Abstract
Gene expression is controlled by the action of transcription factors that bind to DNA and influence the rate at which a gene is transcribed. The quantitative mapping between the regulator concentrations and the output of the gene is known as the cis-regulatory input function (CRIF). Here, we show how the CRIF shapes the form of the joint probability distribution of molecular copy numbers of the regulators and the product of a gene. Namely, we derive a class of fluctuation-based relations that relate the moments of the distribution to the derivatives of the CRIF. These relations are useful because they enable statistics of naturally arising cell-to-cell variations in molecular copy numbers to substitute for traditional manipulations for probing regulatory mechanisms. We demonstrate that these relations can distinguish super- and subadditive gene regulatory scenarios (molecular analogs of AND and OR logic operations) in simulations that faithfully represent bacterial gene expression. Applications and extensions to other regulatory scenarios are discussed.
Keywords: gene expression, mathematical modeling, noise analysis inference, flow cytometry
Transcription factors regulate the expression of genes by binding to specific sites on the DNA that are typically spatially close to, sometimes even in, sequences that code for proteins. A group of such binding sites is collectively known as a cis-regulatory region. When a gene processes the effects of multiple transcription factors, one can view it as performing a computation. The inputs are the occupancies of the binding sites in the cis-regulatory region and the output is the rate of transcription. The simplest method of describing the relationship between the transcription factors and the output is by using boolean logic (1,2). For example, 2 activators can regulate a gene with AND logic in which both are required or with OR logic in which either transcription factor is sufficient for transcription. Knowing which mode of combinatorial regulation a gene employs can be important for determining its function in regulatory networks. For example, the coherent feed forward loop, one of the most common motifs in gene regulatory networks (3), filters noise in upstream signals differently depending on how one of the participating genes integrates its inputs (4,5).
In general, the output of a gene will not be binary, and will depend on the concentrations of the transcription factors in a complex manner. The notion of logic operations can be generalized by introducing a continuous function that encodes the dependence of the rate of transcription on the concentrations of the inputs. Such “cis-regulatory input functions (CRIFs)” (5) have been evaluated experimentally for the well-studied lac operon. The CRIF of the wild type is complex (6,7), but those of certain mutant operons represent molecular analogs of binary logic gates (8). In higher organisms, many genes are regulated by a large number of transcription factors, often with multiple binding sites for each factor (9–11). It is increasingly clear that the action of many such regulators depends on their molecular context (12,,–19). Because CRIFs are capable of encoding arbitrarily complex modes of regulation, they will likely be even more useful for describing the control of transcription in these systems than in the simpler prokaryotic ones.
To date, the notion of a CRIF has largely been restricted to studying how the average rate of transcription depends on the average concentration of the transcription factor inputs by using bulk measurements. However, experiments performed on single cells reveal that because transcription factors are often present in low copy numbers, stochastic fluctuations in the concentrations of these molecules can have important consequences for gene regulation (20,–25). Although a dialog between theory and experiment has revealed how noise enters the basic processes of transcription and translation (26,27) and propagates through a cascade of genes (23,28,29), the relationship between combinatorial regulation and stochastic fluctuations has received much less attention. In particular, it is unclear how the mode of regulation encoded in the CRIF is related to the statistics that characterize the distribution of protein concentrations across a population\break of cells.
Our approach to this area is motivated by the fact that, at equilibrium, the fluctuations of a property of a material can be related to the response to changes in a conjugate control parameter. A well-known example is the relation between the fluctuations in the energy of a system and its heat capacity. More generally, the fluctuation–dissipation theorem relates fluctuations about equilibrium to the relaxation of the system after it has been perturbed from equilibrium (30). General fluctuation–dissipation relations for linear (or linearized) chemical systems that relate the average values of the chemical concentrations to the second moments can also be derived (31) and have been applied to biological systems(29,32).
