Abstract
Parafibromin is a tumor suppressor protein encoded by HRPT2, a gene recently implicated in the hereditary hyperparathyroidism–jaw tumor syndrome, parathyroid cancer, and a subset of kindreds with familial isolated hyperparathyroidism. Human parafibromin binds to RNA polymerase II as part of a PAF1 transcriptional regulatory complex. The physiologic targets of parafibromin and the mechanism by which its loss of function can lead to neoplastic transformation are poorly understood. We show here that RNA interference with the expression of parafibromin or Paf1 stimulates cell proliferation and increases levels of the c-myc proto-oncogene product, a DNA-binding protein and established regulator of cell growth. This effect results from both c-myc protein stabilization and activation of the c-myc promoter, without alleviation of the c-myc transcriptional pause. Chromatin immunoprecipitation demonstrates the occupancy of the c-myc promoter by parafibromin and other PAF1 complex subunits in native cells. Knockdown of c-myc blocks the proliferative effect of RNA interference with parafibromin or Paf1 expression. These experiments provide a previously uncharacterized mechanism for the anti-proliferative action of the parafibromin tumor suppressor protein resulting from PAF1 complex-mediated inhibition of the c-myc proto-oncogene.
Keywords: HRPT2, hyperparathyroidism, parathyroid cancer, PAF1 complex
The tumor suppressor gene HRPT2 was recently identified by positional candidate cloning (1). Germline mutation of HRPT2 confers susceptibility to the hyperparathyroidism-jaw tumor syndrome (HPT-JT), an autosomal dominant syndrome with high but incomplete penetrance (2). The major features are primary hyperparathyroidism (including 15% of all affected by HPT-JT with parathyroid cancer), jaw tumors, bilateral renal cysts, and, less commonly, solid renal tumors (2, 3). Germline inactivating HRPT2 mutation has also been reported in a minority of kindreds with familial isolated hyperparathyroidism (FIHP) (1, 4) and in up to 30% of patients with apparently sporadic parathyroid cancer (5, 6).
HRPT2 encodes parafibromin, a 531 amino acid, putative tumor suppressor protein. Parafibromin demonstrates weak homology to Cdc73p, a budding yeast protein component of the RNA polymerase II-associated Paf1 complex. Recent evidence suggests that in humans, parafibromin interacts with RNA polymerase II via a human PAF1 complex whose other protein components include human Paf1, CTR9, Leo1 (7–9), and the WD40-repeat protein Ski8 (9). The molecular targets of parafibromin and the mechanism by which its inactivation can lead to neoplastic transformation are poorly understood. We show here that parafibromin, in the context of the PAF1 complex, mediates repression of the c-myc proto-oncogene through both transcriptional and posttranscriptional mechanisms. This finding provides a previously uncharacterized mechanism for the anti-proliferative action of parafibromin and a plausible mechanism by which its loss of function promotes neoplasia.
Results
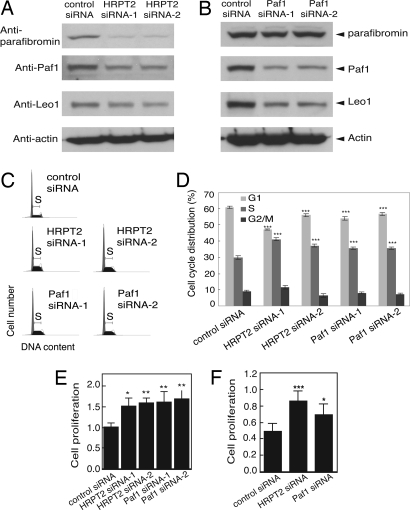
To better understand the normal cellular function of the parafibromin tumor suppressor protein in the context of the transcriptional regulatory PAF1 complex, RNA interference was used to inhibit the expression of endogenous parafibromin and Paf1 protein. The impact of this RNA interference on the expression of several components of the PAF1 complex and cellular proliferation in HeLa cells was determined (Fig. 1). Transfection of cells with small interfering duplex RNAs (siRNA) targeting two different sequences of the HRPT2 gene transcript inhibited the expression of endogenous parafibromin in HeLa cells as previously shown (Fig. 1A) (10). In addition, treatment of cells with HRPT2-targeted siRNAs also inhibited expression of endogenous Paf1 and Leo1 proteins, both also components of the PAF1 transcriptional regulatory complex, compared to control (Fig. 1A) (7–9). Similarly, transfection with Paf1-targeted siRNAs directed against two different Paf1 sequences both inhibited Paf1 (as expected) and Leo1 expression, although the Paf1-targeted gene silencing had little effect on parafibromin expression (Fig. 1B). Thus RNA interference employing HRPT2- and Paf1-targeted siRNA appeared to be an effective strategy for the knockdown of multiple protein components of the endogenous PAF1 complex.
Fig. 1.
