Abstract
Retrieving a large amount of genetic information from extinct species was demonstrated feasible, but complete mitochondrial genome sequences have only been deciphered for the moa, a bird that became extinct a few hundred years ago, and for Pleistocene species, such as the woolly mammoth and the mastodon, both of which could be studied from animals embedded in permafrost. To enlarge the diversity of mitochondrial genomes available for Pleistocene species, we turned to the cave bear (Ursus spelaeus), whose only remains consist of skeletal elements. We collected bone samples from the Paleolithic painted cave of Chauvet-Pont d'Arc (France), which displays the earliest known human drawings, and contains thousands of bear remains. We selected a cave bear sternebra, radiocarbon dated to 32,000 years before present, from which we generated overlapping DNA fragments assembling into a 16,810-base pair mitochondrial genome. Together with the first mitochondrial genome for the brown bear western lineage, this study provides a statistically secured molecular phylogeny assessing the cave bear as a sister taxon to the brown bear and polar bear clade, with a divergence inferred to 1.6 million years ago. With the first mitochondrial genome for a Pleistocene carnivore to be delivered, our study establishes the Chauvet-Pont d'Arc Cave as a new reservoir for Paleogenetic studies. These molecular data enable establishing the chronology of bear speciation, and provide a helpful resource to rescue for genetic analysis archeological samples initially diagnosed as devoid of amplifiable DNA.
Keywords: ancient DNA, pleistocene, Ursus spelaeus
Ancient DNA analysis was initiated >20 years ago by studies performed on the extinct quagga (1) and has long been devoted to the characterization of short nucleotide sequences, most often retrieved from the mitochondrial genome. Cooper et al. (2) raised the field to the genomic level by retrieving complete mitochondrial genomes for the moa, a flightless bird that became extinct a few hundred years ago. Recently, decisive progress has been accomplished by analysis carried out on Pleistocene specimens with a variety of approaches. Complete mitochondrial genomes have been obtained for the woolly mammoth and the mastodon, using either standard or multiplex PCR (3–5). Metagenomic studies performed on the mammoth yielded 13 million base pairs of nuclear DNA and several mitochondrial genomes (6, 7), and a similar approach carried out on Paleo-Eskimo frozen hair provided a mitochondrial genome for a human individual that lived ≈4,000 years ago (8). However, it is still unclear to what extent such approaches are valuable for species that did not benefit from the exceptional preservation conferred by long-term inclusion in permafrost. A genomic analysis of two cave bear (Ursus spelaeus) DNA libraries that contained 1–6% of bear sequences yielded 27 kb of nuclear DNA but did not allow to retrieve any mitochondrial sequence (9). An alternative strategy should therefore be considered to sequence the cave bear mitochondrial genome.
The cave bear, a member of the order of Carnivora, gradually evolved from Ursus deningeri and was present in Europe and the Near East from ≈300,000 to 15,000 years ago, when it became extinct (10). This bear is known from rock art pictures of the late Pleistocene and from skeletal remains that are almost exclusively found in caves. The subterranean milieu ensures stable temperature (12–15°C) conditions, away from UV irradiation, but is still less favorable than permafrost for DNA preservation. Consequently, cave bear mitochondrial genome fragments have only been retrieved as short sequences that up to now could be assembled into a partial control region and a single protein coding gene (11, 12), which together span <10% of the expected 17 kb mitochondrial genome. Phylogenetic analysis carried out using the available sequence information (12–14) supported one hypothesis drawn from morphometric studies of fossil records (10) arguing for an early split of the cave bear from the brown bear lineage. However, considering the accumulating evidence demonstrating that long sequences are often necessary to obtain correct phylogenies (5), it is highly desirable to better characterize the cave bear mitochondrial genome.
In the present study, we collected U. spelaeus bone samples from the Chauvet-Pont d'Arc Cave (Ardèche, France). This cave (44° 23′ N, 4° 26′ E; 240 m above sea level) was discovered in 1994 and contains the oldest rock art pictures ever found, with charcoal drawings dating back to 32,000 years before present (B.P.) (15). The numerous drawings and engravings of the cave are part of a well preserved environment that appears as a reservoir for the analysis of natural (speleothems, grounds), anthropogenic (fireplaces, footprints, carved flints) and animal (bone remains, coprolithes, tracks) material (16). The majority (> 90%) of skeletal pieces belong to the cave bear, with a current record of >4,000 remains dispatched into 130 bone assemblages (17). They belong to a variety of individuals, as shown by the presence of a large number of skulls laying on the ground surface. Osteometric data suggest a homogeneous cave bear population with a predominance of females (17). As part of an interdisciplinary research project, we could collect bone samples in different cave sectors for ancient DNA analysis. Our analytical procedure rested on the design of a series of bear-specific oligonucleotide primers that were used to generate hundreds of overlapping DNA fragments enabling the characterization of a complete cave bear mitochondrial genome.
