Abstract
Orthogonal dipolar interactions between amide C=O bond dipoles are commonly found in crystal structures of small molecules, proteins, and protein–ligand complexes. We herein present the experimental quantification of such interactions by employing a model system based on a molecular torsion balance. Application of a thermodynamic double-mutant cycle allows for the determination of the incremental energetic contributions attributed to the dipolar contact between 2 amide C=O groups. The stabilizing free interaction enthalpies in various apolar and polar solvents amount to −2.73 kJ mol−1 and lie in the same range as aromatic-aromatic C–H⋯π and π–π interactions. High-level intermolecular perturbation theory (IMPT) calculations on an orthogonal acetamide/N-acetylpyrrole complex in the gas phase at optimized contact distance predict a favorable interaction energy of −9.71 kJ mol−1. The attractive dipolar contacts reported herein provide a promising tool for small-molecule crystal design and the enhancement of ligand–protein interactions during lead optimization in medicinal chemistry.
Keywords: mutant cycle, protein folding, torsion balance, medicinal chemistry
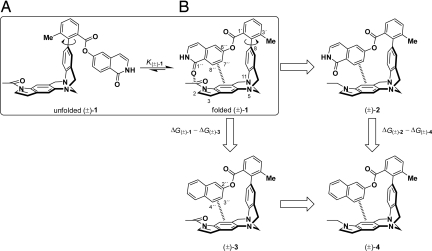
Dipolar contacts involving organic bond dipoles and a carbonyl group, or a pair of carbonyl groups are commonly found both in chemistry and biology (1). On the basis of an analysis of crystal structures of small molecules, proteins, and protein–ligand complexes, it is theorized that, owing to steric constraints, these dipoles preferentially adopt an orthogonal alignment at closest contact distance (Fig. 1). For their expected weakness, the contributions of these interactions have previously been neglected in the study of inter- and intramolecular contacts. Recently, we have been able to determine the free interaction enthalpy between a C–F and a C=O bond dipole ranging between −0.8 kJ mol−1 and −1.2 kJ mol−1 (2, 3). Despite the evident importance of orthogonal interactions between carbonyl dipoles for crystal packing (4–9), peptide secondary structure (10–12), and medicinal chemistry (13), no experimental quantification of the energetic contributions of such interactions has been reported so far. Intermolecular perturbation theory (IMPT) calculations performed by Allen et al. (9) assessed the interaction energy for a perpendicular propan-2-one dimer at an optimal contact distance of dO⋯C=O = 3.02 Å to amount to −7.6 kJ mol−1, comparable to the strength of weak hydrogen bonds. To provide experimental proof for the favorable energetic contributions attributed to orthogonal dipolar carbonyl–carbonyl interactions, we embarked on their determination. We chose to investigate a monomolecular model system, based on the Wilcox molecular torsion balance (14–16), providing for the determination of weak interactions with a greater accuracy and geometric control than given by the study of bimolecular chemical or biological complexes. The primary noncovalent interaction, an aromatic–aromatic edge-to-face C–H⋯π contact, prearranges the functional groups bearing the interacting carbonyl groups rather than enforcing a discrete geometry. The incremental energetic contribution attributed to the orthogonal dipolar C=O⋯C=O contact is subsequently dissected from the primary interaction by applying a chemical double-mutant cycle approach popularized by Hunter and coworkers (17). The work presented herein details the design, the synthesis, and the evaluation of a set of indole-extended molecular torsion balances ((±)-1 to (±)-4 in Fig. 2), mimicking the orthogonal amide carbonyl–amide carbonyl interaction geometries observed in protein and small-molecule crystal structures. The thermodynamic double-mutant cycle depicted in Fig. 2 is constructed and the individual folding free enthalpies were determined in C6D6, CDCl3, C2D2Cl4, and CD2Cl2 solution. The difference in folding free enthalpy between the vertical/horizontal mutations (Eq. 1) provides the incremental free enthalpy attributed to the interaction between the 2 amide carbonyl groups in (±)-1.
