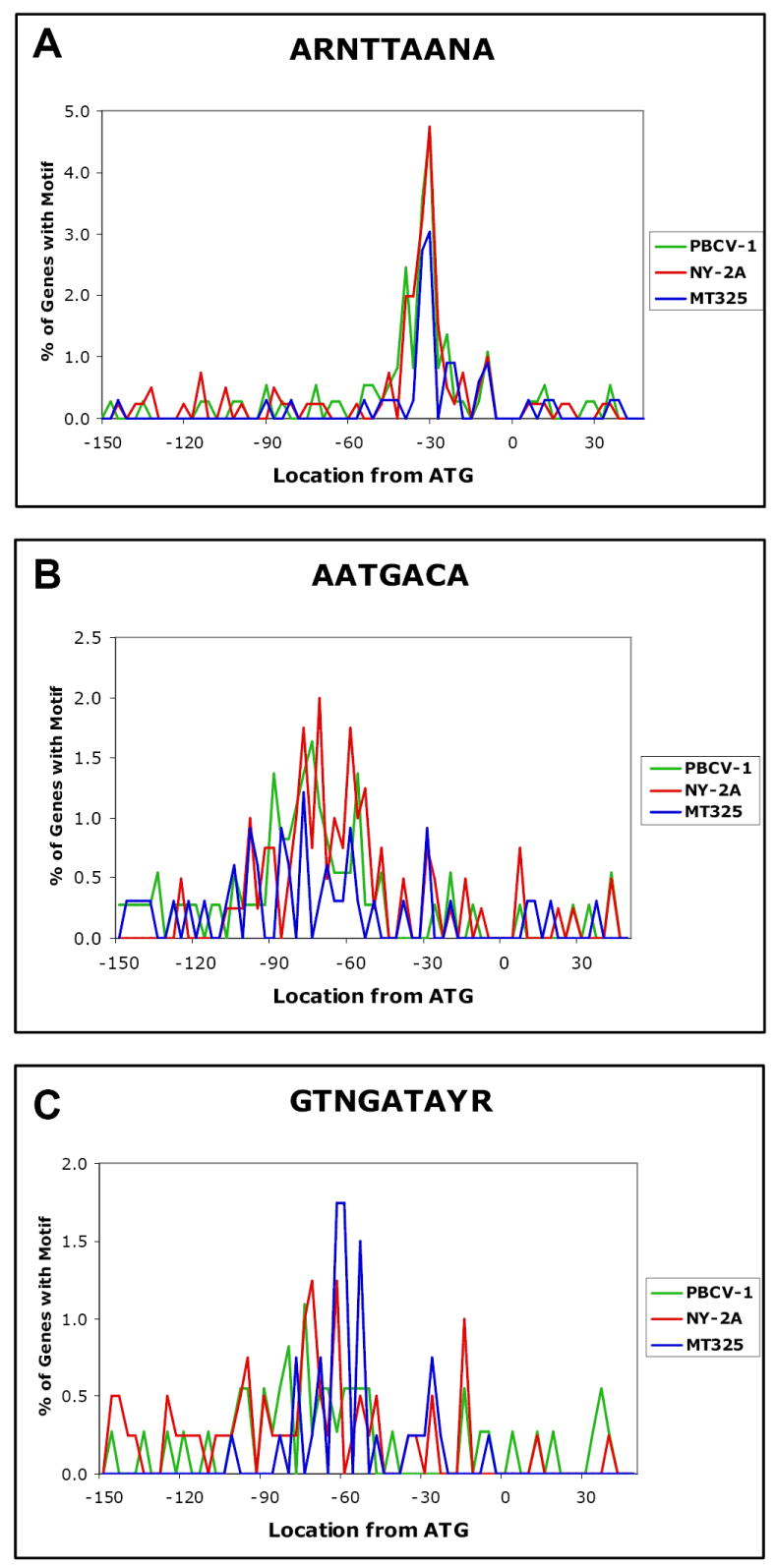
The positional distributions of three conserved nucleotide sequences in the gene promoter region of chlorella viruses PBCV-1, NY-2A, and MT325 with respect to the ATG translation start codon.