Here, we use a stochastic model of a gene regulated by an arbitrary number of transcription factors to show that logic operations are most closely related to the derivatives of the CRIF. In turn, the derivatives can be related to higher moments of the distribution of input and output copy numbers in analogy to molecular systems at equilibrium. These facts suggest means for inferring regulatory synergies from measurements that report on single cells. Using simulations of simple constructs with reasonable parameters, we demonstrate that the signatures of combinatorial logic should be detectable with presently available experimental methods. This is useful because proximity in DNA binding is not sufficient to infer combinatorial interactions, and they cannot be readily probed by traditional methods (e.g., knockouts) or high-throughput expression assays (e.g., microarray data). Broader implications for the work are discussed.
Results
In this section, we develop a model of gene expression and use it to derive a general expression relating the distribution of protein copy numbers to the derivatives of the CRIF. We then illustrate the utility of this relationship by considering idealized logic gates. Finally, predictions are made for an actual system to enable validation of the theory and its assumptions.
Fluctuation Relations for a Gene.
We model a gene regulated by an arbitrary number of transcription factors by using chemical reactions for the production and degradation of the transcription factor inputs and the output:
where the {Xm} for m = 1 to n are the transcription factor inputs to the logic gate, X0 is the measured output of the gene of interest, and arrows from and to ∅ denote synthesis and degradation, respectively. The rate of production of X0 is determined by the concentrations of the transcription factors and is encoded in the (dimensionless) cis-regulatory input function, which we denote symbolically by f(N1,N2,…,Nn), where Nm is the number of copies of Xm.
Although f is arbitrary with respect to the mode of regulation, our analytical treatment relies on 2 simplifying assumptions: (i) we model transcription and translation as a single composite step and (ii) we employ a quasi-steady-state approximation to absorb the association and dissociation of the regulators into the CRIF. The latter assumption is justified because those reactions are significantly faster than protein production and degradation (33). Although these assumptions enable the derivation of analytical results, we show with simulations that the conclusions of the theory hold when these elementary steps are considered explicitly.
To relate the CRIF to the noise properties of the system, we describe the dynamics of the gene by a master equation (31):
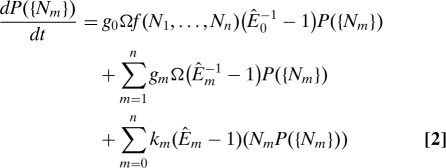 |
where Nm denotes the set of copy numbers of the inputs and the reporter, and P is the probability of observing a particular combination of protein copy numbers. Êm is a step operator defined such that Êmh(Nm) = h(Nm + 1) and Êm−1h(Nm) = h(Nm − 1), where h represents an arbitrary function of Nm. Eq. 2 simply tracks P({Nm}) assuming the protein copy numbers evolve according to the reactions in Eq. 1. The first 2 terms on the right-hand side of Eq. 2 represent the creation of the output and the inputs, respectively, and the last term represents degradation of all the species.
The parameter Ω in Eq. 2 quantifies the size of the system; it simply reflects the choice of units of the kinetic parameters. We include it explicitly to facilitate expansion of the master equation (31). To this end, motivated by the fact that fluctuations are expected to scale as Ω, we perform a change of variables from the {Nm} to {ξm} defined such that Nm = Ωϕm(t) + Ωξm. Here, φm(t) is the solution to the deterministic rate equations, and ξm represents the fluctuating part of Nm. Although Nm is discrete, both φm and ξm are continuous. The steady-state value of φm is equal to the steady-state average concentration of species Xm Xm(φm = 〈Nm〉 / Ω). As a consequence, one can view ξm as the difference between the instantaneous value of the concentration of Xm and its average, or, equivalently, the difference between the value in a single cell and the average across the population.
After this change of variables, we expand the master equation in powers of Ω– and include as many terms as necessary to derive the lowest order nonzero contribution to the moment of interest. Mathematical details can be found in supporting information (SI) (Appendix). We obtain a general expression that relates the moments of the distribution to the derivatives of the CRIF at steady state:
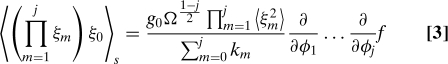 |
The left-hand side of Eq. 3 is the (j + 1)-point correlation function involving any j of the n total regulators and the output. The right-hand side is a product of the jth mixed partial derivative of the CRIF with respect to the concentrations of the same j regulators and a collection of system-specific constants. Because these constants are all positive, the correlation function and the derivative in Eq. 3 have the same sign. In the one regulator case (j = 1), the relationship shows that an activator and its target are positively correlated, whereas a repressor and its target are negatively correlated. This statement is trivial, but its generalization to the case of 2 or more regulators (j ≥ 2) allows us to understand qualitatively how the noise statistics are shaped by combinatorial regulation. Below we illustrate this point explicitly for the case of 2 regulators.