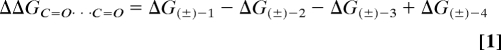 |
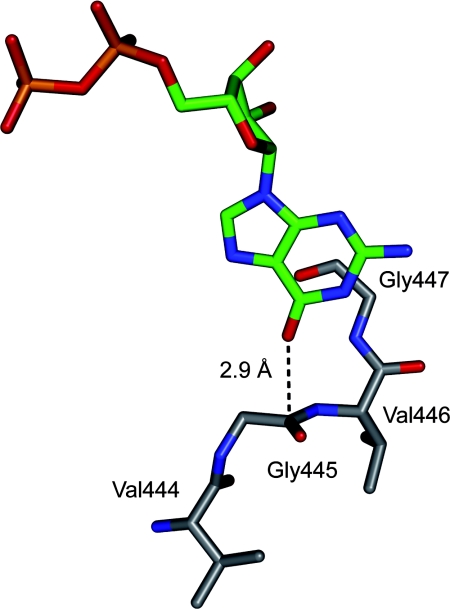
Fig. 1.
Section of the X-ray crystal structure of the complex between adenylosuccinate synthetase and GDP (PDB ID code 1lny; 2.20-Å resolution, EC 6.3.4.4) showing an orthogonal dipolar C=O⋯C=O interaction (18). Color code: ligand skeleton, green; C, gray; O, red; N, blue; P, orange.
Fig. 2.
A set of indole-extended molecular torsion balances for the measurement of orthogonal dipolar C=O⋯C=O interactions. The numbering scheme used throughout this work is represented in (±)-1 and (±)-3. (A) Atropisomerism around a biaryl-type bond (C8–C2′) interconverts the unfolded into the folded conformation. (B) Thermodynamic double-mutant cycle used to assess the incremental contribution of the C=O⋯C=O interaction to the folding free enthalpy of (±)-1.
Results and Discussion
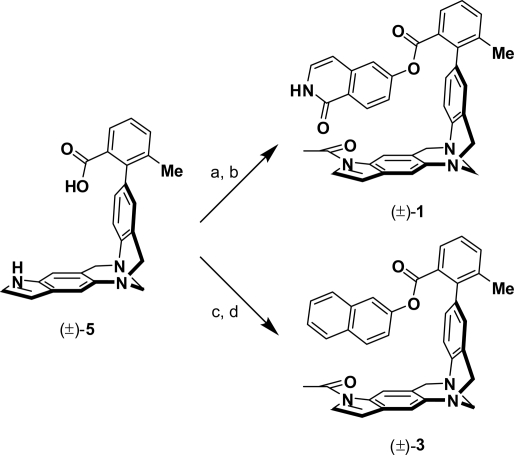
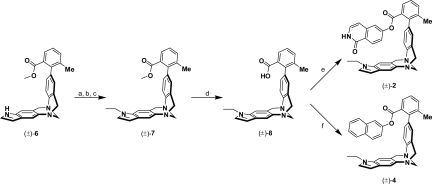
The molecular torsion balances (±)-1 and (±)-3 were synthesized starting from the common carboxylic acid precursor (±)-5 (Fig. 3 and supporting information (SI) Fig. S1) (3). Acylation of the indole N atom, followed by esterification (19) of the carboxylic acid with 6-hydroxyisoquinolin-1(2H)-one furnished a racemic mixture of (±)-1 bearing 2 amide C=O bond dipoles in an orthogonal alignment in the folded conformation. An inverted sequence comprising an esterification of (±)-5 with naphthalen-2-ol and a subsequent acylation of the indole provided the torsion balance (±)-3 in fair yields. To ensure the construction of a valid thermodynamic double-mutant cycle, the indole N atom in the mutant torsion balances (±)-2 and (±)-4 had to be substituted for a noninteracting alkyl chain. Standard protocols for the alkylation of the indole N atom by using various strong or moderate bases and alkyl halides gave predominantly mixtures of quaternary Tröger base (20) salts and the elimination products thereof. Thus, we relied on a stepwise procedure (Fig. 4 and Fig. S2) comprising a selective N-acylation of the methyl benzoate (±)-6, followed by treatment of the N-acetylindole with Lawesson reagent, and subsequent reduction of the thioamide to the N-ethylindole (±)-7 by using a surplus of Raney Nickel (RaNi) in dry THF. Saponification of the methyl ester provided the carboxylic acid (±)-8 which was coupled with 6-hydroxyisoquinolin-1(2H)-one or naphthalen-2-ol under Castro conditions (19) to give the desired molecules (±)-2 and (±)-4, respectively.