Sequence ARNTTAANA
The sequence ARNTTAANA occurs in the promoter region in 91 of the 366 PBCV-1 genes (25%), in 90 of the 404 virus NY-2A genes (22%), and in 40 of the 329 MT325 genes (12%) (Table 2). Relative to the entire genome, this sequence is present within the 200-nucleotide promoter region 44% of the time in PBCV-1, 49% of the time in NY-2A, and 37% of the time in MT325. Furthermore, the location of the sequence is biased to nucleotide position -15 to -45, relative to the ATG translation start codon (64% in PBCV-1, 66% in NY-2A, and 65% in MT325) (Fig. 3A). Thus the region between nucleotides -15 and -45 is a hotspot for the ARNTTAANA sequence.
Table 2.
General characteristics of three conserved, putative promoter elements in three chlorella viruses.
| Percentage of genes with motif in promoter region | Ratio of motif hits in the promoter region to total hits in the genome | Predicted promoter location | Percentage of promoter motifs found within predicted promoter location | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PBCV-1 | NY-2A | MT325 | PBCV-1 | NY-2A | MT325 | (nt from ATG start) | PBCV-1 | NY-2A | MT325 | |
| ARNTTAANA | 25% | 22% | 12% | 0.44 | 0.49 | 0.37 | -15 to -45 | 64% | 66% | 65% |
| AATGACA | 16% | 18% | 8% | 0.54 | 0.53 | 0.25 | -60 to -90 | 44% | 37% | 33% |
| GTNGATAYR | 13% | 14% | 11% | 0.28 | 0.22 | 0.21 | -50 to -80 | 39% | 38% | 70% |
Sequence AATGACA
The sequence AATGACA occurs in the promoter region in 60 of the 366 PBCV-1 genes (16%), in 74 of the 404 NY-2A genes (18%), and 25 of the 329 MT325 genes (8%) (Table 2). Relative to the entire genome, this sequence is present within the 200-nucleotide promoter region in 54% of the PBCV-1 genes, 53% of the NY-2A genes, and 25% of the MT325 genes. Furthermore, the AATGACA sequence is biased to nucleotide position -60 to -90, relative to the ATG initiation codon (44% in PBCV-1, 37% in NY-2A, and 33% in MT325) (Fig. 3B). These results indicate that the region between nucleotides -60 and -90 is a hotspot for the AATGACA sequence. This sequence resembles the consensus -35 element (TTGACA) in E. coli promoters.
Sequence GTNGATAYR
The sequence GTNGATAYR occurs in the promoter region in 49 of the 366 PBCV-1 genes (13%), 58 of the 404 NY-2A genes (14%), and 36 of the 329 MT325 genes (11%) (Table 2). Relative to the entire genome, this sequence is found specifically within the 200-nucleotide promoter region in 28% of the PBCV-1 genes, 22% of the NY-2A genes, and 21% of the MT325 genes. The location of the sequence is biased to nucleotide positions -50 to -80, relative to the ATG initiation codon (39% in PBCV-1, 38% in NY-2A, and 70% in MT325) (Fig. 3C). These results indicate that the region between nucleotides -50 and -80 is a hotspot for the GTNGATAYR sequence.
Occurrence of conserved sequences in PBCV-1 allowing a one base mismatch
The presence of these three conserved sequences in the promoter regions was also determined with one base mismatch in PBCV-1. Under complete stringency, 48% of the 366 PBCV-1 gene promoter regions contain at least one of the three conserved sequences. With one base-pair mismatch, ARNTTAANA occurs in 306 (84%) of the gene promoter regions, AATGACA occurs in 204 (56%) of the gene promoter regions, and GTNGATAYR occurs in 155 (42%) of the PBCV-1 366 promoter regions. [Note: some of the genes have two sequences located one or more times in the same promoter region (supplement 1).] Allowing a one base mismatch, one of these three motifs is present in all but 15 of the 366 PBCV-1 promoter regions. The locations of the sequences with one base pair mismatch relative to the ATG translation start codon are similar to the locations under complete stringency (results not shown).