RNA interference with HRPT2 or Paf1 expression in human cells leads to down-regulation of endogenous Paf1 and Leo1 expression and increased cell proliferation. (A and B) Immunoblotting analysis of parafibromin (Top Panel), Paf1 (Second Panel), Leo1 (Third Panel) in HeLa cells following RNAi-mediated depletion of HRPT2 or Paf1 transcripts. Cells were treated with control siRNA or one of two HRPT2- or Paf1-specific siRNAs and analyzed as described in Methods. The expression of β-actin is shown as loading control (Fourth Panel). (C and D) Inhibition of parafibromin or Paf1 expression by RNAi resulted in increased S phase cell population. HeLa cells were treated with control siRNA or one of two HRPT2- or Paf1-specific siRNA and subjected to FACS analysis as described in Methods. (D) The histogram shows the distribution of different cell cycle population; values are representative of two independent experiments with similar results. Significance of the differences on S phase and G1 phase were evaluated by Student's t test (***, P < 0.001). (E and F) Proliferation analysis. Cultured HeLa (E) or NHDF cells (F) were treated as above for 48 h. Cell proliferation was estimated by a colorimetric cell proliferation assay. Presented are data representative of three individual repeats. Significance of the differences were evaluated by Student's t test (*** P < 0.005, **, P < 0.01, *, P < 0.05).
The effect of anti-HRPT2- and anti-Paf1-directed siRNA treatment on cell proliferation was studied in two different assays. Exponentially dividing HeLa cells treated with control, HRPT2-, or Paf1-directed siRNAs and stained with propidium iodide were analyzed by fluorescence-activated cell sorting (FACS) and the proportion of cells in G1, S, and G2/M phases estimated (Fig. 1 C and D). Treatment with HRPT2 siRNA-1 or -2 or with Paf1 siRNA-1 or -2 construct caused a significant increase in the proportion of cells in S phase with a corresponding decrease in the number of cells in G1 (Fig. 1D). These results confirm the findings of Yart, et al. (8). An independent assay of cell proliferation was used to compare the control, HRPT2-, and Paf1 siRNA-treated cells and demonstrated that the siRNA-mediated knockdown of either PAF1 complex component increased the rate of cellular proliferation over control in both HeLa cells (Fig. 1E) and primary human fibroblasts (Fig. 1F). Treatment of human osteosarcoma-derived U2-OS cells with either HRPT2 or Paf1 siRNA also stimulated cell proliferation [supporting information (SI) Fig. S1]. These results were consistent with previous findings that transfection of wild-type, but not clinically significant patient-derived mutants of, parafibromin strongly inhibited cell growth in proliferation assays (11, 12).
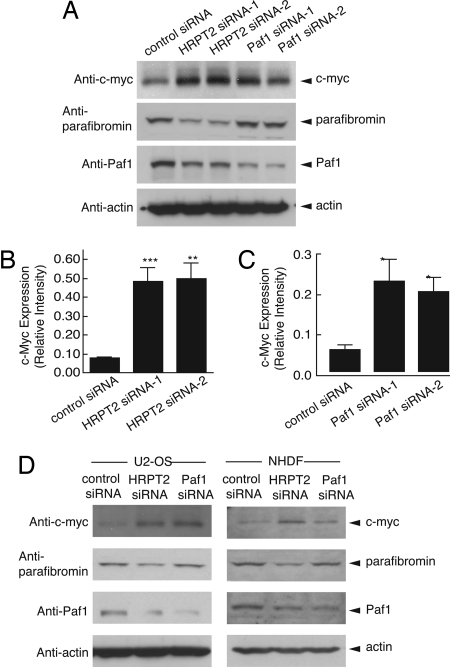
The c-myc proto-oncogene encodes a transcription factor of the basic helix–loop–helix/leucine zipper family strongly implicated in the control of cell growth. Because siRNA-mediated knockdown of both HRPT2- and Paf1-stimulated cellular proliferation, we checked the effect of such siRNA treatment on cellular levels of the c-myc proto-oncogene by immunoblotting of HeLa cell lysates (Fig. 2A). Both pairs of HRPT2- and Paf1-directed siRNAs elevated the level of c-myc protein above the control (Fig. 2 A–C). C-myc protein levels were also up-regulated in human U2-OS cells or primary human fibroblasts (Fig. 2D) treated with either HRPT2- or Paf1-targeted siRNAs.
Fig. 2.
Up-regulation of c-myc protein in HeLa cells, U2-OS cells and NHDF cells following RNAi-mediated inhibition of HRPT2 or Paf1 expression. (A) HeLa cells were treated with control siRNA or one of two HRPT2- or Paf1-specific siRNA for 48 h and cells were then subjected to immunoblot analysis using anti-c-myc (Top Panel), anti-parafibromin (Second Panel), anti-Paf1 (Third Panel). (B and C) Based on Western blot analysis as presented in Fig. S7, quantification of c-myc expression was carried out by quantitative ECL and then normalized to actin expression. Significance of the differences were evaluated by Student's t test (***, P < 0.005, **, P < 0.01, *, P < 0.05). (D) U2-OS cells and NHDF cells were treated with control siRNA, HRPT2- or Paf1-specific siRNA as described in Methods. 48 h after treatment, cells were analyzed for expression of c-myc (Top Panel), parafibromin (Second Panel), Paf1 (Third Panel). In A and D the expression of β-actin is shown as a loading control.