Results and Discussion
After searching for cave bear skeletal elements that could be analyzed for DNA content in the Chauvet-Pont d'Arc cave, we identified a bone sample that reproductively yielded robust PCR amplifications. Us18 laid along the track of human footprints that extends from the Gallery of the Cross-Hatches to the Chamber of the Skull (Fig. 1). It consists of a sternebra that was radiocarbon dated to 31,870 (+300, −270) years B.P. (Groningen AMS sample number: GrA-28194).
Fig. 1.
Bear bone sample and archaeological context. (A) Geographical localization of the Chauvet-Pont d'Arc Cave. (B) Cave topography. Red and black characters refer to the color of rock art pictures in the entry and deep sectors, respectively. (C) The sector of the Gallery of the Cross-Hatches from which Us18 (purple arrow) was retrieved.
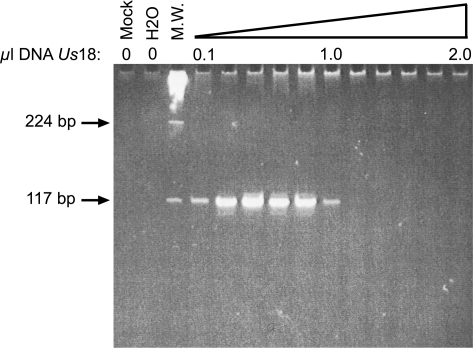
The Chauvet-Pont d'Arc Cave is expected to contain cave bear rather than brown bear remains (16, 17). We nevertheless initiated the molecular characterization of Us18 using primers which, although encompassing a highly variable portion of the mitochondrial control region, are conserved enough to allow DNA amplification from a variety of cave bear and brown bear mitochondrial haplotypes. These primers proved to be highly efficient for PCR amplification (Fig. 2), and generated a DNA fragment that displayed seven substitutions with the closest brown bear sequence, but was identical to sequences for the cave bear B haplogroup obtained in Scladina (40,000 to 45,000 years-old samples) (13). These data therefore strongly support the notion that we have retrieved authentic cave bear DNA. In addition, the amount of Us18 DNA extract allowing successful amplification (0.1 μl, or 0.05% of the total amount) was low enough to plan a large series of experiments. Ancient DNA samples usually contain DNA polymerase inhibitors that prevent from using large volume of extracts in the PCR (18, 19), as was indeed observed here. Nevertheless, the range of suitable DNA amounts spanned one order of magnitude, indicating that robust PCR conditions could be easily defined.
Fig. 2.
Gel electrophoresis analysis of mitochondrial genome fragments generated by PCR from Us18. Variable amounts of the DNA extract (from 0.1 to 2.0 μl) were amplified using primer pair # 236, predicting a 117-bp DNA fragment. The total reaction volume was electrophoresed through an acrylamide gel stained with SYBR Green I. Negative controls included reactions carried out on a mock extract (Mock) or in the absence of any extract (H2O). Molecular weight marker (M.W.) corresponds to λBstEII digest.
Because independent replication is a prerequisite for the study of ancient DNA (18, 19), a second extract was obtained and analyzed by another group of investigators from a different Institute (see Methods). The same and another overlapping pair of primers confirmed the sequence initially obtained, further corroborating that the sample was a reliable source of cave bear DNA. Subsequent experiments were carried out on both extracts, using 0.05 to 0.1% of the ancient DNA sample in each PCR.
To decipher the complete cave bear mitochondrial genome, we first selected a series of 147 primer pairs targeting conserved sequence motifs scattered throughout the brown bear and polar bear mitochondrial genomes. Because large DNA fragments are very rarely obtained from Pleistocene specimens, except for animals conserved in permafrost (3–7), most primer pairs were designed to amplify 150- to 180-bp DNA sequences. This first round of whole mitochondrial genome screening yielded 7.2 kb of DNA sequence, i.e., less than half of the mitochondrial genome. Considering that unsuccessful PCRs resulted from the use of primers that may not perfectly match the cave bear genome, we performed a screening iterative procedure using the cave bear sequence to design much more specific primers. We used a total of 245 primer pairs to retrieve a complete mitochondrial genome [supporting information (SI) Table S1].