The conserved motifs are not specific to direction or location within the genome
None of the three conserved sequences exhibit a preference for direction or location within the three viral genomes. This finding is not surprising because genes classified as early or late occur throughout the PBCV-1 genome (Yanai-Balser et al., unpublished results) and they are approximately equally positioned in both orientations.
Homologous virus genes often share similar motif patterns at conserved locations
Viruses PBCV-1, NY-2A, and MT325 share many homologs (~80% of the genes are conserved among the three viruses). Therefore, we examined the occurrence of the conserved sequences among homologs in the three viruses. Homologous gene products often share similar motif patterns at conserved locations relative to the ATG translation start site (supplement 1). For example, a putative VLTF2-type transcription factor is a gene product encoded by all three viruses. Homologs in each of the viruses have the same motif (ARNTTAANA) in a similar location (-32, -31, and -33 nucleotides from the ATG translation start codon in viruses PBCV-1, NY-2A and MT325, respectively). Furthermore, if a specific motif occurs outside of the expected promoter region (e.g. outside of the -50 and -80 region for GTNGATAYR), homologous genes contain the motif in a similar location. For example, a gene encoding a putative PBCV-1 replication factor C protein subunit (a417l) contains the GTNGATAYR sequence beginning at -146. The homologous gene in NY-2A contains the same sequence beginning at nucleotide −142. This sequence is not present in the MT325 replication factor C gene homolog.
Conserved sequences in the promoter region of homologous genes often contain identical nucleotides at degenerate positions
In addition to conserved sequences in their promoter regions, homologous genes often have similar nucleotide preferences at degenerate nucleotide positions within those sequences. Two of the three conserved nucleotide sequences (ARNTTAANA and GTNGATAYR) have degenerate nucleotide positions, and conserved nucleotide preferences occur among homologs for each of these two sequences. For example, the promoter region of the ribonucleotide reductase large subunit gene (a629r in PBCV-1 and b832r in NY-2A) contains the sequence GTNGATAYR, a sequence with three degenerate nucleotide positions. At the ‘N’ position, the PBCV-1 and NY-2A homologs contain a cytidine residue. At the ‘Y’ position, both viral genomes contain a cytidine residue and at the ‘R’ position, both viral genomes contain an adenine residue (Table 3). Not surprisingly, the nucleotide conservation at degenerate positions is more frequent between PBCV-1 and NY-2A than between either of these viruses and MT325.
Table 3.