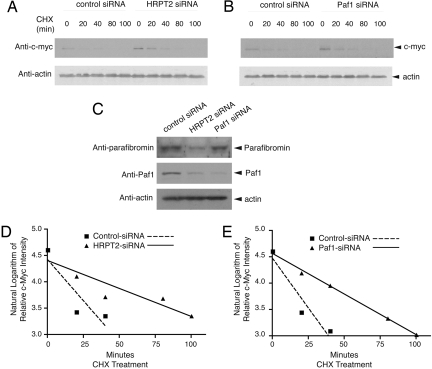
The c-myc protein is normally short-lived but can be stabilized by mitogenic stimuli like growth factors (13) and p21 Ras (14). To determine if c-myc protein up-regulation in response to knockdown of HRPT2 and Paf1 might result from protein stabilization, the half-life of c-myc was estimated in HeLa cells treated with cycloheximide and either control, HRPT2- or Paf1-directed siRNAs (Fig. 3 A and B). The expression of c-myc protein was examined at multiple time points between 0 and 100 min and normalized by reference to the level of actin in each sample. As expected at t = 0, c-myc proteins levels were higher in cells treated with HRPT2- or Paf1-directed siRNAs compared to control (Fig. 3 A and B), and the siRNAs were effective at reducing their respective targeted proteins (Fig. 3C). The natural logarithm of the actin-normalized c-myc protein expression level (relative to t = 0) was plotted over time, and linear regression analysis used to estimate the c-myc protein half-life assuming first-order kinetics (Fig. 3 D and E). The estimated half-life of c-myc was greater upon treatment with either HRPT2- or Paf1-directed siRNAs compared to control (Fig. 3D; control siRNA: r2 = 0.79, c-myc half-life 16 ± 15 min; HRPT2-siRNA: r2 = 0.85, c-myc half-life 47 ± 19 min, p = NS vs. control), (Fig. 3E; control siRNA: r2 = 0.91, c-myc half-life 15 ± 8 min; Paf1-siRNA: r2 = 1.0, c-myc half-life 43 ± 2 min, P < 0.005 vs. control), although the difference between HRPT2-siRNA and its control did not achieve statistical significance (Fig. 3D). Taken together, these data suggest that protein stabilization contributes to the up-regulation of c-myc expression resulting from HRPT2 or Paf1 knockdown.
Fig. 3.
Analysis of c-myc protein stability and half-life in response to inhibition of parafibromin or Paf1 expression. (A and B) Cultured HeLa cells were treated with control siRNA, HRPT2- or Paf1-specific siRNA. At 48 h after transfection, cycloheximide (CHX) was added to the culture medium and the cells were collected at 0, 20, 40, 80, and 100 min. Cell lysates were analyzed by immunoblotting using anti-c-myc or anti-actin antibodies. (C) The expression of parafibromin and/or Paf1 at the zero time point was analyzed by immunoblotting. The expression of β-actin is shown as a loading control. (D and E) Linear regression analysis of the natural logarithm of the relative intensity of c-myc protein expression in control and HRPT2- (D) or Paf1- (E) specific siRNA treated cells at different times of CHX treatment, quantified by quantitative ECL analysis and normalized to β–actin expression, is shown. In this analysis the y-value at time = 0 for all plots was 4.61 (the natural log of 100 [%] relative expression), and a y-value of 3.91 (indicating a relative expression of 50 [%]) maps to the time corresponding to the c-myc protein half-life.
Transcriptional activation of c-myc in response to the siRNA-mediated silencing of HRPT2 or Paf1 is a potential alternative mechanism that could also result in increased c-myc protein expression. This possibility is especially relevant since the human PAF1 complex is physically associated with, and known to regulate the activity of, RNA polymerase II (7–9). Furthermore, other tumor suppressors such as p53 are known to repress the transcription of c-myc (15). Analysis of c-myc transcript levels by quantitative real-time RT-PCR in HeLa cells treated with HRPT2- and Paf1-targeted siRNA at two different times points showed a significant elevation in response to treatment over control (Fig. S2 a and b). The effect of both anti-HRPT2 siRNA treatments was evident at 24 h, while treatment with the anti-Paf1 siRNA did not produce significant elevation of c-myc transcript until 48 h (Fig. S2 a and b). These results would be consistent with transcriptional activation of c-myc resulting from knockdown of parafibromin or Paf1.
The expression of c-myc is controlled in many cell types by a block in the elongation of primary transcripts mediated by transcriptional pause occurring at a locus near the exon 1-intron 1 boundary (16, 17). The Paf1 complex has been implicated in RNA Pol II transcriptional elongation (7, 18). We therefore looked for evidence that the endogenous Paf1 complex might be involved in the transcriptional arrest of c-myc at the exon 1-intron 1 junction by treatment of HeLa cells with HRPT2- and Paf1-targeted siRNAs and quantitative determination of exon 1- and exon 2-containing transcripts by quantitative RT-PCR (Fig. S3 a and b). The level of c-myc exon 1-containing transcripts was significantly increased by either siRNA treatment (Fig. S3a) while the quantity of c-myc exon 2-containing transcripts was increased significantly only by anti-HRPT2 siRNA with the increase by anti-Paf1 siRNA not quite achieving statistical significance (Fig. S3b). The ratio of exon 1- to exon 2-containing transcripts resulting from either HRPT2- or Paf1-targeted siRNA treatment did not differ from control, however, suggesting that relief of the c-myc transcriptional pause was not a major mechanism of c-myc up-regulation resulting from the knockdown of endogenous Paf1 complex components (Fig. S3c).
