Abstract
We used the model nematode Caenorhabditis elegans infected with the human pathogen Staphylococcus aureus to identify components of epithelial immunity. Transcriptional profiling and reverse genetic analysis revealed that mutation of the C. elegans β-catenin homolog bar-1 or the downstream homeobox gene egl-5 results in a defective response and hypersensitivity to S. aureus infection. Epistasis analysis showed that bar-1 and egl-5 function in parallel to previously described C. elegans immune-response pathways. Overexpression of human homologs of egl-5 modulated NF-κB-dependent TLR2 signaling in epithelial cells. These data suggest that β-catenin and homeobox genes play an important and conserved role in innate immune defense.
Keywords: homeobox protein, host-pathogen interactions, Staphylococcus aureus
The use of genetically tractable model hosts, including the nematode Caenorhabditis elegans, has aided the understanding of host immune defenses (1). A variety of human pathogens infect and kill C. elegans, including Pseudomonas aeruginosa, and Staphylococcus aureus, the latter being a growing major public health concern (1, 2). Previous studies have shown that two highly conserved nematode signaling pathways, the daf-2/daf-16 insulin/IGF1 pathway and the nsy-1/sek-1/pmk-1 p38 MAPK pathway, play significant roles in the survival of C. elegans during infection (3–6). The p38 MAPK pathway also plays a key role in the mammalian innate immune response (7).
C. elegans lack a circulatory system and known professional immune cells. The principal site of interaction with ingested pathogenic microbes is the lumen of the gut, which is composed of 20 nonrenewable intestinal epithelial cells. Identification and characterization of the components of the C. elegans immune response is being actively pursued, in part by monitoring gene expression changes in response to infection (6, 8–10). Recent work has characterized the C. elegans transcriptional response to infection by the Gram-negative pathogen P. aeruginosa, and highlighted the role of the nsy-1/sek-1/pmk-1 pathway during that response (6, 10).
In the work reported here, we find a key role for the evolutionarily conserved transcriptional cofactor bar-1/β-catenin in C. elegans intestinal epithelial immunity to S. aureus. bar-1/β-catenin was previously implicated in a number of developmental processes in C. elegans as a component of signaling pathways downstream of Wnt secreted glycoprotein ligands (11, 12). We also report here that part of the bar-1-dependent response to S. aureus is mediated by the effector transcription factor EGL-5, a homeobox protein homologous to the Drosophila gene AbdominalB. C. elegans egl-5 controls the development of the posterior end of the animal downstream of Wnt signaling (13–15). Importantly, we also show that human homologs of egl-5, which are induced upon S. aureus infection in human cells, modulate the activity of the innate immune regulator NF-κB.
Results
DAF-16 and PMK-1 are Not Required for the Induction of Selected S. aureus-Responsive Genes.
In experiments described elsewhere, we used Affymetrix whole-genome C. elegans microarrays to identify 186 genes whose transcription was induced in the early response to S. aureus infection, many of which encode proteins predicted to have antimicrobial and detoxification activities (Irazoqui et al., manuscript in preparation). We chose 10 of these S. aureus-responsive genes that represented different levels of up-regulation, different degrees of pathogen specificity, and different predicted antimicrobial functions, and verified their induction by using quantitative real-time PCR (qRT-PCR, Fig. 1 A and C).
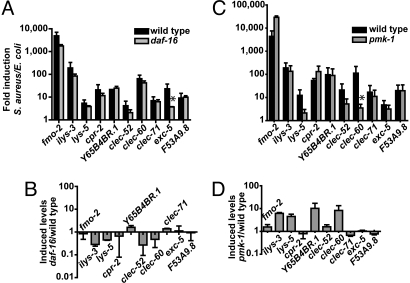
Fig. 1.
Mutation of daf-16 or pmk-1 does not affect the induction of selected S. aureus-induced genes. qRT-PCR analysis showing fold induction (A) and final induced levels (B) after 8 h S. aureus infection in daf-16(mgDf47) mutant animals, and fold induction (C) and final induced levels (D) in pmk-1(km25) mutant animals. Data are the means of two biological replicates, each replicate measured in duplicate and normalized to a control gene, expressed as the ratio of the corresponding S. aureus-induced levels and the basal E. coli levels (A and C; a fold induction of 1 indicates no induction) or expressed as the ratio of the corresponding S. aureus-induced levels in the mutant and the wild type (B and D, a ratio of 1 indicates no difference with wild type). Top and bottom panels are from the same time points. Error bars, SEMs. Asterisks represent P ≤ 0.05.