Several lines of evidence support the conclusion that we deliver a reliable cave bear mitochondrial genome sequence. First, extensive replication was performed, the 245 primer pairs being used to generate 570 PCR fragments (Fig. S1). These PCR fragments were all cloned, and multiple clones were systematically sequenced on both strands to accurately determine a consensus sequence. Second, before assembly, all such consensus sequences were individually analyzed by BLAST to check that the best GenBank match corresponded to an Ursidae sequence. Third, among Ursidae, mitochondrial fragments previously analyzed in the cave bear displayed the best BLAST score with our sequences. As mentioned above, this was initially observed in the control region. The other published cave bear mitochondrial genome fragment concerns the cytochrome B (cytB). Our cytB sequence is identical to that obtained by Loreille et al. (12) for a cave bear from La Balme à Collomb, except for four transitions (0.35% of all cytB nucleotides). Two of these locate at the third base position of codons, and may reveal polymorphisms between cave bear coding sequences. For the two others, we recorded C instead of T residues, suggesting that the Chauvet-Pont d'Arc sample had been better preserved from cytosine deamination, the most frequent damage observed in ancient DNA (18, 19). The cave bear sample from La Balme à Collomb was also analyzed for a highly variable fragment of the control region (13), in which it displays two differences with Chauvet Us18. This supports the notion that the two cave bear specimens correspond to different haplotypes.
The length of the cave bear mitochondrial genome (16,810 bp) is in the range of those reported for extant bear genomes, which vary between 16,723 (Ursus maritimus) and 17,044 bp (Ursus thibetanus formosanus). The length differences between the bear genomes mostly come from the control region, which displays a highly variable number of repeats for a 10-bp motif. This specific domain of the control region could not be retrieved through a single PCR from the Chauvet-Pont d'Arc cave bear sample. We therefore used two primer pairs to separately target its 5′ and 3′ ends, and assembled all fragments into a repeat region of 350 bp. This is likely a minimal estimate, since the same approach carried out on another cave bear sample yielded a 360-bp sequence for this domain. The G + C nucleotide content of the cave bear mitochondrial genome (40.5%) is quite similar to that reported (40.4 to 41.6%) for extant bears (20). The cave bear and extant bear mitochondrial genomes all contain 13 protein coding genes, 22 tRNA genes, and 2 rRNA genes. The 13 protein coding genes predict polypeptides of similar size in all bear species, except for ND5, in which three additional codons are present in U. spelaeus and U. thibetanus formosanus. For three protein coding genes (COX3, ND3, and ND4), the stop codon is absent in the cave bear genome, being created by polyadenylation. This phenomenon, widely present in vertebrate mitochondrial genomes, is observed on the same genes in all extant bear genomes.
Because the previously published Ursus arctos mitochondrial genome (21) clusters into the brown bear eastern lineage described by Taberlet et al. (22), it was essential for a comprehensive phylogeny to also make available a complete mitochondrial genome sequence for the brown bear western lineage. This was accomplished by analyzing a brown bear from a French Pyrenean site (Guzet, Ariège) (Table S2 and Fig. S2). To exclude the possibility of contaminations in future analysis, DNA extraction from the modern brown bear sample was performed in a building different from that were cave bears DNA are extracted and stored.
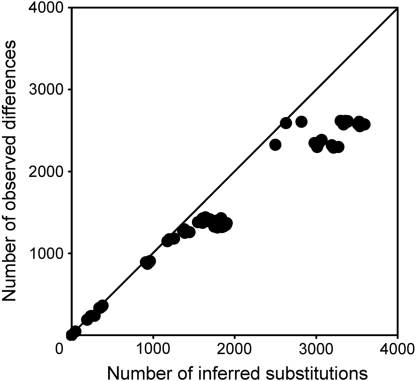
The Ursidae phylogeny was inferred using the newly obtained U. spelaeus and U. arctos sequences, 10 previously published mitochondrial genomes for extant bears, and the giant panda that served as an outgroup. To estimate the mutational saturation of this dataset, we plotted the genetic distance against the patristic distance for each pair of species (Fig. 3). These distances are almost equal, indicating that mutational saturation is weak. Hence, considering the low extent of homoplasy, these mitochondrial genomes convey an information that can securely be used to analyze phylogentic relationships.
Fig. 3.
Mutational saturation analysis of the complete mitochondrial genome dataset. The y axis shows the observed number of differences between pair of species sequences. The x axis shows the inferred number of substitutions between the same two sequences in a Maximum Parsimony tree determined using Patristic software. The straight line represents the case for which there is no saturation, with no reversion occurring in the sequences.