Examples of homologous proteins of known function containing promoter motifs with conserved nucleotides at degenerate positions. Bold nucleotides represent non-degenerate positions. A (-) denotes either a homolog does not exist or a homologous protein does not contain the motif. Degenerate positions are as follows: N = A/C/G/T, R = A/G, and Y = C/T
| 100% similar promoter motifs in at least two genomes | |||
|---|---|---|---|
| PBCV-1 | NY-2A | MT-325 | |
| Ribo. Reductase (large) | GTCGATACA | GTCGATACA | - |
| 6-phosphofructokinase | AACTTAAGA | AACTTAAGA | - |
| GDP-D-mannose dehydrogenase | AACTTAACA | AACTTAACA | - |
| Glycosyltransferase | AACTTAAGA | AACTTAAGA | - |
| PCNA | AACTTAAGA | AACTTAAGA | - |
| RNA triphosphatase | AACTTAACA | AACTTAACA | AGCTTAACA |
| Rnase III | AATTTAAGA | AATTTAAGA | AACTTAATA |
| TFIID | AATTTAAAA | AATTTAAAA | - |
| VLTF2-type transcription factor | AATTTAAGA | AATTTAAGA | AACTTAACA |
| Only one difference in a degenerate position in at least two genomes | |||
| PBCV-1 | NY-2A | MT-325 | |
| Replication factor C | GTCGATACG | GTCGATATG | - |
| dUTP pyrophosphatase | GTTGATACG | GTTGATATA | GTCGATATA |
| Coat protein-like | AACTTAAAA | AAATTAATA | AACTTAAGA |
| Fructose -2,6 bisphosphatase | AATTTAAAA | - | AACTTAAAA |
| Fucose synthase | AATTTAAGA | AACTTAAGA | - |
| Ubiquitin C-terminal hydrolase | AGCTTAACA | AGTTTAACA | - |
| UDP-glucose dehydrogenase | AGATTAACA | AATTTAACA | - |
| Two or more differences in degenerate positions | |||
| PBCV-1 | NY-2A | MT-325 | |
| Adenine DNA methylase | AAGTTAATA | AATTTAAAA | - |
| ATPase (AAA+ Class) | AAATTAATA | AATTTAAGA | - |
| ATPase (DNA repair) | AAATTAATA | - | AATTTAACA |
| Histidine decarboxylase | AAATTAATA | AATTTAAGA | - |
| Transposase | AACTTAAGA | AATTTAATA | AATTTAACA |
Motif AATGACA is strongly associated with PBCV-1 early gene expression
To determine if there is a correlation between time of expression and the presence of a putative promoter sequence in the PBCV-1 genes, we constructed a microarray containing probes from each gene in the genome. Competitive hybridization experiments were conducted employing cDNA from poly A-containing viral RNAs obtained from cells at 20, 40, 60, 90, 120, 240, and 360 min p.i., which allows us to follow global transcription of PBCV-1 replication.
The microarray results established that PBCV-1 transcripts fall into two groups: (i) early genes (59%), expressed before 60 min p.i. (the beginning of DNA synthesis) and (ii) late genes (41%), expressed after 60 min p.i. However, transcripts of 42% of the early genes are also present at late times after infection, referred to as early/late genes in Fig. 4 (Yanai-Balser et al., unpublished results).
Fig. 4.
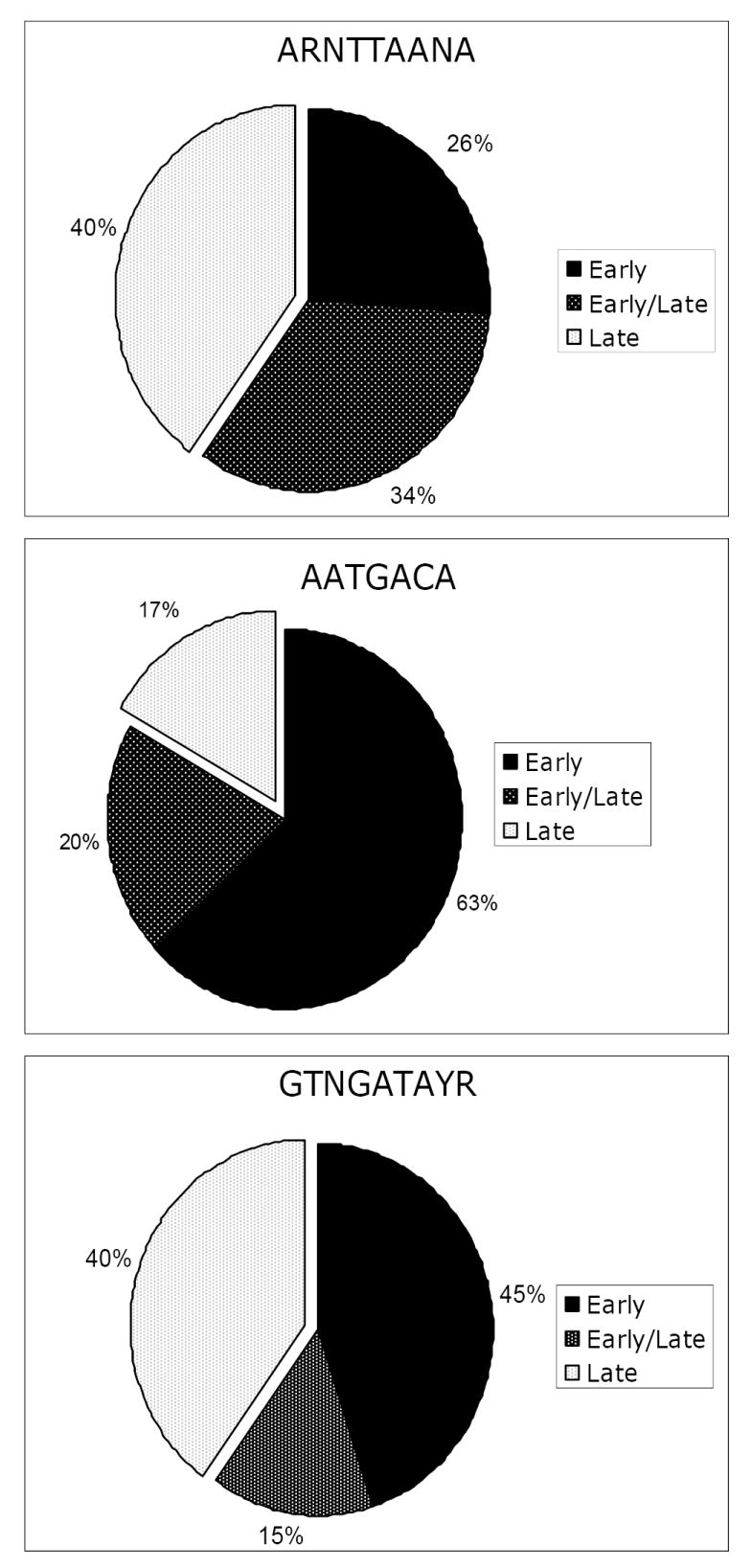