We used the set of 10 selected S. aureus-induced genes as biomarkers to determine whether two previously identified immune-response pathways, the pmk-1 p38 MAPK pathway and the daf-2/insulin receptor–daf-16/FoxO insulin pathway, are involved in the C. elegans response to S. aureus. Both of these pathways are important for the host response to P. aeruginosa, but appear to act in parallel (6). daf-16 mutants were not affected in the fold induction of any of the 10 selected biomarker genes, except exc-5 (Fig. 1A), which was constitutively de-repressed in daf-16 mutants (data not shown). Moreover, there were no statistically significant differences in induced levels between wild-type and daf-16 animals (Fig. 1B). These data are similar to previous results showing daf-16 independence of P. aeruginosa-induced genes (6) and consistent with the fact that three different daf-16 mutants are no more susceptible to S. aureus killing than wild type ((4), data not shown). Furthermore, the daf-16 target genes lys-7, sod-3, and mtl-1 were not induced upon S. aureus infection and DAF-16 failed to translocate into the nucleus during S. aureus (or P. aeruginosa) infection [data not shown; (6, 10)], suggesting that DAF-16 is not involved in the host response to these pathogens.
Because nsy-1, sek-1, and pmk-1 mutants, corresponding to three kinases in the p38 MAPK cascade, are more susceptible to S. aureus infection than wild-type animals [(5); also see below], it was surprising that mutation of pmk-1 did not significantly affect the fold induction (except clec-60, Fig. 1C), or the induced levels (Fig. 1D) of most of the 10 biomarker genes compared with wild type.
Role of BAR-1/β-catenin and the HOX Gene egl-5 in the Host Response to S. aureus.
Because of the potential for cross-talk between immune- and stress-response pathways, our laboratory has explored the role of various stress-related signaling pathways in the C. elegans immune response (16). The β-catenin gene bar-1 was recently shown to be involved in the C. elegans response to oxidative stress (17). As an initial step to identify novel immunity-related pathways, we tested the induction of the 10 S. aureus-activated biomarker genes described above in a bar-1 mutant background. We found that for seven of the 10 biomarker genes, the fold induction in bar-1 mutants was lower compared with wild type (Fig. 2A). This defective response resulted in dramatically lower induced transcript levels in the bar-1 mutant for six of the 10 biomarker genes (Fig. 2B). The lack of induction of clec-60 and F53A9.8 was also observed using bar-1 RNAi to knock down bar-1 expression in GFP transcriptional reporter strains (Fig. 2 E and F, data not shown). Consistent with the gene expression defect shown in Fig. 2, bar-1 mutant animals were significantly more sensitive to S. aureus-mediated killing than wild type (Fig. 3A), and S. aureus-elicited anal swelling was absent in bar-1 mutants (data not shown). The requirement for bar-1 activity appeared to be at least in part post-developmental, because RNAi-mediated knockdown of bar-1 in adult animals caused a modest but significant increase in susceptibility to S. aureus-mediated killing (see supporting information (SI) Fig. S1D). Moreover, a functional BAR-1::GFP fusion construct (18) was transiently induced in the posterior gut early during S. aureus infection (Fig. S1 A–C).
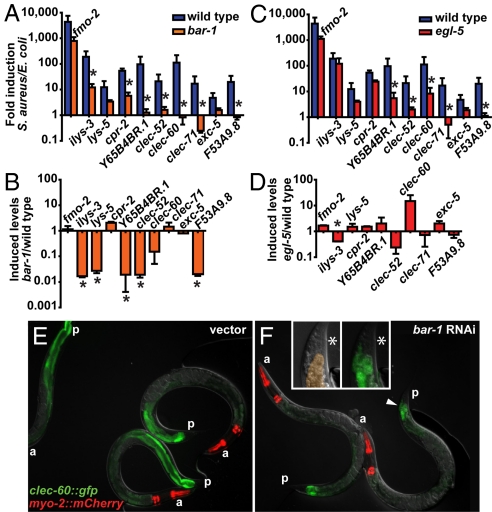
Fig. 2.
bar-1 and egl-5 are required for the host response to S. aureus infection. qRT-PCR analysis of wild type and mutant animals infected with S. aureus for 8 h. (A) Fold induction levels and (B) final expression levels after infection in bar-1(ga80) versus wild type. (C) Fold induction levels and (D) final expression levels after infection in egl-5(n486) versus wild type. Asterisks denote P ≤ 0.05. Top and bottom panels are from the same time points. (E and F) Induction of clec-60::gfp is bar-1-dependent. clec-60::gfp animals were treated with empty vector (E) or bar-1 RNAi (F) and subsequently infected during 12 h with S. aureus. Inserts show a higher magnification of the area indicated with an arrowhead. Left panel, DIC micrograph showing intestinal cells highlighted in orange. Right panel, overlay DIC and fluorescence micrograph showing GFP leakage into the body cavity. Asterisk indicates the anus. (a) anterior end. (p) posterior end.