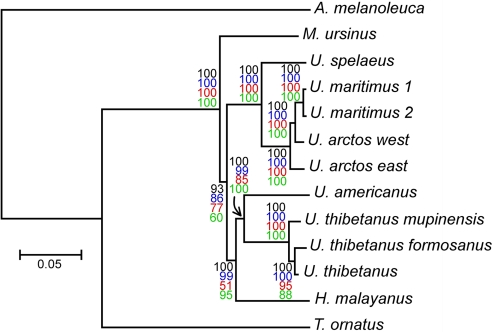
Using complete mitochondrial genomes, we obtained trees of similar topology for analysis performed with different reconstruction methods (Fig. 4). The transition/transversion ratio is 23:1 (Table S3), confirming that saturation is low (4). The alpha parameter calculated by PhyML and MrBayes are 0.20 (without invariable site) and 1.60 (with 57% of invariable sites), respectively. Albeit different, these values that are approximations of the same phenomenon, both indicate that rate heterogeneity among sites is important. As shown in Fig. 4, the cave bear clusters with the brown and polar bears with maximal bootstrap and posterior probability values. Besides, the sequencing of a western brown bear mitochondrial genome confirms Ursus arctos as a paraphyletic taxon with respect to U. maritimus (23). However, the brown and polar bears group is not paraphyletic toward the cave bear, showing that the cave bear lineage appeared before the diversification within the brown bears. For other species, the topology is the same as the one reported by Yu et al. (20): The Ursinae form a monophyletic group, from which M. ursinus diverged early. Then Ursinae split into two branches, one leading to the U. spelaeus, U. arctos, and U. maritimus group, and the other leading to the H. malayanus, U. americanus, and U. thibetanus group. The analysis of the amino acid sequence of the concatenated protein-coding genes do not give exactly the same topology, but the sister group relationship between the cave bear and the brown and polar bears is still robustly supported (Fig. S3).
Fig. 4.
Molecular phylogeny inferred from complete mitochondrial genome sequences. Tree construction was performed by MrBayes analysis, using the giant panda (Ailuropoda melanoleuca) as an outgroup. The posterior probability value (×100) of each node is indicated in black, and the scale for genetic distance is shown at the bottom of the figure. The same tree topology was obtained using three other methods, and bootstrap values are indicated with colored characters for PhyML (blue), maximum parsimony (red), and neighbor joining (green) analysis. The Ursus spealeus and Ursus arctos western lineage (west) sequences are from this study. GenBank accession numbers (from top to bottom) for the other sequences are as follows: EF196663, EF196662, AJ428577, AF303111, AF303110, AF303109, DQ402478, EF076773, EF196661, EF196664, and EF196665.
We also conducted a gene-by-gene strategy analysis (Figs. S4 and S5). The current and previously published (12) cave bear CYTB sequences cluster together. The topologies derived from individual genes vary in the branching of several species (especially U. americanus, U. thibetanus, H. malayanus, and M. ursinus). The sister group relationship between U. spelaeus and U. arctos–U. maritimus is found in every tree, the sole exception being the tree generated with ND6 sequence, a gene with weak phylogenetic information (20). It is worth emphasizing that bootstrap values are lower in the gene-by-gene analysis than in the whole genome tree. Only five genes give a 100% bootstrap value (16S rRNA, Cox1, ND2, ND4, and ND5) for the monophyly of the U. spelaeus, U. arctos, and U. maritimus group. Therefore, increasing the amount of data yields more robust trees, demonstrating the relevance of analyzing a complete mitochondrial genome.
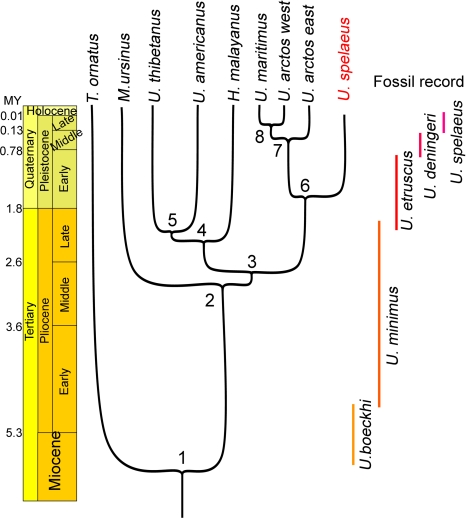
The robust phylogeny obtained with complete mitochondrial genomes offers the opportunity of evaluating the divergence dates between species. To perform this analysis, we set the split between the giant panda and Ursidae to 12 MY (24). The split between Ursinae and Tremarctinidae was set to 6 MY, which corresponds to the most ancient fossil record for Plionarctos, i.e., the first tremarctine bear (25). Consistent with this calibration, the bayeasian analysis returned a divergence date between Tremarctinidae and Ursinae centered to 6.3 MY (Fig. 5), a few hundred thousand years before the appearance of U. minimus boeckhi, the first representative of the ursine lineage (26).