Distribution of three putative promoter motifs relative to when PBCV-1 genes are expressed. Transcripts of genes classified as early/late are detected prior to the beginning of DNA replication and are present after DNA synthesis begins.
Most of the genes with the AATGACA sequence are expressed early during infection (83%); transcripts from 24% of these early genes are also present after virus DNA synthesis begins (Fig. 4). The remaining 17% of the genes containing the AATGACA sequence are expressed late.
The other two sequences, ARNTTAANA and GTNGATAYR, have a no correlation with expression time. Sixty percent of the genes with the sequence ARNTTAANA are transcribed early; transcripts from 56% of these early genes are also present after virus DNA synthesis begins. The remaining 40% of the genes with the ARNTTAANA sequence are expressed late. Likewise, 60% of the genes with the sequence GTNGATAYR in the promoter region are transcribed early and 25% of these genes produce transcripts that are also detected late during infection. The remaining 40% of the genes containing the GTNGATAYR sequence are expressed late. However, since 60% of the total genes are expressed early and 40% of the total genes are expressed late, there is no correlation with time of expression and these two sequences.
Promoter elements in related viruses
This is the first attempt to identify promoter elements by bioinformatic procedures in the phycodnaviruses. However, two previous reports described conserved nucleotide sequences in promoter regions that are associated either with a single chlorella virus gene, a gene encoding a potassium ion channel protein (Kang et al., 2004), or with 23 immediate early expressed genes in chlorella virus CVK2 (Kawasaki et al., 2004). The motif identified in the immediate early genes by Kawasaki et al. (ATGACAA) is similar to a motif identified in this manuscript (AATGACA), which also correlated with early transcripts.
The phycodnaviruses probably share a common evolutionary ancestry with the poxviruses, iridoviruses, asfarviruses, and the mimivirus (Iyer et al., 2001; Iyer et al., 2006; Raoult et al., 2004). All of these viruses have nine gene products in common and at least two of these viral families have an additional 41 homologous ORFs (Iyer et al., 2006). Collectively, these large dsDNA viruses are referred to as nucleocytoplasmic large DNA viruses (Iyer et al., 2001).
A bioinformatics study on mimivirus identified an eight-nucleotide sequence, AAAATTGA, which occurs in the putative promoter regions (-150 to 0) of 403 of the 911 (45%) mimivirus ORFs (Suhre et al., 2005). This element is specific to the mimivirus lineage and the authors suggest that the element may correspond to an ancestral promoter structure predating the radiation of the eukaryotic kingdom.