Fig. 3.
bar-1 and egl-5 mutants are more susceptible to S. aureus-mediated killing than wild type. (A) S. aureus-mediated killing. bar-1 med. surv. = 33 h, n = 102, P < 0.0001; wild type med. surv. = 51 h, n = 105. (B) S. aureus-mediated killing. egl-5 med. surv. = 36 h, n = 98, P < 0.0001; wild type med. surv. = 60 h, n = 99. (C) S. aureus killing after knockdown of cdc-25 to prevent progeny production. Wild type med. surv. = 87 h, n = 94; bar-1 med. surv. = 51 h, n = 100/12, P < 0.0001; egl-5 med. surv. = 51 h, n = 113, P < 0.0001. (D) Lifespan on heat-killed P. aeruginosa PA14 with FUDR. Wild type med. surv. = 12.83 d, n = 91/5; egl-5 = 14.83 d, n = 98, P < 0.0001; bar-1 = 13.88 d, n = 89, P = 0.0079; pmk-1 = 12.83 d, n = 105, P = 0.3211; daf-16 = 11.96 d, n = 100, P < 0.0001 for comparison with wild type.
To determine whether the transcriptional activity of bar-1 is required for defense, we examined loss of function mutants in three C. elegans homeobox (HOX) transcription factor genes, lin-39, mab-5, and egl-5, previously identified as downstream mediators of bar-1-dependent signaling (18, 19). egl-5 null mutants (but not lin-39 or mab-5 mutants) were more susceptible to S. aureus killing than wild type (Fig. 3B and Fig. S2 A and B) and exhibited reduced induction of five of the 10 biomarker genes (Fig. 2C). However, the induction defect in the mutant resulted in statistically significant decreased induced expression only of ilys-3 (and maybe clec-52, Fig. 3D), because of higher constitutive expression of the other four genes in the mutant (data not shown).
Pathogenicity-Related Phenotypes of bar-1 and egl-5 Mutants.
Mutants with defects in pharyngeal grinding or defecation exhibit increased susceptibility to killing by P. aeruginosa (20). In contrast, the susceptibility of bar-1 and egl-5 mutants to S. aureus is not likely a consequence of a grinding defect or constipation. bar-1 and egl-5 mutant animals variably accumulated S. aureus more rapidly than wild type in their intestinal lumina (Fig. S3 A–C), but did not accumulate E. coli (Fig. S3D). Defecation-defective mutants aex-2(sa3) and unc-25(e156) were not more susceptible than wild type to S. aureus infection (Fig. S2C), and aex-2(sa3) did not accumulate more S. aureus than wild type (data not shown).
Epifluorescence and transmission electron microscopy of bar-1 mutants infected with S. aureus revealed leakage of cytoplasmic contents into the body cavity (Fig. 2F Inset) and severely damaged intestinal cells (Fig. 4A), suggesting more severe S. aureus-elicited gut degradation than in wild type (Fig. 4E). The microvilli in bar-1 animals were significantly shortened or were completely absent on the intestinal cell apical surface. In contrast to wild type, we did not observe lysed bacterial cells in the intestinal lumen in the bar-1 mutants; on the contrary, the bacteria appeared to be undergoing active division, as evidenced by readily observable septa (Fig. 4A). These phenotypes were not a consequence of gross malformations of the intestine in the bar-1 mutants, because bar-1 animals feeding on E. coli exhibited a normal overall intestinal ultrastructure (Fig. 4B).
Fig. 4.
bar-1 mutant animals exhibit more severe degradation of intestinal cells 12 h after infection with S. aureus. Transmission electron micrographs (TEM) of transversal midbody sections of (A) a bar-1(ga80) mutant animal infected with S. aureus, (B) an E. coli-fed bar-1(ga80) mutant animal, (C) a pmk-1(km25) mutant animal infected with S. aureus, (D) an E. coli-fed pmk-1(km25) mutant animal, (E) a wild-type animal infected with S. aureus for 12 h, and (F) an E. coli-fed wild type animal. In all E. coli-fed controls, occasionally bacterial cells were observed in the lumen mostly occupied by debris. MV, Microvilli; IC, intestinal cells; TW, terminal web; A, live and D, dead bacterial cells; L, intestinal lumen. (Scale bars, 500 nm.)
Interestingly, the phenotype of infected bar-1 mutants was more severe than that of infected pmk-1 mutants (compare Fig. 4 A and C). In contrast to bar-1 animals, pmk-1 animals contained lysed as well as live dividing bacterial cells. This correlates with the observation that bar-1 and egl-5 mutants were more susceptible to S. aureus than pmk-1 mutants (Fig. 5 A and B). This relationship was reversed during infection by P. aeruginosa (Fig. S2D), indicating that the pmk-1/p38 MAPK pathway is more important for conferring resistance to P. aeruginosa than it is for conferring resistance to S. aureus, and vice versa for the bar-1/egl-5 pathway.