To further examine the mechanism of transcriptional activation of c-myc upon knockdown of parafibromin or Paf1, three fragments of human genomic DNA comprising various lengths of upstream regulatory sequence and extending through the 1st intron of the c-myc gene were subcloned into the pGL3 Basic luciferase reporter construct to generate the −836/+2193, −63/+2193 and + 613/+2193 c-myc promoter reporters (Fig. S2c). Position −63 is just upstream of the P1 and P2 promoter start sites, while position +613 is just downstream of the transcriptional pause sites at the exon 1-intron 1 junction. Treatment of primary human fibroblasts with HRPT2-directed siRNA significantly elevated the promoter activity of the −836/+2193 and −63/+2193 c-myc luciferase reporters over control but had no effect on the low basal activity of the +613/+2193 luciferase reporter (Fig. S2d). Knockdown of endogenous Paf1 gave similar results except that the activation of the −63/+2193 c-myc luciferase reporter did not achieve statistical significance (Fig. S2d). In HeLa cells, as in primary fibroblasts, both HRPT2- and Paf1-directed siRNA treatments activated the −836/+2193 c-myc luciferase reporter (Fig. S2e). No effect of the siRNA treatments was seen on the promoter-less pGL3 Basic vector alone (Fig. S4). The results with the luciferase reporters suggest the principal effects of the endogenous PAF1 complex on c-myc transcription occur in the upstream regulatory region distant from the transcriptional pause site.
Activation of the c-myc promoter that results from knockdown of either HRPT2 or Paf1 implies a transcriptional inhibitory role for the endogenous PAF1 complex likely resulting from physical association of the complex with c-myc promoter elements. To test this hypothesis, the occupancy of the c-myc promoter and internal gene regions by parafibromin was evaluated by chromatin immunoprecipitation (ChIP) analysis. ChIP with control rabbit IgG gave no signal with primers targeting either a region of the c-myc promoter just 3′ to the transcription start site (P2, +77), or a more internal region of c-myc (+1572) in human HeLa cells (Fig. S5a). Specific signal with both pairs of c-myc- directed primers was detected, however, when ChIP used anti-parafibromin antibodies (Fig. S5a). Anti-Paf1 and anti-Leo1 antibodies also gave a strong positive signal in the ChIP analysis employing c-myc- directed primers in HeLa cells (Fig. S5b). In contrast, the signal with PCR primers to a region approximately 2 kB upstream of c-myc was either weak (with anti-parafibromin antibody; Fig. S5a) or absent (with anti-Paf1 and anti-Leo1 antibodies; Fig. S5b). These results confirm the occupancy of the human c-myc promoter by the endogenous PAF1 complex.
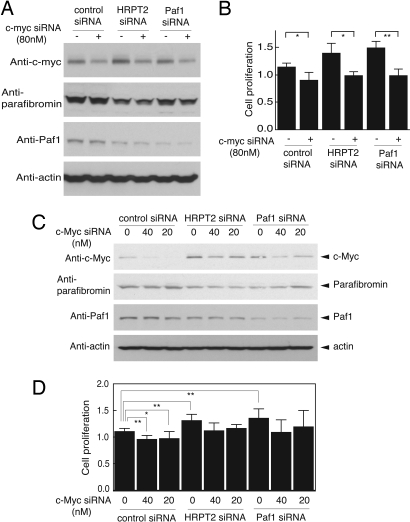
Although knockdown of HRPT2 and Paf1 by targeted siRNA treatment-stimulated cell proliferation and up-regulated c-myc protein levels, it is unclear to what extent the former effect depends on the latter. To check for the c-myc dependence of the proliferative response to knockdown of HRPT2 or Paf1, the proliferative rate of HeLa cells was estimated after treatment with control, HRPT2-, or Paf1-targeted siRNAs without or with concurrent c-myc siRNA treatment (80 nM). The c-myc siRNA treatment was sufficient to reduce c-myc protein levels relative to control under all conditions and did not interfere with the knockdown of parafibromin or Paf1 protein resulting from treatment with the respective siRNA constructs (Fig. 4A). The c-myc siRNA treatment did inhibit the basal cell proliferative rate in control siRNA-treated cells and abolished the cell proliferative response to HRPT2- or Paf1 knockdown (Fig. 4B). Since treatment with 80 nM c-myc siRNA was sufficient to inhibit c-myc expression below basal levels even after knockdown of parafibromin and Paf1, this result does not exclude the possibility that c-myc and the Paf1 complex work in parallel pathways to control cell proliferation. To better examine this issue, the experiment was repeated using graded amounts of 0, 20, and 40 nM c-myc siRNA so as to find conditions that would permit expression of near-control levels of c-myc protein even after knockdown of the Paf1 complex components. Immunoblots of cells treated under these conditions showed a general dose-dependence of the c-myc knockdown that did not interfere with the HRPT2- and Paf1-siRNA-mediated knockdown of parafibromin and Paf1, respectively (Fig. 4C). Using the smaller doses of c-myc siRNA, however, the level of residual c-myc protein under conditions of parafibromin- and Paf1-knockdown was equivalent to, or even slightly higher than, the basal, control c-myc protein level (Fig. 4C, Top Panel). Nevertheless, the more modest doses of 20 and 40 nM c-myc siRNA still inhibited the basal cell proliferative rate in control siRNA-treated cells and abolished the cell proliferative response to HRPT2 or Paf1 knockdown (Fig. 4D). These results demonstrate that RNA interference with the expression of the c-myc proto-oncogene can prevent cell proliferation in response to knockdown of parafibromin and Paf1 components of the PAF1 complex, even when basal levels of c-myc protein are maintained. Treatment with c-myc siRNA also abolished the cell proliferative response to HRPT2 or Paf1 knockdown in human U2-OS cells (Fig. S6).