Fig. 3.
Synthesis of the molecular torsion balances (±)-1 and (±)-3 starting from the common carboxylic acid precursor (±)-5. (a) Ac2O, DMAP, NEt3, CH2Cl2, 24 °C, 72 h, 60%. (b) 6-Hydroxyisoquinolin-1(2H)-one, BOP, NEt3, CH2Cl2, 24 °C, 45 h, 50%. (c) Naphthalen-2-ol, BOP, NEt3, CH2Cl2, 24 °C, 72 h, 72%. (d) Ac2O, DMAP, NEt3, CH2Cl2, 24 °C, 72 h, 68%. DMAP, 4-N,N-(dimethylamino)pyridine; BOP, benzotriazol-1-yloxy-tris(dimethylamino)phosphonium hexafluorophosphate.
Fig. 4.
Synthesis of the molecular torsion balances (±)-2 and (±)-4 starting from the common methyl benzoate precursor (±)-6. (a) Ac2O, DMAP, NEt3, CH2Cl2, 24 °C, 16 h, 86%. (b) Lawesson reagent, PhMe, 100 °C, 20 h, 63%. (c) RaNi, THF, 24 °C, 25 min, 98%. (d) LiOH, MeOH, H2O, 50 °C, 17 h, 93%. (e) 6-Hydroxyisoquinolin-1(2H)-one, BOP, NEt3, CH2Cl2, 24 °C, 45 h, 29%. (f) Naphthalen-2-ol, BOP, NEt3, CH2Cl2, 24 °C, 72 h, 48%. RaNi, Raney nickel.
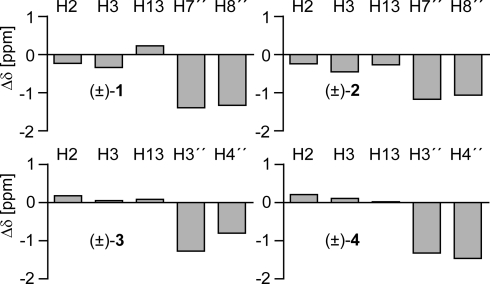
The structure of the molecular torsion balances in solution could be ascertained by extensive 1- and 2-dimensional NMR spectroscopy. In the folded conformation of all 4 torsion balances, the electron-rich indole moiety interacts with the 6-hydroxyisoquinolin-1(2H)-one moiety in (±)-1 and (±)-2 or the naphthalene-2-ol moiety in (±)-3 and (±)-4 in an aromatic edge-to-face C–H⋯π interaction. The H atoms on C7″ and C8″ in (±)-1 and (±)-2 and the respective protons adjacent to C3″ and C4″ in (±)-3 and (±)-4 come to rest within the shielding cone of the indole ring. Consistent with the observations in previous torsion balances, the respective 1H NMR signals in C6D6 are shifted by Δδ = −0.8 to −1.5 ppm to higher field on folding (Fig. 5). The tautomeric 1-hydroxyisoquinoline/isoquinolin-1(2H)-one equilibrium lies predominantly on the side of the latter (21); thus, the anisotropic effect of the carbonyl group in (±)-1 and (±)-2 shifts the resonance signals of the protons on C2 and C3 of the indole to higher field with respect to the unfolded conformation. The deshielding cone of the naphthyl moiety instead induces a weak downfield shift in the indole hydrogens. The observation of a significant 1H–1H NOE between the protons of the acetyl group on the indole N atom in the folded conformation of torsion balance (±)-1 and the hydrogen on N2″ of the isoquinolin-1(2H)-one moiety (CH3⋯HN distance <4.5 Å) indicates of a sub van der Waals contact distance between the interacting C=O bond dipoles (see Fig. S3). The overall geometry of the folded state is further corroborated by X-ray crystal structures of related torsion balances (2, 3).
Fig. 5.
Shifts of the 1H NMR signals of selected protons in the folded conformation relative to the unfolded torsion balances.
On the NMR spectroscopy timescale, the interconversion between the rotational atropisomers is slow (ΔG‡ ≥ 67 kJ mol−1 at 273 K) (14), thus, 2 separate resonance signals for the methyl group on C3′ of the rotor, corresponding to the folded and the unfolded conformation of the torsion balances, can be observed. The 1H NMR spectra recorded at 298 K in C6D6, CDCl3, C2D2Cl4, and CD2Cl2 (see Fig. S4) were line-fitted to Lorentz functions and integrated to determine the relative population of both conformations at thermodynamic equilibrium. The resulting folding free enthalpies derived from the equilibrium constants at 298 K are summarized in Table 1.