Fig. 5.
Phylogeny and divergence times determined using the mitochondrial genome sequence of the cave bear and of eight extant bears. Divergence times were calculated using BEAST software with the splits between the giant panda and Ursidae and between Ursinae and Tremarctinidae set to 12 and 6 million years (MY), respectively. Age for each node and 95% credibility intervals are as follows: 1, 6.3 MY (5.4–7.2); 2, 3.0 MY (2.2–3.8); 3, 2.8 MY (2.1–3.5); 4, 2.4 MY (1.7–3); 5, 2.1 MY (1.4–2.7); 6, 1.6 MY (1–2.1); 7, 0.6 MY (0.3–0.8); and 8, 0.4 MY (0.2–0.5).
The radiation of the ursine bears (node 2 to 5) occurred in a brief lapse of time between 2 and 3 MY ago, thus explaining the difficulties in obtaining congruent phylogenetic relationships between these four species. This radiation occurred during the Pliocene when U. minimus, the assumed ancestor of the cave, brown and Asiatic black bears, was the most common bear species in Europe. We date the split between the arctoid and speleoid lineages back to 1.6 MY, during the Villafranchian stage. The divergence between eastern and western brown bear lineages occurred 550,000 years ago, probably during the Mindel glacial stage, and the split between the western lineage and the lineage leading to the polar bear occurred 350,000 years ago. A recent analysis conducted on northeastern European brown bears suggested that their last common ancestor is 174,000 years old (27).
To explore the robustness of the divergence times deduced from our data, we performed similar analyses using sequences of various lengths (Fig. S6). Strikingly, a sequence of 1 kb provides different node ages and much wider credibility intervals than those conveyed by longer sequences. Up to sequence stretches of 5 kb, the divergence date between the cave bear and arctoid lineages had not yet stabilized to the value of 1.6 MY obtained with the whole genome dataset.
The availability of a cave bear mitochondrial genome opens a wave of possibility. First, it is expected to help in the analysis of species that preexisted to the cave bear, such as Ursus deningeri. The sequence information provided by extant bears may not be sufficient to efficiently design experiments aiming at the retrieval of DNA fragments from such an ancient species. Second, the cave bear mitochondrial genome makes feasible to better explore archeological specimens ascribed to this species. Such a possibility was evaluated for Chauvet-Pont d'Arc bear samples that failed to yield any DNA when queried for the control region. Targeting another portion of the mitochondrial region with primers designed from the current cave bear genome (Fig. S7) rescued a series of samples for genetic analysis, providing successful amplification for 48% instead of 17% of the 23 Chauvet-Pont d'Arc samples analyzed so far. Together with the observation that cave bear intrusions extended from at least 37,000 to 29,000 years B.P (28), these samples indicate that exploring genetic diversity and variation through time is feasible at Chauvet-Pont d'Arc.
In conclusion, we provide a mitochondrial genome sequence for the extinct cave bear. This mitogenomic analysis definitely assesses the cave bear as a sister taxon to the brown bear and polar bear clade, and displays the tempo of bear history during the Pliocene and Pleistocene. Our study also demonstrates the feasibility of retrieving complete mitochondrial genomes from the subterranean milieu, an environment that contains remains for a variety of extinct species, and points to the painted cave of Chauvet-Pont d'Arc as a reservoir for paleogenetic investigations.
Methods
DNA Sequence Authentication.
To guarantee the authenticity of the cave bear mitochondrial sequence, we followed previous recommendations for works performed on ancient DNA (18, 19). First, to avoid contaminations from previous and current analyses, pre-PCR steps (i.e., DNA extraction and set-up of PCRs) were carried out in a building where no other molecular work on bear DNA had been performed previously, and handling of amplified products was done in a different building. Second, negative controls included mock extracts and PCR blanks (where water was added instead of DNA), which always failed to yield any amplification product. Third, we selected oligonucleotide primers that display weak homology with non-bear DNA sequences and checked by BLAST analysis that the best hit for each DNA fragment was a recorded cave bear sequence (when available in GenBank) or a sequence for another Ursidae. Fourth, we observed an expected molecular behavior for the ancient DNA extracts, with successful amplifications mainly for short (< 180 bp) DNA fragments, whereas sequences >200 bp were exceptionally obtained (4.2% of attempts). Fifth, reproducibility was assessed using the same and a second DNA extract. Sixth, we systematically designed PCR primers generating overlapping fragments. This strategy allowed us to read 8,498 nt (50.6% of the genome) from DNA fragments obtained with different PCR primers. As outlined in refs. 4, 18, and 19, this procedure allows to conclude that numts are unlikely to be present in our sequence. Seventh, to detect errors induced by DNA damage and deduce a reliable consensus sequence, we cloned each PCR fragment and systematically sequenced at least 12 clones on both strands. As a whole, the redundancy achieved through PCR replicates, overlaps between fragments and sequencing of multiple clones provided a mean number of 93 reads for each nucleotide of the cave bear mitochondrial genome. Eighth, DNA extracts obtained in each team (i.e., Saclay and Marseille) and analyzed by different investigators, using their own batch of reagents yielded the same cave bear DNA sequence, which demonstrated that the results could be independently replicated. Finally, to prevent from cross-contaminations, the brown bear sample was handled in a building different from those where the cave bear DNA had been extracted and analyzed, and experiments on the brown bear DNA were initiated once those on the cave bear samples have been completed.