Fig. 4.
c-Myc is downstream of the Paf1 complex in the control of cell proliferation. (A) c-myc expression in HeLa cells treated with 80 nM of c-myc siRNA in the presence of control, HRPT2- or Paf1-specific siRNA. Efficacy of RNAi-mediated gene silencing of c-myc (Top Panel), HRPT2 (Second Panel) and Paf1 (Third Panel) were assessed by immunoblot analysis. (B) For the cell proliferation assay, HeLa cells were treated with control siRNA, HRPT2- or Paf1-specific siRNA in the absence or presence of 80 nM of c-myc-specific siRNA. Forty-eight hours after treatment, proliferation was estimated by a colorimetric assay as described in Methods. Data are representative of three individual repeats with similar results. (C) c-Myc expression in HeLa cells treated with control-, HRPT2- or Paf1-specific siRNA in the presence of 0, 20, or 40 nM of c-myc-specific siRNA. Gene silencing efficiency of c-myc (Top Panel), HRPT2 (Second Panel) and Paf1 (Third Panel) are monitored by Western blot analysis. (D) Cell proliferation of HeLa cells after treatment for 48h with HRPT2- or Paf1-specific siRNA in the presence of 0, 20, or 40 nM of c-myc-specific siRNA was estimated by a colorimetric assay. Data are representative of three independent experiments with similar results. Significance of the differences were evaluated by Student's t test (**, P < 0.01, *, P < 0.05). In A and C the expression of β-actin is shown as a loading control.
Discussion
Most clinically significant sporadic or germline mutations of HRPT2 predict loss of parafibromin expression or function (1, 5, 6, 19–25), supporting the view that parafibromin is a tumor suppressor protein. The mechanism(s) by which loss of parafibromin function promotes neoplasia are nonetheless largely unknown. Neither the recognition that parafibromin is a component of the PAF1 complex (7–9) nor the observation that parafibromin binds to β-catenin and promotes Wnt signaling (26) has given insight into the critical pathway(s) in which loss of parafibromin function facilitates tumor development.
Anti-proliferative properties are common to many tumor suppressor proteins, as are pro-apoptotic actions such as that recently shown for endogenous parafibromin (10). The c-myc proto-oncogene has a well-established role in promoting cell proliferation and c-myc transcript levels correlate with the proliferative response of primary cultures of parathyroid cells to serum (27). Given the inhibition of c-myc expression by endogenous parafibromin and Paf1 demonstrated here, the inactivation of parafibromin in human parathyroid and other tumors is likely to promote neoplasia at least in part because of such loss-of-function. The interaction between parafibromin and c-myc demonstrated here expands a list of previously established functional interactions between tumor suppressors and oncogenes, including those between tuberin and p21 Ras (28) and between PTEN and PI3K- Akt (29).
Up-regulation of c-myc message and protein has been observed in a variety of benign primary and secondary parathyroid tumors (30). This implies that disinhibition of c-myc expression resulting from parafibromin loss of function would be insufficient in itself to promote the development of malignancy. So beyond the PAF1 complex-mediated inhibition of the c-myc proto-oncogene demonstrated here, given the strong link between HRPT2 inactivation and parathyroid cancer (1, 5, 6), there are likely additional pathways by which loss of parafibromin function critically facilitates malignant transformation.
Methods
Cell Culture and Transfection.
Human HeLa and U2-OS cells were maintained at 37 °C and 5% CO2 in Dulbecco's Modification of Eagle's Medium (DMEM) (Mediatech) with 10% FBS (HyClone), 1% penicillin/streptomycin, and 2 mM glutamine. Normal human primary dermal fibroblast (NHDF) cells (Lonza, Inc.) were maintained in FBM fibroblast basal medium supplemented with hFGF-B, insulin, FBS, and gentamicin/amphotericin-B in the presence of 5% CO2. For transfection, HeLa cells, NHDF cells, and U2-OS cells were transfected at 60–70% confluence 24 h after plating by Lipofectamine 2000 transfection reagent (Invitrogen) in accordance with the manufacturer's instructions. For luciferase assay, NHDF cells were transfected 24 h after seeding by FuGENE 6 transfection reagent (Roche Applied Science).
Cloning the c-myc Promoter Region and Plasmid Construction.