Fig. 5.
bar-1, egl-5 function in parallel to pmk-1. (A and B) S. aureus killing assays. (A) Med. surv. pmk-1;bar-1 = 27 h, n = 88/6; bar-1 = 41 h, n = 89/15, P < 0.0001; pmk-1 = 41 h, n = 93/2, P < 0.0001 for comparison with double mutant. Although slightly in this assay, bar-1 mutants were significantly more susceptible than pmk-1 mutants in most assays (data not shown). (B) Med. surv. egl-5;pmk-1 = 27 h, n = 91/5; egl-5 = 32 h, n = 90/2, P < 0.0001; pmk-1 = 41 h, n = 93/2, P < 0.0001 for comparison with double mutant). Wild type and pmk-1 data are the same as in A. (C) Western blot of whole animal lysates from wild type (WT), pmk-1(km25), bar-1(ga80), and egl-5(n486) animals. Similar results as those obtained with the pmk-1 mutant were also obtained with the sek-1(ag1) mutant, as previously published (data not shown). Left, anti-phospho-p38 immunoblot. Right, total PMK-1 immunoblot using anti-PMK-1 antiserum. Arrows indicate the 44 kDa band corresponding to PMK-1. Other bands are nonspecific background. (D) qRT-PCR in wild type, bar-1(ga80), egl-5(n486), and pmk-1(km25) strains, normalized to a control gene, and presented as ratios with wild type. Asterisks indicate P ≤ 0.05.
In previous studies, it was noted that many bacterial pathogens elicit an egg-laying-defective (Egl) phenotype, causing some eggs to hatch within the mother's uterus and resulting in matricide, known as “bagging” (3, 5). However, the enhanced susceptibility of bar-1 and egl-5 mutants to pathogen infection is not simply a consequence of constitutive bagging. Both egl-5 and bar-1 mutants were more susceptible than wild type to P. aeruginosa-mediated killing in the presence of FUDR, which blocks embryonic development and allows the evaluation of phenotypes masked by bagging [Fig. S2D, (21)]. Because FUDR inhibits S. aureus growth and prevents S. aureus-mediated killing, for S. aureus killing assays we blocked bagging by sterilizing bar-1 and egl-5 worms with cdc-25 RNAi (10) and found that they were significantly more susceptible to S. aureus killing than equally treated wild type (Fig. 3C). Conversely, RNAi knockdown of bar-1 in several sterile mutant backgrounds, including spe-9;fer-15, caused markedly increased susceptibility to S. aureus (Fig. S2E). Finally, the decreased lifespan of bar-1 and egl-5 mutants on nonpathogenic E. coli because of bagging (Fig. S4A) was rescued almost entirely in the presence of FUDR (Fig. S4B) and bar-1 and egl-5 mutants had approximately wild-type longevity when feeding on heat-killed P. aeruginosa in the presence of FUDR (Fig. 3D). These experiments confirm that the egg-laying defect in bar-1 and egl-5 mutants is separable from their defect in innate immunity.
Interaction of the BAR-1 and DAF-2 Pathways.
We next used epistasis analysis to determine whether bar-1 and egl-5 are components of the previously studied insulin DAF-2/DAF-16 pathway. bar-1 RNAi completely suppressed the daf-2(-) resistance to S. aureus down to the level of the bar-1 RNAi-treated wild type (Fig. S4C), suggesting that bar-1 may function downstream of daf-2. However, because loss of daf-16 by mutation or RNAi only suppressed daf-2 resistance to the level of wild type [Fig. S4C, (4)], because mutation of bar-1 or egl-5 did not cause major changes in daf-16 target gene expression in a daf-2(+) background (Fig. S4D), and because daf-16 did not affect the induction of S. aureus-response genes (Fig. 1 A and B), we conclude that it is unlikely that bar-1 and egl-5 function in a linear pathway with daf-16. This conclusion is similar to previously published data showing that although loss of pmk-1 suppressed the resistance phenotype of daf-2 mutants, pmk-1 and daf-2/daf-16 function in parallel pathways (6). As in the case of pmk-1, it is possible that activation of other defense pathways is insufficient for resistance to S. aureus in the absence of bar-1.
BAR-1 and EGL-5 Act in Parallel to PMK-1/p38 MAPK.
We also carried out epistasis analysis to determine whether bar-1 and egl-5 function as part of the tir-1/nsy-1/sek-1/pmk-1 p38 MAPK signaling pathway. pmk-1;bar-1 double mutants were more susceptible than either single mutant (Fig. 5A). Likewise, the phenotype of the egl-5;pmk-1 double mutant was stronger than that of either single mutant (Fig. 5B). These results suggest that bar-1 and egl-5 act in parallel to the pmk-1 pathway.