DNA Extraction.
DNA was extracted from the bone cortex. One gram of bone powder was incubated 40 h at 42°C under constant agitation in 10 ml of extraction buffer consisting of 0.45 M EDTA, 10 mM Tris·HCl (pH 8.0), 0.1% SDS, 65 mM DTT, and 0.5 mg/ml proteinase K. Atfer centrifugation, the supernatant was recovered, extracted once with one volume of phenol, once with a phenol-chloroform-isoamylalcohol (25:24:1) mixture, and once with chloroform. The aqueous phase was then concentrated using Centricon YM-30 (Millipore), and the column was washed five times with distilled water. The DNA extract was subsequently recovered as a ≈200-μl sample volume.
Primer Design.
PCR primers were designed with the help of Oligo 6.0 software (Medprobe). For experiments on cave bear DNA, we aligned the Ursus arctos and U. maritimus mitochondrial genomes and selected 147 primer pairs targeting conserved sequences. Sixty-four (44%) of these pairs were successfully used in PCR experiments that yielded 7.2 kb of the cave bear mitochondrial genome. We subsequently used this sequence information to iteratively design new series of primer pairs to generate PCR fragments that allowed to fill the gaps. As expected, these subsequent series of primers increased the success rate, with 181 of 250 pairs (72.4%) allowing the amplification of cave bear mitochondrial DNA fragments. For experiments carried out on brown bear DNA, we used 52 primer pairs to retrieve a complete mitochondrial genome sequence.
DNA Amplification and Analysis.
PCR was performed in a 50-μl reaction volume containing mock or ancient DNA extracts, 300 pM sense and antisense primers, 200 μM dNTP, 2.5 mM MgCl2, 2.5 μg of T4 gene 32 protein (USB), 5 μl of GeneAmp 10X PCR buffer II, and 2.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems). The amount of DNA to be used was tested in initial experiments and consisted of 0.2–0.4 μl and 0.1 μl of the cave bear and brown bear DNA extracts, respectively. An equivalent volume of water was substituted to the DNA sample in PCR blanks. After an activation step (95°C, 8.5 min), a single round of 45 PCR cycles (95°C for 15 s, 50–60°C (according to primers Tm) for 20 s, and 70°C for 1 min) was performed in 9600, 7000 or 7300 Applied Biosystems thermal cyclers. The full reaction volume was loaded onto an 8% polyacrylamide gel. To increase the sensitivity of our PCR assay, we used Sybr Green I (Invitrogen) instead of ethidium bromide to stain the gel. PCR amplicons were eluted from the gel and inserted into pCR4-TOPO (Invitrogen). Plasmid minipreparations of the clones were sequenced on ABI 377XL or 3130 XL DNA sequencers, using BigDye 3.1 terminator chemistry (Applied Biosystems). We systematically analyzed a minimum of 12 colonies for each cloned fragment and sequenced both DNA strands using M13 forward and T3 primers.
Phylogenetic Analyses.
The complete mitochondrial genomes of Ursidae and of the giant panda were aligned using ClustalW with the default parameters. The domain of the control region containing the 10-bp repeat motif was removed before all analyses. Phylogenetic analyses were carried out on different datasets : complete mitochondrial genomes, concatenated nucleotide sequences of protein-coding genes, amino acid sequences of individual proteins, concatenated tRNA genes, and rRNA genes. All of the genes were aligned individually before concatenation. As some of them are overlapping, a few nucleotides are duplicated in our concatenations. Phylogenetic trees were constructed from these datasets with Maximum Likelihood (ML), Maximum Parsimony (MP), and Neighbor-Joining (NJ) methods, using PhyML (29, 30), MrBayes 3.1.2 (31), and Mega 3.1 (32) program packages, as appropriate.