Three regions from the human c-myc promoter were amplified by PCR from normal human genomic DNA using primers as follows: 3.028-Kb, 5′-ACGCGTGGAACAGGCAGACACATCTCAGGG-3′ (forward) and 5′-AAGCTTCGTCGCGGGAGGCTGCTGGAGCGG-3′ (reverse = R2193); 2.255-kb, 5′- ACGCGTGATCCTCTCTCGCTAATCTCCGCC-3′ (forward) and R2193 (reverse); 1.579-kb, 5′-ACGCGTGAGTCGAATGCCTAAATAGGGTG-3′ (forward) and R2193 (reverse) (the MluI and HindIII restriction sites are underlined). The amplified PCR products were cloned directly into the pCR-TOPO vector (Invitrogen) to give pCR-TOPO-c-myc promoters. The sequence of the inserts were confirmed by DNA sequencing. The three fragments were subcloned into the MluI/HindIII sites of pGL3-Basic luciferase reporter vector (Promega) to give the −836/+2193, −63/+2193 and + 613/+2193 pGL3 human c-myc promoter constructs.
RNA Interference.
For gene silencing of endogenous HRPT2 or Paf1, siRNA targeting sequence specific for HRPT2 used was 5′-TTCAGTGTCATACCATGGTAGGTTC-3′ (HRPT2 siRNA-1) and 5′-GGGCACTGCAATTAGTGTTACAGTA-3′ (HRPT2 SiRNA-2); for Paf1 the sequences used were 5′-ATCACCTGAGCACATGGATTGATCC-3′ (Paf1 siRNA-1) and 5′-TAATCATGGCCTGAGACATCATCTC-3′ (Paf1 siRNA-2), respectively, and the duplex RNAi were synthesized (Invitrogen). Cultured HeLa, U2-OS, and NHDF cells were transfected with each of above siRNAs, control siRNA (Stealth RNAi Negative Control, Invitrogen) or transfected in the presence or absence of c-myc siRNA (Santa Cruz). Forty-eight hours after transfection, cells were subjected to specific experiments, as indicated.
Fluorescence-Activated Cell Sorting Analysis and Cell Proliferation Assay.
For analyzing cell cycle distribution, HeLa cells grown in 6 cm cell culture dishes were transfected with control-, HRPT2-, or Paf1-specific siRNA in quadruplicate. Forty-eight hours after transfection, cells were harvested by brief centrifugation, fixed in 70% (vol/vol) ethanol for more than 4 h at −20 °C. Cells were then treated with propidium iodide (50 μg/ml) and RNase A (50 μg/ml) (Sigma Chemical Co.). Stained cells were run on FACScan machine (Becton Dickinson). Twenty thousand cells were collected and the cell cycle distribution was analyzed with ModFit Software (Verity Software). Cell proliferation in vitro was measured using the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega). Briefly, cells were plated into the wells of a 96-well plate at 5,000 per well in sextuplicate. Cells were allowed to grow for 24 h and then transfected with control-, HRPT2-, or Paf1-specific siRNA. 48 h later, 20 μl/well of combined 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium/phenazine ethosulfate solution was added. After incubation of 1 h at 37 °C in a humidified 5% CO2 atmosphere, the absorbance at 490 nM was recorded by using a microplate reader.
Immunoblot Analysis.
Transfected cells were washed twice with PBS and lysed in ice-cold lysis buffer (25 mM Tris·Cl, pH 7.5, 137 mM NaCl, 2.7 mM KCl, 1% Triton X-100, and 1 mM PMSF) containing protease inhibitor mixture (Roche Applied Science). After a brief sonication, whole cell lysates were centrifuged at 15,000 rpm for 5 min to remove insoluble materials. The protein concentration of the supernatants was determined using the BIO-RAD protein assay reagent. Protein extracts were run on SDS/PAGE, transferred to nitrocellulose membrane (Invitrogen) and probed with antibodies for parafibromin, Paf1 and Leo1 (BETHYL Labs), c-myc (Santa Cruz) and β-actin (Sigma). The secondary antibodies used were horseradish peroxidase conjugated (Amersham). Signal detection was carried out with an enhanced chemiluminescence detection system (ECL Plus, Amersham). Quantification of protein band intensities on nitrocellulose membranes was directly acquired and analyzed by quantitative ECL using AlphaImager™ 3400 imaging system and software. The relative intensity of c-myc protein was calculated after normalization with the β-actin band from the same lysate.
Quantitative Real-Time RT-PCR Analysis.
Cultured HeLa cells were transfected with control siRNA, HRPT2- or Paf1-specific siRNA for 24 h or 48 h. At each time point following transfection, total RNA was isolated using RNeasy Mini kit (Qiagen). Quantitative real-time RT-PCR was performed using the Brilliant SYBR Green QRT-PCR Master Mix Kit, 1-Step (Stratagene) and specific primers for human c-myc and β-actin. Relative mRNA expression of c-myc were normalized to the values of β-actin mRNA for each reaction. The following primers were used: c-myc exon 1, 5′-CACGAAACTTTGCCCATAGC-3′ (forward) and 5′-GCAAGGAGAGCCTTTCAGAG-3′ (reverse); c-myc-exon 2, 5′- CCCTCAACGTTAGCTTCACC-3′ (forward) and 5′- AGCAGCTCGAATTTCTTCCA-3′ (reverse); c-myc exon 3, 5′-CAGATCAGCAACAACCGAAA-3′ (forward) and 5′-GGCCTTTTCATTGTTTTCCA-3′ (reverse), β-actin, 5′-GATGCAGAAGGAGATCACTGC-3′ (forward) and 5′-CATACTCCTGCTTGCTGATCC-3′ (reverse). Primer sequence was obtained using Primer3 program and checked for gene specificity by BLASTN.