In the iridovirus, Chilo iridescent virus (CIV), 5 nucleotides (AAAAT) located between -19 and -15 have been described as essential for promoter activity (Nalcacioglu et al., 2007). Interestingly, this promoter sequence is not only in the putative promoter regions of other CIV genes but also in other iridoviruses. Conserved nucleotide sequences in the promoter regions of the poxviruses (Moss, 2007) and the asfarvirus, African swine fever virus (Garcia-Escudero et al., 2000), have also been reported.
CONCLUSIONS
This study identified three conserved 7 to 9 nucleotide sequences that probably function as promoter elements in the chlorella viruses. One of these sequences is associated primarily with early viral gene transcription and is likely to serve as a promoter for early genes. One way to test these predictions is to place one or more of these suspected early gene promoter regions in front of a late virus gene and determine if the “late” gene is now expressed early. Unfortunately, these experiments are not possible at the present time because procedures for manipulating the chlorella virus genomes are lacking.
MATERIALS AND METHODS
Bioinformatics
The genome sequences and annotations for viruses PBCV-1, NY-2A, and MT325 are available from GenBank under accession numbers U42580, DQ491002, and DQ491001, respectively. The same material is also located at http://greengene.uml.edu. For this study, the promoter region was defined as the region encompassing 150 nucleotides upstream of the ATG translation initiation codon and 50 nucleotides downstream of the ATG translation start codon.
AlignAce software (Roth et al., 1998) was used to identify conserved motifs in the promoter regions of the 366 PBCV-1 genes. Three conserved sequences were initially identified (Fig. 2). Two of the sequences were 10-mers and one was a 12-mer. These sequences were optimized and shortened (from each end) one base at a time by trial and error to generate the highest ratio of sequence hits in the promoter region relative to total sequence hits in the PBCV-1 genome (Table 1). The PBCV-1, NY-2A, and MT325 genomes were then searched for the occurrence of the three optimized sequences under complete stringency; the locations of the sequences within the promoter region were identified for each gene. The position of each sequence was then plotted with respect to the ATG translation initiation codon (Fig. 3). In addition, the PBCV-1 genome was searched for the three conserved sequences allowing one nucleotide mismatch and plotted.
RNA isolation
Infected chlorella cells (m.o.i. of 5) were collected at 20, 40, 60, 90, 120, 240, and 360 min p.i. Cells were disrupted with glass beads in the presence of Trizol (Invitrogen, Carlsbad, CA) and RNA was isolated using the Absolutely RNA Miniprep kit (Stratagene, LaJolla, CA), according to the manufacturer’s instructions. RNA integrity was verified in denaturing 1% agarose gels where intact host cytoplasmic and chloroplast rRNAs were visualized.
Microarrays fabrication and hybridization
A microarray containing 50-mer oligonucleotide probes representing each ORF in the PBCV-1 genome was constructed by MWG Biotech (Ebersberg, Germany) and the Microarray Core Facility (University of Nebraska Medical Center). For each time point, 20 mg of total RNA was reverse-transcribed using oligo(dT) as primers and cDNA was labeled with Cy3 or Cy5-dUTP (GE Healthcare, Piscataway, NJ) with the aid of a SuperScript Indirect cDNA Labeling System (Invitrogen, Carlsbad, CA) following the supplier’s directions. Competitive hybridization experiments were conducted for each time point against a pool of transcripts representing every gene isolated in the time course.
Microarrays Analysis
Results from three independent biological hybridizations were analyzed using the GenePix Pro v.6.0 software (Molecular Devices, Sunnyvale, CA) and TIGR microarray software suite (TM4) (Saeed et al., 2003). Many transformations were performed to eliminate low quality data, to normalize the measured intensities using Lowess algorithm, and to regularize the standard deviation of the intensity of the Cy3/Cy5 ratio across the blocks. Genes that displayed statistically significant modulation were identified by a one-way analysis of variance, using P values of <0.01 as a cutoff. Genes with similar expression profiles were grouped into 10 different clusters using a K-means algorithm.
Supplementary Material
Putative PBCV-1, NY-2A and MT325 genes are grouped by their functional categories. If a conserved nucleotide motif has been identified within the promoter region of the gene, the sequence and location are noted.