For nucleotide analysis, PhyML and MrBayes were used with the general time reversible (GTR) + 4Γ + I model, and, for the NJ method, we used the Tamura 3-parameter and the gamma-distribution shape parameter estimated with PhyML and MrBayes. For amino acid, PhyML and MrBayes analyses were conducted with a gamma substitution rate model and a mammalian mitochondrial model of substitutions (MtMam), and NJ analysis was performed using a gamma substitution rate model and a Jones–Taylor–Thornton (JTT) matrix of substitution. Bayesian analyses were run using four Metropolis coupled Markov Chain Monte Carlo for at least 1 million generations, sampling trees every 100 generations. MP analyses were run with the Mega 3.1 default parameters.
To estimate the robustness of the phylogenetic inferences, we used the bootstrap method (2,000 replicates for NJ and MP, 500 replicates for PhyML). For Bayesian analyses, posterior probabilities of the nodes in the consensus tree were estimated. To evaluate possible bias introduced by saturation, we tested the substitution saturation for the complete mitochondrial genome dataset, using Patristic 12.0.0 software (33).
Divergence times were estimated using complete mitochondrial genomes with BEAST software (34). We used as calibration points the divergence between the giant panda and Ursidae and between Ursinae and Tremarctinidae, set at 12 ± 1 MY (24) and 6 ± 0.5 MY (25), respectively, considering a normal distribution. We chose a GTR + 4Γ + I substitution model, a relaxed uncorrelated lognormal molecular clock, and a Yule process of speciation (35). We performed two independent chains that each consisted of 10,000,000 points. Data were collected every 1,000 points, and the burn-in was set to 10,000.
To test the impact of sequence length on estimated divergence times, we randomly created alignments of various length from whole mitochondrial genome sequences, and calculated node ages using the parameters described above.
Supplementary Material
Acknowledgments.
We thank D. Baffier for giving us the authority to collect cave bear samples, M.-C. Gaillard for help in initiating the project, J. L. Orengo for the brown bear sample, all investigators of the Chauvet scientific team for fruitful discussions, A. Martel for bioinformatics support, M. Azéma and N. Kidman for artwork, P. Legrain for encouragement and helpful suggestions for an early version of the manuscript. This work was supported by grants from the Commissariat à l'Energie Atomique and the French Ministère de la Culture et de la Communication. C.B. received PhD funding from the Commissariat à l'Énergie Atomique.
Note Added in Proof.
This work was under review and in the publication process when a cave bear mitochondrial genome sequence was obtained from a bone sample found in Gamssulzen Cave, Austria (36). The Chauvet and Gamssulzen cave bear mitochondrial genome sequences are highly homologous and locate at similar positions in a phylogenetic tree. The divergence dates between ursine lineages deduced from the two studies display however a number of differences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ227308, EU327344, and EU497665).
This article contains supporting information online at www.pnas.org/cgi/content/full/0806143105/DCSupplemental.
References
- 1.Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC. DNA sequences from the quagga, an extinct member of the horse family. Nature. 1984;312:282–284. doi: 10.1038/312282a0. [DOI] [PubMed] [Google Scholar]
- 2.Cooper A, et al. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature. 2001;409:704–707. doi: 10.1038/35055536. [DOI] [PubMed] [Google Scholar]
- 3.Rogaev EI, et al. Complete mitochondrial genome and phylogeny of Pleistocene mammoth Mammuthus primigenius. PLoS Biol. 2006;4:e73. doi: 10.1371/journal.pbio.0040073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krause J, et al. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature. 2006;439:724–727. doi: 10.1038/nature04432. [DOI] [PubMed] [Google Scholar]
- 5.Rohland N, et al. Proboscidean mitogenomics: Chronology and mode of elephant evolution using mastodon as outgroup. PLoS Biol. 2007;5:e207. doi: 10.1371/journal.pbio.0050207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poinar HN, et al. Metagenomics to paleogenomics: Large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert MT, et al. Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science. 2007;317:1927–1930. doi: 10.1126/science.1146971. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert MT, et al. Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science. 2008;320:1787–1789. doi: 10.1126/science.1159750. [DOI] [PubMed] [Google Scholar]