Table 1.
Folding free enthalpies ΔG of torsion balances (±)-1 to (±)-4 in C6D6, CDCl3, C2D2Cl4, and CD2Cl2
| Compound | ΔG, kJ mol−1* |
|||
|---|---|---|---|---|
| C6D6 | CDCl3 | C2D2Cl4 | CD2Cl2 | |
| (±)-1 | −3.00 | −1.15 | −0.40 | −1.54 |
| (±)-2 | −0.92 | 0.15 | 1.05 | −0.40 |
| (±)-3 | −0.85 | −0.48 | −0.04 | −0.84 |
| (±)-4 | −1.50 | −0.69 | −0.29 | −0.93 |
*Determined by integration of line-fitted 1H NMR (500 MHz) spectra of 2 mM solutions at 298 K. Uncertainty: ±0.12 kJ mol−1. For an error analysis see the SI Text.
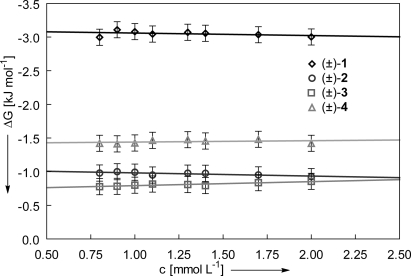
The atropisomeric equilibrium of all torsion balances in C6D6 and CD2Cl2 lies predominantly on the side of the folded conformation, reflected in the negative values of ΔG. In CDCl3 and C2D2Cl4, however, the population of the folded conformation is substantially decreased. We attribute this observation to interactions between solvent molecules and the Tröger base cleft, shifting the atropisomeric equilibrium toward the unfolded conformation of the torsion balances. The introduction of a functional group in (±)-1 and (±)-2, namely the isoquinolin-1(2H)-one moiety, capable of interacting via hydrogen bonds with the solvent or other solute molecules, imposes an uncertainty on the accuracy of the determination of the respective absolute folding free enthalpies. For steric reasons a dimerization of the torsion balances in solution is only possible in the unfolded conformation. As a consequence, the hydrogen bonding interaction would most certainly shift the atropisomeric equilibrium of (±)-1 and (±)-2 toward the unfolded conformation, thereby underestimating the interaction between the amide carbonyl dipoles. The effect should be particularly strong in C6D6 and weaker in CDCl3, as observed for the dimerization of isoquinolin-1(2H)-one (Kdim = 333 L−1 in C6D6, Kdim = 57 L−1 in CDCl3; see Fig. S5). To assess the influence of intermolecular hydrogen-bonding interactions on the folding free enthalpy ΔG of the torsion balances (±)-1 to (±)-4, a dilution study in C6D6 was performed (Fig. 6). Within the experimental errors, essentially no concentration dependence on the folding equilibrium could be observed.
Fig. 6.
Concentration dependence of the folding equilibrium of the torsion balances (±)-1 to (±)-4 in C6D6 solution acquired at 298 K.