Describing Combinatorial Regulation
Consider a CRIF that is a function of the concentration of 2 transcription factors f(φ1,φ2). How does the output respond to simultaneous changes in the concentrations of the inputs? Note that here we are not envisioning stochastic fluctuations, but small changes to the average concentrations due to perturbations (either natural or artificial cues) to the system. Eq. 3 then relates the response to these perturbations to the intrinsic noise that can be observed even when the system is unperturbed. By Taylor expanding, we find:
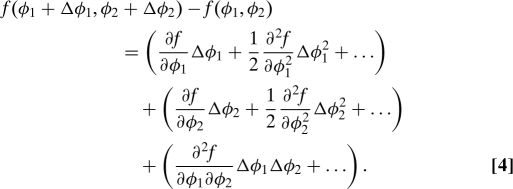 |
The first and second terms give the responses to changes in only X1 and only X2, respectively. The third term represents the synergistic contributions, and the mixed partial derivative is thus the lowest-order term that reports on them. The mixed partial derivative can be accessed through the 3-point correlation function 〈ξ0ξ1ξ2〉, as shown by the specific form Eq. 3 takes in the case of 2 regulators (j = 2):
where C is a positive constant. Thus, Eqs. 4 and 5 taken together suggest that information about synergistic regulation is available from the 3-point correlation function. A pedagogical discussion of the intuitive meaning of this result can be found in SI Appendix, and Figs. S1 and S2.
Idealized Two-Regulator Systems.
Although Eqs. 3 and 5 are general and apply regardless of the mode of regulation, it is instructive to consider a simple model for the CRIF. Consider the 2 activator system depicted schematically in Fig. 1. This system has 4 possible states, one corresponding to each combination of bound transcription factors. If the binding and unbinding of the transcription factors to the DNA are taken to be fast, one obtains (34)
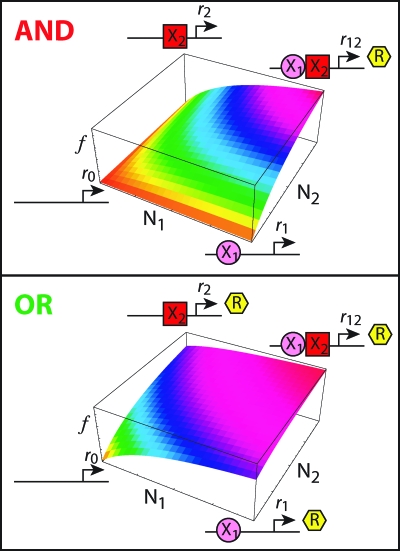
where Ki is the Michaelis constant for the binding of the ith transcription factor and rα is the rate of transcription when the system is in state α (α = 0, 1, 2, 12). For an AND gate both regulators must be bound to initiate transcription so r0 = r1 = r2 = 0 and r12 = r, whereas for an OR gate, binding of either regulator enables maximum production so r0 = 0 and r1 = r2 = r12 = r. The f functions for these idealized systems are plotted in Fig. 1. The differences between the AND and OR gates are most visible at the corners of the plots, where one activator is expressed strongly and the other is expressed weakly. The gene activities vary continuously with the levels of the activators because they reflect averages over the states of the cis-regulatory regions sampled as the transcription factors associate and dissociate from the DNA.
Fig. 1.
cis-regulatory input functions (CRIFs). Each surface maps the levels of the activators of a gene to its expression. The two transcription factors are denoted X1 and X2 and their copy numbers are denoted N1 and N2. The diagrams at the corners of the plots indicate the likely states of the cis-regulatory region for the indicated concentrations and whether the measured output (which can be mRNA or protein) is produced in each state; rα is the net rate of expression when the cis-regulatory region is in configuration α.