Dual Luciferase Assay.
Cultured NHDF cells and HeLa cells (5 × 104 cells/well in a twelve-well plate) were acutely cotransfected with a constant amount of the −836/+2193, −63/+2193 and +613/+2193 c-myc promoter luciferase reporters in pGL3, pRL-TK encoding Renilla luciferase with control siRNA, HRPT2- or Paf1-specific siRNA. Forty-eight hours after transfection, cells were lysed and assayed for luciferase activity using the Dual-Luciferase reporter assay system (Promega Corporation) according to the manufacturer's recommendations. The transfection efficiency was normalized using pRL-TK reporter activity.
Protein Decay Rate Analysis.
HeLa cells were transfected with control siRNA, HRPT2- or Paf1-specific siRNA. At 48 h after transfection, cells were treated with cycloheximide (Sigma) at a final concentration of 50 μg/ml. At the indicated time points after adding cycloheximide, cells were collected, and whole cell lysates were subjected to immunoblotting using anti-c-myc or anti-β-actin antibodies, with quantification by ECL as described above. Linear regression analysis of the natural logarithm of the relative actin-normalized c-myc protein band intensity versus time was performed using Prism software version 4.0c (GraphPad Software, Inc.) (31).
Calculation of c-myc Half-Life.
According to the linear regression analysis described above, the y-value at time = 0 for all plots was 4.61 (the natural log of 100[%] relative expression), and a y-value of 3.91 (indicating a relative expression of 50[%]) maps to the time corresponding to the c-myc protein half-life. The Prism software program calculated the slope (m) ± SEM, y-intercept (b) ± SEM, r2 value, and x-value at y = 3.91 (half-life) of the best-fit lines as part of the linear regression analysis. To calculate the standard error of the half-life, the relation y = mx+b was first re-ordered as x = (y−[b ± SEM])/m ± SEM, where x = the half-life, y = 3.91 (the natural log of 50(%)), b = the mean y-intercept, and m = the mean slope of the best-fit line. The standard error of the half-life (SEMx) under each condition was then estimated using the following equation, assuming b and m to be Gaussian variables: SEMx = ([3.91−b]/m) × (31). The unpaired Student's t test was used to evaluate for statistical significance.
Chromatin Immunoprecipitation (ChIP).
ChIP assays were performed employing a kit provided by Upstate Biotechnology, Inc according to manufacturer's instructions. The chromatin solution extracted from HeLa cells was immunoprecipitated overnight with normal rabbit IgG, anti-parafibromin, anti-Paf1 and anti-Leo1 polyclonal antibodies at 4 °C, followed by incubation with protein A agarose/salmon sperm DNA for an additional 1 h at 4 °C. After washing, the immune complexes were eluted with elution buffer (1% SDS and 0.1M NaHCO3) and formaldehyde cross-links were reversed by adding 5M NaCl and heating at 65 °C for 4 h. DNAs of the immunoprecipitates and control input DNA were purified using a QIAquick PCR purification kit (Qiagen) and then analyzed by quantitative real-time PCR using human c-myc-specific primers. The primer sequences targeted the c-myc gene, at a position approximately 2 kB upstream (UP-2K) (5′-TCACGTTTGCCATTACCGGTTC -3′ [forward] and 5′-TTTCAGGTTGGCTGCA G A AGGT-3′ [reverse]; 171 bp PCR product), at + 77 (5′-CAGGGCTTCTCAGAGGCTTGG-3′ [forward] and 5′-CTGCTCGCCCGGCTCTTCCACC-3′ [reverse]; 162 bp PCR product) or at + 1572 (5′-CAGATCAGCAACAACCGAAA-3′ [forward] and 5′-GGCCTTTTCATTGTTTTCCA-3′ [reverse]; 167 bp PCR product) positions.
Supplementary Material
Acknowledgments.
We are grateful to Sunita Agarwal and Stephen Marx for encouragement and helpful discussion. This research was supported by the Intramural Research Program of the NIH, NIDDK to W.F.S.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710725105/DCSupplemental.