Further support for this conclusion was provided by analysis of PMK-1 activation via phosphorylation (3). Significantly, loss of neither bar-1 nor egl-5 caused a significant change in PMK-1 activation in whole animal lysates, suggesting that bar-1 and egl-5 do not act upstream of pmk-1 (Fig. 5C). As an additional measure of PMK-1 activity, we determined the transcript levels of several genes that require PMK-1 for expression (6). Consistent with the western blot analysis, the expression levels of these genes were reduced in the pmk-1 but not the bar-1 and egl-5 mutants (Fig. 5D). Furthermore, loss of bar-1 or egl-5 did not affect the P. aeruginosa-elicited induction of pmk-1-dependent genes (C32H11.1, F49F16, and K08D8.5), but modestly affected the pmk-1-independent gene T24C4.4 (Fig. S4E). These results suggest that neither bar-1 nor egl-5 are required for signaling downstream of pmk-1. It is also unlikely that pmk-1 functions upstream of bar-1 and egl-5, because loss of pmk-1 did not affect the S. aureus-elicited induction of any bar-1- or egl-5-dependent genes (Figs. 1D and 3 A and C). Thus, we conclude that bar-1/egl-5 function in parallel to pmk-1 during the response to infection.
Human Homologs of EGL-5 Regulate NF-κB Signaling.
To identify human homologs of EGL-5 potentially associated with innate immunity in the context of S. aureus infection, we evaluated publicly available microarray data of human polymorphonuclear neutrophils (PMNs) infected with S. aureus in vitro (22). We found that S. aureus infection induced a number of key genes of the inflammatory and innate immune responses, including proinflammatory cytokines and chemokines (e.g., IL1B, IL8, IL12B, and TNF) and components of the Toll (e.g., TLR2, IRAK1, NFKBIA, and NFKB1) and MAP kinase pathways (e.g., MAP2K3 and DUSP3) (Fig. S5A). Additionally, a number of Wnt signaling genes (e.g., WNT2B, FZD3, FZD6, TCF2, TCF4, TCF7L1, and TCFL5) and HOX genes (HOXA5, HOXA9, HOXB5, HOXB9, HOXC10, and HOXD1) exhibited similar expression profiles (Fig. S5A). Network analysis using protein interaction data suggested the possible involvement of the EGL-5 homolog HOXA9 in the Toll-like receptor 2 (TLR2) signaling pathway, which is essential for S. aureus recognition (Fig. S5B).
To determine the function of HOX expression in TLR2 signaling, we measured Pam3csk4-elicited NF-κB activation in human epithelial-TLR2 reporter cell lines expressing HOXA9 (see Methods). Overexpression of HOXA9 inhibited Pam3csk4-induced transcription of an NF-κB reporter (Fig. 6A) and IL-8 (Fig. 6B). This suggested that HOXA9 may dampen NF-κB -dependent TLR2 signaling, consistent with a recent report (23). Furthermore, the induction of the NF-κB reporter by constitutively active IKKβ was inhibited by HOXA9 in a dose-dependent manner (Fig. 6C), suggesting that HOXA9 functions at the level of or downstream of NF-κB. We observed similar results with HOXC10 (Fig. 6C), showing the potential for multiple egl-5 homologues to modulate NF-κB redundantly. These data suggest that multiple human HOX proteins may play a role in the innate immune response to S. aureus infection as modulators of NF-κB -dependent transcription.
Fig. 6.
Up-regulation of HOX genes inhibits NF-κB -dependent TLR2 signaling. Overexpression of human HOXA9 inhibits Pam3csk4-elicited NF-κB reporter (A) and IL-8 (B) induction. HEK293-TLR2 cells transfected with NF-κB reporter vector were stimulated (or mock-stimulated) with Pam3csk4 for 6 h. Data are mean values of relative luciferase activity normalized to Renilla luciferase acitivity from duplicate experiments ± SD (A) or mean values from qRT-PCR measurements of IL-8 transcript levels (relative to GAPDH and the unstimulated vector control) from duplicate experiments ± SD. (C) Overexpression of human HOXA9 or HOXC10 inhibits NF-κB activity induced by constitutively active IKKβ. HEK293 cells were transfected with 3xFLAG-HOXA9 or HOXC10, constitutively active IKKβ, NF-κB-luciferase (NF-κB-luc), and empty vectors as indicated. All samples were also transfected with Renilla luciferase vector for normalization. Bottom panel, western blot detection of 3xFLAG-tagged HOXA9 and HOXC10, FLAG-IKKβ, and actin in lysates used for the reporter assay.
Discussion
We show here that bar-1/β-catenin, which is involved in many processes during C. elegans development, is also required postdevelopmentally to induce genes that respond to S. aureus infection. egl-5, a HOX gene previously shown to act downstream of bar-1 in the development of the nematode posterior end, is also required for the proper response to S. aureus infection. Mutation of bar-1 or egl-5 results in enhanced susceptibility to pathogenic attack. Importantly, bar-1 and egl-5 appear to operate in a novel pathway that is distinct from the C. elegans daf-2/daf-16 insulin and the tir-1/pmk-1 p38 MAPK signaling pathways.