Above, we showed that the mixed-partial derivative reports on synergies (Eq. 4). For this model, this derivative is
Because K1 and K2 are positive, the sign of the right-hand side of this equation is determined by the quantity in square brackets. This is the rate when both regulators are bound (r12) less the novel contributions from having one regulator bound (r1−r0 and r2−r0 for regulators 1 and 2, respectively) and the basal rate (r0). Thus, this mixed derivative reports on the novel effects that emerge when 2 regulators are bound. Substitution of the values defining the AND and OR gates shows that the mixed partial derivative is always positive for the former and always negative for the latter. There is a simple physical interpretation of these results. An AND gate requires that both transcription factors be present for the gene to be expressed, so the effect of having both proteins is greater than the sum of the individual effects (no output). In the case of an OR gate, once one transcription factor is bound, no additional activity is obtained from the binding of the second, and the net effect is therefore less than the sum of the individual effects of the regulators.
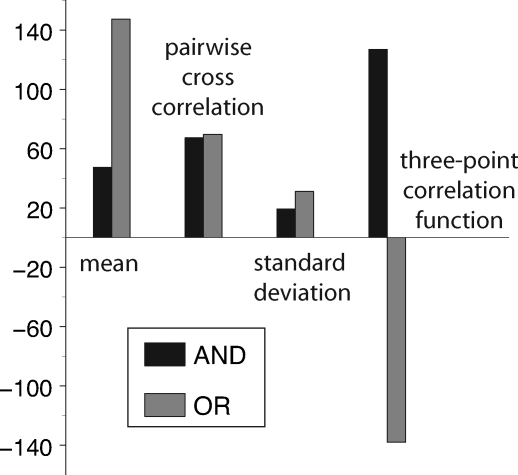
Eq. 5 shows that the sign of the mixed-partial derivative is the same as the sign of the 3-point correlation function involving both regulators and the output. Because our analytical results were derived with simplifying assumptions as discussed above, we performed stochastic simulations (35) of genes regulated by AND and OR logic in which the processes of transcription, translation, DNA binding and unbinding, and protein and mRNA degradation were treated explicitly (Fig. 2). Consider the mean concentration of the output for genes regulated by AND and OR logic gates. Although the mean for the OR gate is higher because either activator can initiate transcription, a change in the kinetic parameters could yield an AND gate with a mean output concentration identical to that for the OR gate and vice versa. Thus, there is no unique signature of the regulatory mechanism present in this statistic. Similarly for the variance of the output and the cross-correlation involving either individual input and the output, it is possible to tune the kinetic parameters for either the AND or the OR gate to achieve any specific positive value of these statistics. In contrast, the 3-point correlation function is positive for AND gates and negative for OR gates regardless of the parameter values. Thus, these simulations support the notion that the mode of combinatorial regulation is unambiguously reflected in this correlation function. It will be positive for synergistic (AND-like) regulation, negative for subadditive (OR-like) regulation, and zero for additive regulation. Although the above discussion has focused on a gene regulated by 2 activators, the 3-point correlation function also distinguishes between qualitatively different modes of combinatorial regulation in other situations as summarized in Table 1.
Fig. 2.
Calculated moments of the distribution of regulator and output copy numbers for genes that function as logic gates. In these simulations no equilibrium assumptions were made, and reactions for dimerization, DNA binding, transcription, translation, and degradation were simulated explicitly (SI Appendix, Table S1).
Table 1.
Three-point correlation function for idealized 2-regulator systems
| Regulators | Sign | Interpretation |
|---|---|---|
| a a | + | Superadditive (synergistic) regulation (an AND gate) |
| a a | − | Subadditive regulation (an OR gate) |
| r r | + | Subadditive regulation (an OR gate) |
| r r | − | Superadditive (synergistic) regulation (an AND gate) |
| a r | + | Activator dominant |
| a r | − | Repressor dominant |
a, activator, r, repressor; sign, the sign of the 3-point correlation function.
Practical Issues for the Detection of Noise Signatures.