With the differences in folding free enthalpy ΔG determined for each torsion balance, the double-mutant cycle depicted in Fig. 2 can be applied. The horizontal mutations [(±)-1→(±)-2 and (±)-3→(±)-4, respectively] account for the changes in electron density in the π-system of the indole moiety, on substitution of an electron-withdrawing acetyl group on N1 by an electron-donating ethyl residue. The vertical mutations (±)-1→(±)-3, and (±)-2→(±)-4 reflect the perturbation of the electronic structure of the edge component induced by the lactam moiety. The computed (Eq. 1) interaction free enthalpy ΔΔG298K between 2 amide C=O bond-dipoles is strongest in C6D6 (−2.73 ± 0.25 kJ mol−1). In chlorinated solvents, such as CDCl3 (−1.50 ± 0.25 kJ mol−1), C2D2Cl4 (−1.70 ± 0.25 kJ mol−1), and CD2Cl2 (−1.22 ± 0.25 kJ mol−1), the dipolar interaction is substantially weaker. This observation can be attributed to a stronger interaction of dipolar solvents with both amide groups in (±)-1 shifting the thermodynamic equilibrium toward the unfolded conformation. A closer analysis of the thermodynamic data in Table 1 reveals that the mutation of an N-ethyl-substituted indole to an N-acylated indole [(±)-4→(±)-3] brings about a small unfavorable change in folding free enthalpy of +0.09 to +0.65 kJ mol−1 in C6D6 and the chlorinated solvents. The electron density of the indole decreases because of the introduction of an electron-withdrawing substituent, and thus weakens the primary edge-to-face interaction. The vertical mutation [(±)-4→(±)-2] accounts for the difference in folding free energy between a naphthyl and an isoquinolin-1(2H)-one ester. On introduction of the latter, the folding free energy decreases. Despite the fact that the electron-withdrawing effect of the amide group stabilizes the C–H⋯π interaction, the repulsion between the lone pairs of the carbonyl O atom and the p-orbital of the indole N atom dominates the interaction. As a consequence, the equilibrium is shifted toward the unfolded conformation. The effect is weakest in C6D6 (+0.58 kJ mol−1) and CD2Cl2 (+0.53 kJ mol−1), whereas it amounts to +0.84 kJ mol−1 and +1.34 kJ mol−1 in CDCl3 and C2D2Cl4, respectively.
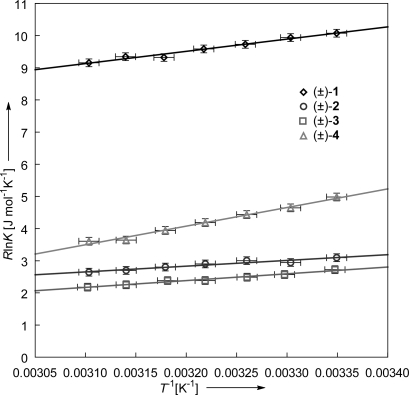
In an effort to gain a deeper insight into the driving force of the folding process, we dissected the overall folding free enthalpy ΔG into the enthalpic ΔH and the entropic ΔS components. Variable temperature 1H NMR experiments in C6D6 were performed to determine the temperature dependence of the folding equilibrium in a range from 298 K to 322 K. On heating 2 mM solutions of the torsion balances (±)-1, (±)-2, (±)-3, and (±)-4 in C6D6 (see Fig. S6), a shift of the folding equilibrium toward the unfolded conformation can be observed in the 1H NMR spectra. As a consequence, the folding free enthalpy ΔG at 322 K decreases by +0.06 kJ mol−1 to +0.33 kJ mol−1. The individual equilibrium constants at different temperatures were plotted as RlnK against T−1 (van′t Hoff plot), and are depicted in Fig. 7. Both the enthalpy ΔH and the entropy ΔS were determined by fitting linear equations to the experimental data points. The goodness of fit (r2) exceeds 0.98 for (±)-1, (±)-3, (±)-4, and 0.93 for (±)-2.
Fig. 7.
Van′t Hoff plot of RlnK against T−1 for the torsion balances (±)-1 to (±)-4 in C6D6. The goodness of fit (r2) for the linear equations exceeds 0.93 in all cases.
Table 2 summarizes the individual thermodynamic parameters. The folding process is enthalpy-driven for all torsion balances with the strongest contribution determined for (±)-1 and (±)-4 (ΔH = −3.81 kJ mol−1 and ΔH = −5.79 kJ mol−1, respectively). The folding enthalpy for (±)-2 and (±)-3 is weaker, reflecting the observations in the double-mutant cycle. The horizontal mutation (±)-4→(±)-3 reduces the electron density of the indole, therefore weakening the edge-to-face interaction, reflected in a decrease in folding enthalpy (δΔH = ΔH(±)-3 − ΔH(±)-4 = +3.68 kJ mol−1). The opposite effect can be observed for the substitution of the ethyl residue on N1 in (±)-2 by an acetyl group ((±)-2→(±)-1). Despite the electron density in the indole being reduced on introduction of a COCH3 group, the folding free enthalpy increases (δΔH = ΔH(±)-1 − ΔH(±)-2 = −2.03 kJ mol−1). It is intriguing to relate this difference in enthalpy (ΔΔH = ΔH(±)-1 − ΔH(±)-2 − ΔH(±)-3 + ΔH(±)-4 = −5.71 kJ mol−1) to the interaction between the orthogonal amide C=O bond-dipoles. However, it cannot be concluded from the data whether this is the unique contribution. Solvent–solvent, as well as solvent–solute interactions, or a cooperative effect enforcing an optimized primary edge-to-face interaction have to be considered. In the case of torsion balance (±)-4, the stronger enthalpic component ΔH is mirrored in a decrease in entropy ΔS, indicative of a tighter interaction in the folded conformation. This efficient enthalpy–entropy compensation levels out the overall differences in folding free enthalpy in Table 1.