Although the molecular mechanism underlying bar-1 and egl-5 function during the response to infection is unknown, one possibility is that bar-1-mediated transcriptional activation of immune effector genes, including egl-5, results directly in enhanced resistance. bar-1 RNAi in adult animals caused modest sensitivity to S. aureus and BAR-1 protein levels increased transiently in adults upon infection, supporting this view. Unfortunately, heat-shock induced overexpression of bar-1 or egl-5 negatively affects viability, precluding a direct experimental test of this hypothesis [data not shown, (15)]. Alternatively, bar-1/egl-5 could be involved in the development of a tissue that is required for the detection of S. aureus infection. These two scenarios are not mutually exclusive.
When comparing biomarker expression on S. aureus vs. E. coli, the profiles of the bar-1 and egl-5 mutants are similar (Fig. 2 A and C), which supports the notion that they function in the same pathway. The levels of most biomarker genes were strikingly reduced in bar-1 mutants exposed to S. aureus (Fig. 2B), but were relatively unaffected in egl-5 animals (Fig. 2D), even though bar-1 and egl-5 animals are equally susceptible to S. aureus. The reason for this apparent discrepancy is that most of these biomarker genes are constitutively derepressed in the egl-5 mutant (data not shown), suggesting that egl-5 may act as a negative regulator, although it is also possible that egl-5 is required for the expression of immune effectors not included in our biomarker set. These complex gene expression patterns imply sophisticated regulatory mechanisms and likely involve several signaling pathways acting in concert (Fig. S6). Recent work showed that egl-5 is required for the swelling response to the nematode-specific pathogen Microbacterium nematophilum, although the molecular mechanism of egl-5 action remains unclear (24). This result is in agreement with our results showing an important role for egl-5 in the host response to infection.
We found that a BAR-1::GFP translational fusion is faintly expressed throughout the body, with highest expression in the posterior gut, consistent with reports that egl-5 also is expressed and functions in the posterior end (14, 19, 25). Interestingly, the three bar-1-dependent biomarker genes we tested (clec-52, -60, and F53A9.8) are more highly expressed in the posterior intestine. Thus, bar-1/egl-5 may act in response to S. aureus infection in a local manner to induce the expression of genes in the posterior end of the animal. Unfortunately, our attempts to determine the tissue-specific requirements of bar-1 and egl-5 in immunity by rescuing the bar-1 mutant using various tissue-specific promoters have so far been unsuccessful, despite clear rescue of the vulval morphogenesis phenotype by ectopic and hypodermal expression constructs (data not shown).
Seventy percent of 186 genes up-regulated by S. aureus infection did not overlap with genes up-regulated during either P. aeruginosa or M. nematophilum infection (Irazoqui et al. manuscript in preparation). This indicates that a large component of the transcriptional host response to infection is pathogen-specific, consistent with recent reports using other approaches (8, 26). Our data show a correlation between the severity of host-response defects and susceptibility to killing, such that bar-1 and egl-5 mutants are more susceptible to S. aureus, whereas pmk-1 mutants are more susceptible to P. aeruginosa. Thus, C. elegans may defend itself against infection via pathogen-specific defense response pathways (Fig. S6).
Recent work has shown that β-catenin is transiently activated in human epithelia after stimulation by bacteria (27, 28), but the functional consequences of this induction remain unclear. Our work and that of others show that the long-term stabilization of β-catenin can lead to down-regulation of NF-κB, causing the loss of expression of some NF-κB target genes; for example, IL-8 and κB-element based reporters, but not others (e.g., TRAF1, FAS, and CRP) [data not shown, (28, 29)]. Moreover, the data from our C. elegans model suggest the involvement of β-catenin in the regulation of pathogen-response genes independently of NF-κB [C. elegans does not have an NF-κB ortholog (30)] and recent evidence suggests this may also be the case in mammalian intestinal epithelia (31–34).
Like egl-5 in nematodes, HOX proteins downstream of β-catenin (35) may be required for the induction of pathogen-response genes in humans. Given the greatly expanded HOX gene clusters in humans, it is possible that several HOX proteins exert this function redundantly. We show that HOXA9 and HOXC10, two HOX proteins induced by stimulation with S. aureus, can modulate NF-κB in human epithelial cells, consistent with recent reports (36). Previous data also show that HOXA9 is required for the expression of immunity-related genes (37, 38).
Finally, because a fully functional NF-κB pathway is absent in nematodes and because nematodes and humans share a conserved immune function for β-catenin and HOX signaling, we hypothesize that β-catenin-dependent innate immune pathways may have evolved earlier than NF-κB signaling pathways. Subsequently, with the acquisition of the TLR/NF-κB axis in arthropods, the β-catenin/HOX pathway may have been recruited to modulate NF-κB signaling.
Materials and Methods
Strains.
C. elegans strains used in this study are detailed in Table S1. Bacterial strains are detailed in Table S2.
Infection and Lifespan Assays.