Given single-cell data for the joint distribution of copy numbers of X0 and the transcription factors X1 and X2, we compute the 3-point correlation function 〈ξ0ξ1ξ2〉 as follows. As discussed above, up to a factor involving the volume, ξm is the difference between the value of the concentration of Xm in a single cell and its average across the population of cells. We thus calculate the average level of each species in the instrument response units (〈Nm 〉, for m = 0,1,2), subtract the average from the level of the species in each cell to obtain a deviation (δNm = Nm − 〈Nm〉), multiply the 3 deviations for each cell together (δN1δN2δ0), and average the resulting product over the population of cells (〈δN1δN2δN0〉); 〈ξ1ξ2ξ0〉 = Ω– 〈δN1δN2δN0〉, but there is no need to distinguish between these quantities when working in arbitrary response units. In situations in which the regulators exist in a variety of forms, the concentrations should ideally be taken to be the concentrations of their active form(s); to the extent that they are approximately proportional to their total concentrations, this distinction is unnecessary.
The data needed for this analysis can be obtained from flow cytometry, in which several molecular species are simultaneously fluorescently tagged and detected to estimate their concentrations in many individual cells. Although more reliable data can be obtained by engineering fusions to fluorescently active proteins, antibodies coupled to dyes can often be used and obviate molecular biological manipulation. A limitation of flow cytometry is that the measurement errors can be quite large. Nonetheless, because determining the qualitative mode of combinatorial regulation only requires determining the sign of the 3-point correlation function, it is relatively robust to such errors. Fig. S3 in SI Appendix shows that numbers of cells routinely accessible 106 are sufficient to determine the sign of the correlation function with high certainty even when the measurement errors are of the same magnitude as the fluorescence signal.
A more challenging issue is that fluctuations that affect gene expression globally [e.g., changes in the number of ribosomes or RNA polymerase (RNAP) molecules, variations in cell sizes, cell cycle stages] can create correlations between molecular species which are independent of transcriptional regulation. In the literature, such variability is termed “extrinsic noise” and is distinguished from variability directly associated with the processes of gene expression under consideration (“intrinsic noise”) (20,36). To explore this issue, we performed simulations of the idealized logic gates in which we explicitly included fluctuations in RNAP. As expected, correlations were observed even between species that had no causal relationship (i.e., the 2 inputs). The fluctuations in RNAP led to a decrease in the 3-point correlation function because they increased the output less than what was expected from summing the effects of the concomitant increases in the individual inputs. Variations in cell size can be detected by using forward scatter in a flow cytometry experiment; other extrinsic fluctuations can be quantified by measuring an additional species without any (direct or indirect) regulatory relation to the species of interest. Associated spurious correlations can be suppressed by sorting the data into bins for the forward scatter or the concentration of the additional species, computing the statistics of interest separately for each bin, and then pooling the results SI Appendix, Fig. S4). The main limitation of this procedure is that one must be able to detect sufficient variations in the species of interest beyond those arising from the extrinsic noise.
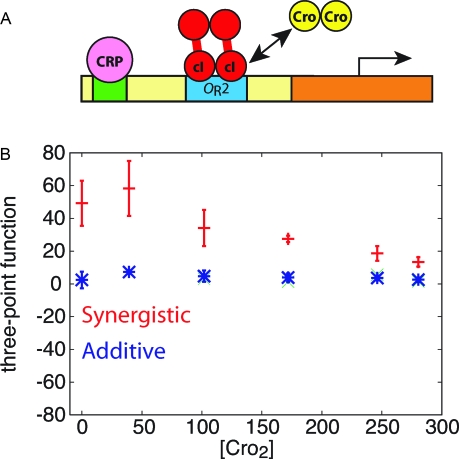
Phage-λ Construct.
To assess whether signatures of combinatorial regulation would be detectable experimentally, we consider the genetic circuit based on the phage-λ operon from ref. 37. In this system, the PRM promoter and OR2 binding site are in their natural locations and an additional binding site for the Escherichia coli lac activator CRP is located upstream; the cI dimer activates transcription by binding to OR2 (Fig. 3A). The authors also created a second construct that only contained the PRM promoter and OR2 binding site. By measuring the expression from these 2 constructs in the presence and absence of cI protein, they were able to assay the activities resulting from each of the four possible combinations of transcription factors bound: none, just CRP, just cI, or both. These had activities of 21, 206, 158, and 1,420 units, respectively, showing that these protein factors function synergistically (1,420 units > 158 + 206 units). The authors also identified a mutant that served as a positive control (pc). In that case, the combination of cI and CRP was nearly additive (293 units).