Table 2.
Folding enthalpies ΔH and folding entropies ΔS determined graphically
| Compound | ΔH, kJ mol−1* | ΔS, J mol−1 K−1* | TΔS298K, kJ mol−1 |
|---|---|---|---|
| (±)-1 | −3.81 ± 0.26 | −2.67 ± 0.86 | −0.80 ± 0.26 |
| (±)-2 | −1.78 ± 0.22 | −2.86 ± 0.71 | −0.85 ± 0.21 |
| (±)-3 | −2.11 ± 0.14 | −4.36 ± 0.44 | −1.30 ± 0.13 |
| (±)-4 | −5.79 ± 0.28 | −12.46 ± 0.90 | −4.31 ± 0.27 |
*Errors were determined graphically.
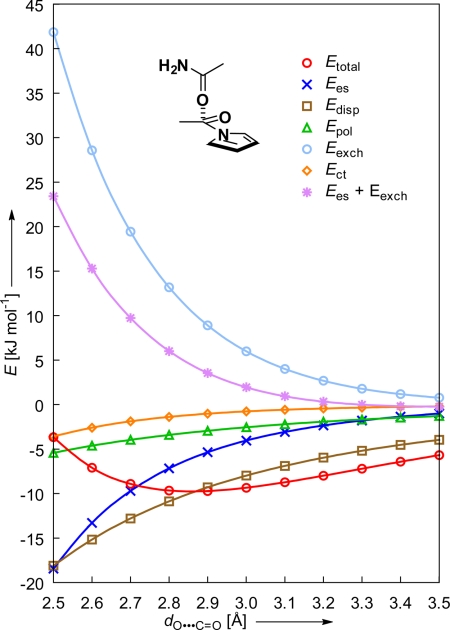
Intermolecular perturbation theory (IMPT) calculations (see Computational Methods for full details) were performed on an acetamide/N-acetylpyrrole model mimicking the orthogonal dipolar interaction geometry in (±)-1 (see Fig. S7). This geometry was chosen based on extensive analyses of C=O⋯C=O (9) and C≡N⋯C≡N (22) interactions in crystal structures obtained from the CSD (23). The Etotal profile illustrated in Fig. 8 is dominated by attractive contributions from dispersion Edisp and electrostatic Ees interactions, counterbalanced by the exchange repulsion term Eexch. Both the polarization Epol and the contributions attributed to charge transfer interactions Ect are smaller in magnitude. The interaction energy Etotal between the 2 amide groups is minimized (−9.71 kJ mol−1) at a O⋯C=O contact distance of 2.9 Å. With respect to the propan-2-one dimer (Etotal,min = −8.02 kJ mol−1, dO⋯C=O = 3.0 Å; see Fig. S8) both an increase in polarization and dispersion interactions can be observed for the acetamide/N-acetylpyrrole model. These changes can be accredited to an increase in dipole moment and the extended delocalization of the amide N atom lone pair with the C=O group.
Fig. 8.
Ab initio-based molecular orbital calculations (6–31G**) using intermolecular perturbation theory (IMPT) applied to an orthogonal interaction between an acetamide and an N-acetylpyrrole mimicking the dipolar interaction in torsion balance (±)-1.
In summary, we have presented the design, synthesis, and evaluation of a set of molecular torsion balances, providing experimental evidence for the existence of attractive orthogonal dipolar interactions between 2 amide carbonyl groups. The measured contributions of this dipolar contact to the overall folding free enthalpy ΔG of (±)-1 lie in the range of −2.73 kJ mol−1 (C6D6) to −1.22 kJ mol−1 (CD2Cl2). Because of crystal packing effects, solvent effects, and sterically constrained molecular geometries, an ideal interaction reflecting the calculated values will most certainly never be observed in crystal structures or in solution. Yet, the enthalpic (ΔΔH) contribution to the dipolar interaction of torsion balance (±)-1 in C6D6 amounts to almost 60% of the calculated interaction energy of an orthogonal acetamide/N-acetylpyrrole complex in the gas phase at optimized contact distance. Aside from providing a promising tool for the enhancement of small-molecule and ligand–protein interactions in crystal design as well as in medicinal chemistry, the study of attractive orthogonal dipolar interactions between carbonyl groups is expected to contribute new insights into the process of protein folding and the stabilization of secondary protein structures.