References
- 1.Carpten JD, et al. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat Genet. 2002;32:676–680. doi: 10.1038/ng1048. [DOI] [PubMed] [Google Scholar]
- 2.Jackson CE, et al. Hereditary hyperparathyroidism and multiple ossifying jaw fibromas: A clinically and genetically distinct syndrome. Surgery. 1990;108:1006–1012. [PubMed] [Google Scholar]
- 3.Chen JD, et al. Hyperparathyroidism-jaw tumour syndrome. J Intern Med. 2003;253:634–642. doi: 10.1046/j.1365-2796.2003.01168.x. [DOI] [PubMed] [Google Scholar]
- 4.Simonds WF, et al. Familial isolated hyperparathyroidism is rarely caused by germline mutation in HRPT2, the gene for the hyperparathyroidism-jaw tumor syndrome. J Clin Endocrinol Metab. 2004;89:96–102. doi: 10.1210/jc.2003-030675. [DOI] [PubMed] [Google Scholar]
- 5.Shattuck TM, et al. Somatic and germ-line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N Engl J Med. 2003;349:1722–1729. doi: 10.1056/NEJMoa031237. [DOI] [PubMed] [Google Scholar]
- 6.Cetani F, et al. Genetic analyses of the HRPT2 gene in primary hyperparathyroidism: Germline and somatic mutations in familial and sporadic parathyroid tumors. J Clin Endocrinol Metab. 2004;89:5583–5591. doi: 10.1210/jc.2004-0294. [DOI] [PubMed] [Google Scholar]
- 7.Rozenblatt-Rosen O, et al. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yart A, et al. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol Cell Biol. 2005;25:5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu B, et al. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin L, Czapiga M, Nini L, Zhang JH, Simonds WF. Nuclear localization of the parafibromin tumor suppressor protein implicated in the hyperparathyroidism-jaw tumor syndrome enhances its proapoptotic function. Mol Cancer Res. 2007;5:183–193. doi: 10.1158/1541-7786.MCR-06-0129. [DOI] [PubMed] [Google Scholar]
- 11.Woodard GE, et al. Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene. 2005;24:1272–1276. doi: 10.1038/sj.onc.1208274. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C, et al. Parafibromin inhibits cancer cell growth and causes G1 phase arrest. Biochem Biophys Res Commun. 2006;350:17–24. doi: 10.1016/j.bbrc.2006.08.169. [DOI] [PubMed] [Google Scholar]
- 13.Waters CM, Littlewood TD, Hancock DC, Moore JP, Evan GI. c-myc protein expression in untransformed fibroblasts. Oncogene. 1991;6:797–805. [PubMed] [Google Scholar]
- 14.Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–179. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 15.Ho JS, Ma W, Mao DY, Benchimol S. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol Cell Biol. 2005;25:7423–7431. doi: 10.1128/MCB.25.17.7423-7431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerppola TK, Kane CM. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988;8:4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- 18.Pavri R, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 19.Howell VM, et al. HRPT2 mutations are associated with malignancy in sporadic parathyroid tumours. J Med Genet. 2003;40:657–663. doi: 10.1136/jmg.40.9.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villablanca A, et al. Germline and de novo mutations in the HRPT2 tumour suppressor gene in familial isolated hyperparathyroidism (FIHP) J Med Genet. 2004;41:e32. doi: 10.1136/jmg.2003.012369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonds WF, et al. Familial isolated hyperparathyroidism is rarely caused by germline mutation in HRPT2, the gene for the hyperparathyroidism-jaw tumor syndrome. J Clin Endocrinol Metab. 2004;89:96–102. doi: 10.1210/jc.2003-030675. [DOI] [PubMed] [Google Scholar]
- 22.Kelly TG, et al. Surveillance for early detection of aggressive parathyroid disease: carcinoma and atypical adenoma in familial isolated hyperparathyroidism associated with a germline HRPT2 mutation. J Bone Miner Res. 2006;21:1666–1671. doi: 10.1359/jbmr.060702. [DOI] [PubMed] [Google Scholar]
- 23.Mizusawa N, et al. Genetic analyses in patients with familial isolated hyperparathyroidism and hyperparathyroidism-jaw tumour syndrome. Clin Endocrinol (Oxford) 2006;65:9–16. doi: 10.1111/j.1365-2265.2006.02534.x. [DOI] [PubMed] [Google Scholar]
- 24.Guarnieri V, et al. Diagnosis of parathyroid tumors in familial isolated hyperparathyroidism with HRPT2 mutation: Implications for cancer surveillance. J Clin Endocrinol Metab. 2006;91:2827–2832. doi: 10.1210/jc.2005-1239. [DOI] [PubMed] [Google Scholar]
- 25.Bradley KJ, et al. Parafibromin mutations in hereditary hyperparathyroidism syndromes and parathyroid tumours. Clin Endocrinol (Oxford) 2006;64:299–306. doi: 10.1111/j.1365-2265.2006.02460.x. [DOI] [PubMed] [Google Scholar]
- 26.Mosimann C, Hausmann G, Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 27.Kremer R, Bolivar I, Goltzman D, Hendy GN. Influence of calcium and 1,25-dihydroxycholecalciferol on proliferation and proto-oncogene expression in primary cultures of bovine parathyroid cells. Endocrinology. 1989;125:935–941. doi: 10.1210/endo-125-2-935. [DOI] [PubMed] [Google Scholar]
- 28.Freilinger A, Rosner M, Hanneder M, Hengstschlager M. Ras mediates cell survival by regulating tuberin. Oncogene. 2008;27:2072–2083. doi: 10.1038/sj.onc.1210844. [DOI] [PubMed] [Google Scholar]
- 29.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 30.Bjorklund P, Akerstrom G, Westin G. Accumulation of nonphosphorylated beta-catenin and c-myc in primary and uremic secondary hyperparathyroid tumors. J Clin Endocrinol Metab. 2007;92:338–344. doi: 10.1210/jc.2006-1197. [DOI] [PubMed] [Google Scholar]
- 31.Motulsky H. Intuitive Biostatistics. NY: Oxford Univ Press; 1995. pp. 285–286. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.