Fig. 3.
Simulations of the phage-λ system. (A) Schematic illustration of the genetic construct and its regulators. (B) Results of the stochastic simulations with synergistic (red) or additive (blue) regulation. See SI Appendix, Tables S2 and S3, for reactions and parameters.
We performed realistic stochastic simulations (35) of this circuit by using parameters taken from the literature (37–40). The reactions and parameters that we used can be found in SI Appendix, Tables S2 and S3. We expect these simulations to faithfully reflect the biological system because phage-λ is a well-studied system for which many parameters are measured; comparable models are capable of accurately reproducing distributions of protein concentrations in prokaryotic systems (33,41). To understand how the presence of additional regulators affects the noise signatures, we also include the phage-λ protein Cro which competes with cI for binding to the OR2 binding site and represses transcription from PRM when it is bound.
As expected from the theory (Eq. 5), the 3-point correlation function is always positive for the wild-type construct, which reflects the synergy between CRP and cI (Fig. 3B, red symbols). As more Cro is added, fluctuations in cI have smaller effects because the OR2 binding site is occupied more often by Cro. When the concentration of Cro is high, Cro is nearly always bound, and fluctuations in cI have almost no effect on the output. Consistent with the theoretical predictions, simulations with parameters corresponding to the pc mutant yielded 3-point correlation functions close to zero, which indicates that the activators function nearly independently in that case (Fig. 3B, blue symbols). Thus, the simulations show that synergistic and additive modes of combinatorial regulation can be distinguished by their noise signatures in a realistic situation. The dependence on the concentration of Cro in Fig. 3B shows that the approach is robust to fluctuations in additional, unmeasured species so long as they do not dominate the behavior of the gene.
Discussion
Recently, calculation of the noise properties of biological systems has received much attention. Most studies have focused on the variance of the protein concentration that has been calculated for models of single unregulated genes (26,27,32), simple gene circuits (32), cascades of genes (23), and signaling cascades (28,42,43). A result for the variance of the output of a gene regulated combinatorially by multiple transcription factors has also been reported recently (44), and this statistic, together with the half-autocorrelation time has been suggested to be useful for discriminating between competing models (45). In contrast, we have focused on the cross-correlations between the regulators and the gene product. It is these cross-correlations, rather than the variance, which contain the unambiguous signature of the mode of combinatorial regulation. In particular, we showed that the sign of the 3-point correlation function can distinguish AND- and OR-like logic operations in both idealized and realistic systems with 2 regulators. In the latter case, the theory led to an experimentally testable prediction: the 3-point correlation function will be positive for the natural construct and close to zero for the pc mutant. This prediction can be tested by using flow cytometry to measure the concentrations of the regulators and output (e.g., as in refs. 33 and 46). Although we have focused on the sign of the correlation function and that is independent of units, in some applications it may be useful to utilize a dimensionless quantity. By analogy with the Pearson pair correlation coefficient, the 3-point function can be normalized to give a value between −1 1 (see SI Appendix, Figs. S5 and S6).
Despite the apparent importance of combinatorial regulation (12–19), knowledge of it is presently limited to a few well-studied examples such as the mediation of the response to endocrine signaling by nonsteroid nuclear receptors (12) and the regulation of Oct genes by POU and SOX in pluripotent mammalian cells (13). We believe that the approach presented here will be useful for probing transcription factor synergies and thus expanding our understanding of combinatorial regulation. Beyond the analysis illustrated here for the bacterial construct, one can imagine combining knowledge of the signatures of combinatorial regulation with statistical inference techniques such as Bayesian methods (47). In such an approach, one could use existing statistical methods to characterize the casual relationships and identify genes thought to have more than one input. One could then use the 3-point correlation function to determine the degree to which each such node behaves synergistically. Finally, the casual relationships could be reanalyzed in a Bayesian framework by using conditional probability distributions that account for the combinatorial regulation (i.e., distributions with the appropriate third moments).