- 9.Noonan JP, et al. Genomic sequencing of Pleistocene cave bears. Science. 2005;309:597–599. doi: 10.1126/science.1113485. [DOI] [PubMed] [Google Scholar]
- 10.Kurten B. The Cave Bear Story: Life and Death of a Vanished Animal. New York: Columbia Univ Press; 1976. [Google Scholar]
- 11.Hänni C, Laudet V, Stehelin D, Taberlet P. Tracking the origins of the cave bear (Ursus spelaeus) by mitochondrial DNA sequencing. Proc Natl Acad Sci USA. 1994;91:12336–12340. doi: 10.1073/pnas.91.25.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loreille O, et al. Ancient DNA analysis reveals divergence of the cave bear, Ursus spelaeus, and brown bear, Ursus arctos, lineages. Curr Biol. 2001;11:200–203. doi: 10.1016/s0960-9822(01)00046-x. [DOI] [PubMed] [Google Scholar]
- 13.Orlando L, et al. Ancient DNA and the population genetics of cave bears (Ursus spelaeus) through space and time. Mol Biol Evol. 2002;19:1920–1933. doi: 10.1093/oxfordjournals.molbev.a004016. [DOI] [PubMed] [Google Scholar]
- 14.Hofreiter M, et al. Ancient DNA analyses reveal high mitochondrial DNA sequence diversity and parallel morphological evolution of late pleistocene cave bears. Mol Biol Evol. 2002;19:1244–1250. doi: 10.1093/oxfordjournals.molbev.a004185. [DOI] [PubMed] [Google Scholar]
- 15.Valladas H, et al. Palaeolithic paintings. Evolution of prehistoric cave art. Nature. 2001;413:479. doi: 10.1038/35097160. [DOI] [PubMed] [Google Scholar]
- 16.Clottes J. Chauvet Cave: The Art of Earliest Times. Paris: Seuil; 2001. [Google Scholar]
- 17.Fosse P, Philippe M. Fauna in the Chauvet Cave: Paleobiology and anthropozoology. Bull Soc Prehist Fr. 2005;102:89–102. [Google Scholar]
- 18.Pääbo S, et al. Genetic analyses from ancient DNA. Annu Rev Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 19.Willerslev E, Cooper A. Ancient DNA. Proc Biol Sci. 2005;272:3–16. doi: 10.1098/rspb.2004.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu L, Li YW, Ryder OA, Zhang YP. Analysis of complete mitochondrial genome sequences increases phylogenetic resolution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evol Biol. 2007;7:198. doi: 10.1186/1471-2148-7-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delisle I, Strobeck C. Conserved primers for rapid sequencing of the complete mitochondrial genome from carnivores, applied to three species of bears. Mol Biol Evol. 2002;19:357–361. doi: 10.1093/oxfordjournals.molbev.a004090. [DOI] [PubMed] [Google Scholar]
- 22.Taberlet P, Bouvet J. Mitochondrial DNA polymorphism, phylogeography, and conservation genetics of the brown bear Ursus arctos in Europe. Proc Biol Sci. 1994;255:195–200. doi: 10.1098/rspb.1994.0028. [DOI] [PubMed] [Google Scholar]
- 23.Talbot SL, Shields GF. Phylogeography of brown bears (Ursus arctos) of Alaska and paraphyly within the Ursidae. Mol Phylogenet Evol. 1996;5:477–494. doi: 10.1006/mpev.1996.0044. [DOI] [PubMed] [Google Scholar]
- 24.Wayne RK, van Valkenburgh B, O'Brien SJ. Molecular distance and divergence time in carnivores and primates. Mol Biol Evol. 1991;8:297–319. doi: 10.1093/oxfordjournals.molbev.a040651. [DOI] [PubMed] [Google Scholar]
- 25.Tedford RH, Martin J. Plionarctos, a tremarctine bear (Ursidae: Carnivora) from western North America. J Vertebrate Paleontol. 2001;21:311–321. [Google Scholar]
- 26.Olive F. Evolution of Plio Pleistocene larger Carnivores in Africa and Western Europe. L'Anthropologie. 2006;110:850–869. [Google Scholar]
- 27.Saarma U, et al. Mitogenetic structure of brown bears (Ursus arctos L.) in northeastern Europe and a new time frame for the formation of European brown bear lineages. Mol Ecol. 2007;16:401–413. doi: 10.1111/j.1365-294X.2006.03130.x. [DOI] [PubMed] [Google Scholar]
- 28.Bocherens H, Drucker DG, Billiou D, Geneste JM, van der Plicht J. Bears and humans in Chauvet Cave (Vallon-Pont-d'Arc, Ardèche, France): Insights from stable isotopes and radiocarbon dating of bone collagen. J Hum Evol. 2006;50:370–376. doi: 10.1016/j.jhevol.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 30.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 33.Fourment M, Gibbs MJ. PATRISTIC: a program for calculating patristic distances and graphically comparing the components of genetic change. BMC Evol Biol. 2006;6:1. doi: 10.1186/1471-2148-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drummond AJ, Rambaut A BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krause J, et al. Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene-Pliocene boundary. BMC Evol Biol. 2008;8:220. doi: 10.1186/1471-2148-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.