S. aureus and P. aeruginosa killing assays were conducted at least twice, as described (5, 6). P. aeruginosa assays were supplemented with 80–100 μg/ml 5-fluorodeoxyuridine (FUDR, Sigma). Data are median survival, N (number of deaths/censored) and P value. A P value less than or equal to 0.05 was considered significantly different from control. Lifespan assays were performed at least twice, as described (6). For experiments using heat-killed PA14, overnight cultures of bacteria were concentrated and incubated at 95°C for 30 min. Plates were supplemented with 80–100 μg/ml FUDR and 100 μg/ml kanamycin.
Quantitative RT-PCR Analysis.
Synchronized C. elegans animals were treated essentially as for killing assays described above, omitting FUDR. For S. aureus assays, infected samples were compared with parallel samples feeding on heat-killed E. coli OP50 on the same medium. Total RNA was extracted as described (6) and reverse transcribed by using the SuperScript III kit (Invitrogen). cDNA was subjected to qRT-PCR analysis as described (6). Primer sequences are detailed in Table S3. All values are normalized against the control gene snb-1, which did not vary under conditions being tested. Fold change was calculated by using the Pfaffl method (39). One-sample t tests were performed by using Graphpad Prism 4. A P value less than or equal to 0.05 was considered significant. Total HEK293-TLR2 RNA was reverse-transcribed by using the iScript cDNA synthesis kit (Bio-Rad). Real-time quantitative PCR was performed in the iQ5 detection system (Bio-Rad) by using the iQ-SYBR Green supermix (Bio-Rad). GAPDH was used for normalization.
clec-60 GFP Fusion.
PCR was used to amplify 1,665 bp of DNA upstream of the clec-60 start site. Details of construction can be found in the SI Methods.
RNAi Knockdown.
RNAi was carried out by using bacterial feeding RNAi (see SI Methods for further details).
Western Blot Analysis of PMK-1 Activation.
Synchronized L4 populations of N2 wild type, bar-1(ga80), egl-5(n486), and pmk-1(km25) animals were treated as in (3). Western blot analyses of PMK-1 and activated p38 MAPK were as described (3).
NF-κB Reporter Assays.
Experimental details can be found in the SI Methods.
Supplementary Material
Acknowledgments.
We thank E. Troemel, L. Stanek, S. Margolis, and D. Colón-Ramos for critical reading of the manuscript and J. Powell for help designing qRT-PCR primers. TEM was performed by M. McKee in the Microscopy Core of the Center for Systems Biology supported by National Institutes of Health Grants DK43351 and DK57521. Nematode strains were provided by the Caenorhabditis Genetics Center funded by the National Institutes of Health National Center for Research Resources. This work was funded by National Institutes of Health Grants AI062773 (to R.J.X.) and AI064332 (to F.M.A.) and by postdoctoral fellowships from the Jane Coffin Childs Memorial Fund for Medical Research and the Charles A. King Trust, Bank of America, Co-Trustee (Boston, MA) (to J.E.I.), and from the Crohn's and Colitis Foundation of America (to A.N.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809527105/DCSupplemental.
References
- 1.Sifri CD, Begun J, Ausubel FM. The worm has turned–microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Elston DM. Community-acquired methicillin-resistant Staphylococcus aureus. J Am Acad Dermatol. 2007;56:1–16. doi: 10.1016/j.jaad.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 3.Kim DH, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 4.Garsin DA, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 5.Sifri CD, Begun J, Ausubel FM, Calderwood SB. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect Immun. 2003;71:2208–2217. doi: 10.1128/IAI.71.4.2208-2217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuzawa A, et al. ROS-dependent activation of the TRAF6-ASK1–p38 pathway is selectively required for TLR4-mediated innate immunity. Nat Immunol. 2005;6:587–592. doi: 10.1038/ni1200. [DOI] [PubMed] [Google Scholar]
- 8.Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, Tan MW. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci USA. 2006;103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenmann DM The C. elegans Research Community, editor. Wnt signaling. WormBook. 2005 doi: 10.1895/wormbook. 1.7.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenmann DM, Kim SK. Protruding vulva mutants identify novel loci and Wnt signaling factors that function during Caenorhabditis elegans vulva development. Genetics. 2000;156:1097–1116. doi: 10.1093/genetics/156.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira HB, Zhang Y, Zhao C, Emmons SW. Patterning of Caenorhabditis elegans posterior structures by the Abdominal-B homolog, egl-5. Dev Biol. 1999;207:215–228. doi: 10.1006/dbio.1998.9124. [DOI] [PubMed] [Google Scholar]