The applicability of the analysis in experimental situations is premised on the idea that variations in the output are related to the variations in the regulator levels measured at the same time. If there were a long lag in the detectable response of a gene in comparison with the rates at which the levels of the relevant transcription factors fluctuated within individual cells, single-time multipoint correlation functions would not be informative. However, fluctuations in protein levels in eukaryotic cells can persist for many days and over multiple cell generations (48), so it is likely that single-time measurements can be used in these systems as well as in prokaryotic systems where there are generally fewer layers of regulation between transcription and protein expression and longer lag times in responses are less common.
Although we have focused on the signatures of synergies between 2 regulators that are present in the 3-point correlation function, the general relationship (Eq. 3) shows that synergies between more regulators can be probed by measuring higher-order correlation functions. For example, by extending the 2 regulator model above to include an additional regulator, we found that the 4-point correlation function contains the signature of synergies which are not accounted for by pairwise and single transcription factor effects. Note, however, that the fact that the (j + 1)-point correlation function scales with the system size as means that higher moments will be more difficult to measure in larger systems. By using methods similar to that used to derive Eq. 3, we have also found specific signatures associated with cooperative activation by a single transcription factor and with different orderings of the genes in a cascade (A.W. and A.R.D., unpublished work). By extending the framework to treat systems out of steady state, one could use similar thinking to discover signatures present in the intrinsic noise of developing or even oscillatory systems. Thus, we believe that the general noise-based approach developed here will be applicable to identifying specific probes for a wide variety of biological regulatory situations.
Supplementary Material
Acknowledgments.
We thank Harinder Singh, Ben Gantner, Roger Sciammas, Lawrence Uricchio, and Mark Maienschein-Cline for helpful discussions and critical readings of the manuscript. This work was supported by the National Science Foundation and the Chicago Biomedical Consortium.
Footnotes
This article contains supporting information online at www.pnas.org/cgi/content/full/0809314105/DCSupplemental
References
- 1.Glass L, Kauffman SA. The logical analysis of continuous, non-linear biochemical control networks. J Theor Biol. 1973;39:103–129. doi: 10.1016/0022-5193(73)90208-7. [DOI] [PubMed] [Google Scholar]
- 2.Arkin A, Ross J. Computational functions in biochemical reaction networks. Biophys J. 1994;67:560–578. doi: 10.1016/S0006-3495(94)80516-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milo R, et al. Network motifs: Simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 4.Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- 5.Alon U. Network motifs: Theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 6.Setty Y, Mayo AE, Surette MG, Alon U. Detailed map of a cis-regulatory input function. Proc Natl Acad Sci USA. 2003;100:7702–7707. doi: 10.1073/pnas.1230759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhlman T, Zhang Z, Saier MH, Hwa T. Combinatorial transcriptional control of the lactose operon of Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6043–6048. doi: 10.1073/pnas.0606717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayo AE, Setty Y, Shavit S, Zaslaver A, Alon U. Plasticity of the cis-regulatory input function of a gene. PLoS Biol. 2006;4:555–561. doi: 10.1371/journal.pbio.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: Experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 10.Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 11.Hermsen R, Tans S, ten Wolde PR. Transcriptional regulation by competing transcription factor modules. PLoS Comp Biol. 2006;2:1552–1560. doi: 10.1371/journal.pcbi.0020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 13.Remenyi A, Scholer HR, Wilmanns M. Combinatorial control of gene expression. Nat Struct Mol Biol. 2004;11:812–815. doi: 10.1038/nsmb820. [DOI] [PubMed] [Google Scholar]
- 14.Buchler NE, Gerland U, Hwa T. On schemes of combinatorial transcription logic. Proc Natl Acad Sci USA. 2003;100:5136–5141. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.So CW, Cleary ML. Dimerization: A versatile switch for oncogenesis. Blood. 2004;104:919–922. doi: 10.1182/blood-2004-03-0992. [DOI] [PubMed] [Google Scholar]