Acknowledgments
This investigation was supported in part by Public Health Service grant GM3211 (J.V.E.) and NIH Grant P20RR15635 from the COBRE program of the National Center for Research Resources (J.V.E.). P.T.B. was partially supported by the UNL UCARE program that encourages undergraduate research. The authors would also like to thank David Dunigan, Gary Duncan, and Jim Gurnon for their interest in and support of this project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Fitzgerald LA, Graves MV, Li X, Feldblyum T, Hartigan J, Van Etten JL. Sequence and annotation of the 314-kb MT325 and the 321-kb FR483 viruses that infect Chlorella Pbi. Virology. 2007a;358:459–471. doi: 10.1016/j.virol.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald LA, Graves MV, Li X, Feldblyum T, Nierman WC, Van Etten JL. Sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A. Virology. 2007b;358:472–84. doi: 10.1016/j.virol.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Escudero R, Vinuela E. Structure of African swine fever virus late promoters: requirement of a TATA sequence at the initiation region. J Virol. 2000;74:8176–8182. doi: 10.1128/jvi.74.17.8176-8182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Kang M, Graves M, Mehmel M, Moroni A, Gazzarrini S, Thiel G, Gurnon JR, Van Etten JL. Genetic diversity in chlorella viruses flanking kcv, a gene that encodes a potassium ion channel protein. Virology. 2004;326:150–159. doi: 10.1016/j.virol.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Tanaka M, Fujie M, Usami S, Yamada T. Immediate early genes expressed in chlorovirus infections. Virology. 2004;318:214–223. doi: 10.1016/j.virol.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Li Y, Lu Z, Sun L, Ropp S, Kutish GF. Analysis of 74 kb of DNA located at the right end of the 330-kb chlorella virus PBCV-1 genome. Virology. 1997;237:360–377. doi: 10.1006/viro.1997.8805. [DOI] [PubMed] [Google Scholar]
- Meints RH, Lee K, Burbank DE, Van Etten JL. Infection of a chlorella-like alga with the virus, PBCV-1: ultrastructural studies. Virology. 1984;138:341–346. doi: 10.1016/0042-6822(84)90358-1. [DOI] [PubMed] [Google Scholar]
- Moss B. Poxviridae: the viruses and their replication. In: Knipe PMHDM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. Fifth. WoltersKluwer/Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2905–2946. [Google Scholar]
- Nalcacioglu R, Ince IA, Vlak JM, Demirbag Z, van Oers MM. The Chilo iridescent virus DNA polymerase promoter contains an essential AAAAT motif. J Gen Virol. 2007;88:2488–2494. doi: 10.1099/vir.0.82947-0. [DOI] [PubMed] [Google Scholar]
- Onimatsu H, Suganuma K, Uenoyama S, Yamada T. C-terminal repetitive motifs in Vp130 present at the unique vertex of the chlorovirus capsid are essential for binding to the host chlorella cell wall. Virology. 2006;353:433–442. doi: 10.1016/j.virol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. The 1.2-megabase genome sequence of mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Rylstov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Schuster AM, Girton L, Burbank DE, Van Etten JL. Infection of a chlorella-like alga with the virus PBCV-1: transcriptional studies. Virology. 1986;148:181–189. doi: 10.1016/0042-6822(86)90413-7. [DOI] [PubMed] [Google Scholar]
- Suhre K, Audic S, Claverie JM. Mimivirus gene promoters exhibit an unprecedented conservation among all eukaryotes. Proc Natl Acad Sci USA. 2005;102:14689–14693. doi: 10.1073/pnas.0506465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten JL. Unusual life style of giant chlorella viruses. Ann Rev Genetics. 2003;37:153–195. doi: 10.1146/annurev.genet.37.110801.143915. [DOI] [PubMed] [Google Scholar]
- Yamada T, Onimatsu H, Van Etten JL. Chlorella viruses. Adv Virus Res. 2006;66:293–336. doi: 10.1016/S0065-3527(06)66006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Putative PBCV-1, NY-2A and MT325 genes are grouped by their functional categories. If a conserved nucleotide motif has been identified within the promoter region of the gene, the sequence and location are noted.