- 14.Chamberlin HM, Brown KB, Sternberg PW, Thomas JH. Characterization of seven genes affecting Caenorhabditis elegans hindgut development. Genetics. 1999;153:731–742. doi: 10.1093/genetics/153.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang LI, Sternberg PW. Interactions of EGF, Wnt and HOM-C genes specify the P12 neuroectoblast fate in C. elegans. Development. 1998;125:2337–2347. doi: 10.1242/dev.125.12.2337. [DOI] [PubMed] [Google Scholar]
- 16.Kim D, et al. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc Natl Acad Sci USA. 2004;101:10990–10994. doi: 10.1073/pnas.0403546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- 18.Eisenmann DM, Maloof JN, Simske JS, Kenyon C, Kim SK. The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development. 1998;125:3667–3680. doi: 10.1242/dev.125.18.3667. [DOI] [PubMed] [Google Scholar]
- 19.Maloof JN, Whangbo J, Harris JM, Jongeward GD, Kenyon C. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development. 1999;126:37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- 20.Shapira M, Tan M. Genetic Analysis of Caenorhabditis elegans Innate Immunity. Methods Mol Biol. 2008;415:429–442. doi: 10.1007/978-1-59745-570-1_25. [DOI] [PubMed] [Google Scholar]
- 21.Shaw WM, Luo S, Landis J, Ashraf J, Murphy CT. The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Curr Biol. 2007;17:1635–1645. doi: 10.1016/j.cub.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borjesson DL, Kobayashi SD, Whitney AR, Voyich JM, Argue CM, Deleo FR. Insights into pathogen immune evasion mechanisms: Anaplasma phagocytophilum fails to induce an apoptosis differentiation program in human neutrophils. J Immunol. 2005;174:6364–6372. doi: 10.4049/jimmunol.174.10.6364. [DOI] [PubMed] [Google Scholar]
- 23.Trivedi CM, Patel RC, Patel CV. Homeobox gene HOXA9 inhibits nuclear factor-kappa B dependent activation of endothelium. Atherosclerosis. 2007;195:e50–60. doi: 10.1016/j.atherosclerosis.2007.04.055. [DOI] [PubMed] [Google Scholar]
- 24.Gravato-Nobre MJ, et al. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2005;171:1033–1045. doi: 10.1534/genetics.105.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chisholm A. Control of cell fate in the tail region of C. elegans by the gene egl-5. Development. 1991;111:921–932. doi: 10.1242/dev.111.4.921. [DOI] [PubMed] [Google Scholar]
- 26.Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol. 2007;27:5544–5553. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar A, et al. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J. 2007;26:4457–4466. doi: 10.1038/sj.emboj.7601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, et al. Crosstalk between NF-kappaB and beta-catenin pathways in bacterial-colonized intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2005;289:G129–137. doi: 10.1152/ajpgi.00515.2004. [DOI] [PubMed] [Google Scholar]
- 29.Deng J, et al. beta-catenin interacts with and inhibits NF-kappa B in human colon and breast cancer. Cancer Cell. 2002;2:323–334. doi: 10.1016/s1535-6108(02)00154-x. [DOI] [PubMed] [Google Scholar]
- 30.Kurz CL, Ewbank JJ. Caenorhabditis elegans: An emerging genetic model for the study of innate immunity. Nat Rev Genet. 2003;4:380–390. doi: 10.1038/nrg1067. [DOI] [PubMed] [Google Scholar]
- 31.Cavard C, et al. Overexpression of regenerating islet-derived 1 alpha and 3 alpha genes in human primary liver tumors with beta-catenin mutations. Oncogene. 2006;25:599–608. doi: 10.1038/sj.onc.1208860. [DOI] [PubMed] [Google Scholar]
- 32.van Es JH, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 33.Andreu P, et al. Crypt-restricted proliferation and commitment to the Paneth cell lineage following Apc loss in the mouse intestine. Development. 2005;132:1443–1451. doi: 10.1242/dev.01700. [DOI] [PubMed] [Google Scholar]
- 34.Wehkamp J, et al. The Paneth cell alpha-defensin deficiency of ileal Crohn's disease is linked to Wnt/Tcf-4. J Immunol. 2007;179:3109–3118. doi: 10.4049/jimmunol.179.5.3109. [DOI] [PubMed] [Google Scholar]
- 35.Klapholz-Brown Z, Walmsley GG, Nusse YM, Nusse R, Brown PO. Transcriptional program induced by Wnt protein in human fibroblasts suggests mechanisms for cell cooperativity in defining tissue microenvironments. PLoS ONE. 2007;2:e945. doi: 10.1371/journal.pone.0000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trivedi CM, Patel RC, Patel CV. Differential regulation of HOXA9 expression by nuclear factor kappa B (NF-kappaB) and HOXA9. Gene. 2008;408:187–195. doi: 10.1016/j.gene.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Bei L, Lu Y, Eklund EA. HOXA9 activates transcription of the gene encoding gp91Phox during myeloid differentiation. J Biol Chem. 2005;280:12359–12370. doi: 10.1074/jbc.M408138200. [DOI] [PubMed] [Google Scholar]
- 38.Bandyopadhyay S, Ashraf MZ, Daher P, Howe PH, DiCorleto PE. HOXA9 participates in the transcriptional activation of E-selectin in endothelial cells. Mol Cell Biol. 2007;27:4207–4216. doi: 10.1128/MCB.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.