- 16.Kato M, Hata N, Banerjee N, Futcher B, Zhang MQ. Identifying combinatorial regulation of transcription factors and binding motifs. Genome Biol. 2004;5:R56. doi: 10.1186/gb-2004-5-8-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothenberg EV, Taghon T. Molecular genetics of T cell development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 18.Lemaire V, Lee CF, Lei J, Métivier R, Glass L. Sequential recruitment and combinatorial assembling of multiprotein complexes in transcriptional activation. Phys Rev Lett. 2006;96:198102. doi: 10.1103/PhysRevLett.96.198102. [DOI] [PubMed] [Google Scholar]
- 19.Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 21.Vilar JM, Kueh HY, Barkai N, Leibler S. Mechanisms of noise resistance in genetic oscillators. Proc Natl Acad Sci USA. 2002;99:5988–5992. doi: 10.1073/pnas.092133899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 23.Pedraza J, van Oudenaarden A. Noise propagation in genetic networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 24.Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 25.Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 26.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 27.Pedraza JM, Paulsson J. Effects of molecular memory and bursting on fluctuations in gene expression. Science. 2008;319:339–343. doi: 10.1126/science.1144331. [DOI] [PubMed] [Google Scholar]
- 28.Thattai M, van Oudenaarden A. Attenuation of noise in ultrasensitive signaling cascades. Biophys J. 2002;82:2943–2950. doi: 10.1016/S0006-3495(02)75635-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulsson J. Summing up the noise in genetic networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- 30.Chandler D. Introduction to Modern Statistical Mechanics. New York: Oxford Univ Press; [Google Scholar]
- 31.van Kampen NG. Stochastic Processes in Physics and Chemistry. London: Elsevier Science; 1992. [Google Scholar]
- 32.Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guido NJ, et al. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 34.Bintu L, et al. Transcriptional regulation by the numbers: Applications. Curr Opin Gen Dev. 2005;15:125–135. doi: 10.1016/j.gde.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–2361. [Google Scholar]
- 36.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joung JK, Koepp DM, Hochschild D. Synergistic activation of transcription by bacteriophage λ cI protein and E. coli cAMP receptor protein. Science. 1994;265:1863–1866. doi: 10.1126/science.8091212. [DOI] [PubMed] [Google Scholar]
- 38.Aurell E, Brown S, Johanson J, Sneppen K. Stability puzzles in phage λ. Phys Rev E. 2002;65:051914. doi: 10.1103/PhysRevE.65.051914. [DOI] [PubMed] [Google Scholar]
- 39.Santillan M, Mackey MC. Why the lysogenic state of phage λ is so stable: a mathematical modeling approach. Biophys J. 2004;86:75–84. doi: 10.1016/S0006-3495(04)74085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santillan M, Mackey MC, Zeron ES. Origin of bistability in the lac operon. Biophys J. 2007;92:3830–3842. doi: 10.1529/biophysj.106.101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mettetal JT, Muzzey D, Pedraza JM, Ozbudak EM, van Oudenaarden A. Predicting stochastic gene expression dynamics in single cells. Proc Natl Acad Sci USA. 2006;103:7304–7309. doi: 10.1073/pnas.0509874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shibata T, Fujimoto K. Noisy signal amplification in ultrasensitive signal transduction. Proc Natl Acad Sci USA. 2005;102:331–336. doi: 10.1073/pnas.0403350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanase-Nicola S, Warren PB, ten Wolde PR. Signal detection, modularity, and the correlation between extrinsic and intrinsic noise in biochemical networks. Phys Rev Lett. 2006;97:068102. doi: 10.1103/PhysRevLett.97.068102. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez A, Kondev J. Transcriptional control of noise in gene expression. Proc Natl Acad Sci USA. 2008;105:5081–5086. doi: 10.1073/pnas.0707904105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cox CD, McCollum JM, Allen MS, Dar RD, Simpson ML. Using noise to probe and characterize gene circuits. Proc Natl Acad Sci USA. 2008;105:10809–10814. doi: 10.1073/pnas.0804829105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson LM, Yang H. DNA looping can enhance lysogenic cI transcription in phage lambda. Proc Natl Acad Sci USA. 2008;105:5827–5832. doi: 10.1073/pnas.0705570105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 48.Sigal A, et al. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.