Materials and Methods
Protocols for the determination of the folding free enthalpy of the torsion balances, an error analysis, the procedures for the synthesis of (±)-1 to (±)-4, and the Figs. S1– S8 can be found in the SI Text.
Computational Methods.
The intermolecular perturbation theory (IMPT) of Hayes and Stone (24) as implemented in the CADPAC 6.5 program package (25) was used to calculate intermolecular interaction energies between orthogonal C=O dipoles. The IMPT methodology has been widely used to calculate interaction energies in a variety of systems (26–29). The model systems used in this study were the propan-2-one orthogonal dimer, as described by Allen et al. (9) and the analogous acetamide/N-acetylpyrrole dimer as shown in Fig. 8 and, in more detail, in Fig. S7. The IMPT methodology yields separate interaction energy components (first order, electrostatic and exchange-repulsion energies; second order, polarization, charge-transfer, and dispersion energies) which sum to a total interaction energy (Etotal) that is free from basis-set superposition errors (30). In each set of calculations, the monomers were first geometry optimized separately by using the 6–31G** basis set. The dimer interaction energies were then calculated by using the resulting fixed intramolecular geometries at varying intermolecular C=O⋯C=O distances.
Supplementary Material
Acknowledgments.
F.R.F. and F.D. thank Dr. J. Dunitz, Prof. Dr. W. van Gunsteren, and Prof. Dr. B. Jaun (all at ETH Zurich) for valuable discussions. This work was supported by a grant from the ETH Research Council and the Fonds der Chemischen Industrie.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806129105/DCSupplemental.
References
- 1.Paulini R, Müller K, Diederich F. Orthogonal multipolar interactions in structural chemistry and biology. Angew Chem Int Ed. 2005;44:1788–1805. doi: 10.1002/anie.200462213. [DOI] [PubMed] [Google Scholar]
- 2.Hof F, Scofield DM, Schweizer WB, Diederich F. A weak attractive interaction between organic fluorine and an amide group. Angew Chem Int Ed. 2004;43:5056–5059. doi: 10.1002/anie.200460781. [DOI] [PubMed] [Google Scholar]
- 3.Fischer FR, Schweizer WB, Diederich F. Molecular torsion balances: Evidence for favorable orthogonal dipolar interactions between organic fluorine and amide groups. Angew Chem Int Ed. 2007;46:8270–8273. doi: 10.1002/anie.200702497. [DOI] [PubMed] [Google Scholar]
- 4.Bolton W. The crystal structure of alloxan. Acta Crystallogr. 1964;17:147–152. [Google Scholar]
- 5.Davies DR, Blum JJ. The crystal structure of parabanic acid. Acta Crystallogr. 1955;8:129–136. [Google Scholar]
- 6.Chu SSC, Jeffrey GA, Sakurai T. The crystal structure of tetrachloro-p-benzoquinone (chloranil) Acta Crystallogr. 1962;15:661–671. [Google Scholar]
- 7.Bolton W. The crystal structure of anhydrous barbituric acid. Acta Crystallogr. 1963;16:166–173. [Google Scholar]
- 8.Bolton W. The crystal structure of triketoindane (anhydrous ninhydrin). A structure showing close C=O⋯C interactions. Acta Crystallogr. 1965;18:5–10. [Google Scholar]
- 9.Allen F, Baalham CA, Lommerse JPM, Raithby PR. Carbonyl-carbonyl interactions can be competitive with hydrogen bonds. Acta Crystallogr B. 1998;54:320–329. [Google Scholar]
- 10.Maccallum PH, Poet R, Milner-White EJ. Coulombic interactions between partially charged main-chain atoms not hydrogen-bonded to each other influence the conformations of α-helices and antiparallel β-sheet. A new method for analyzing the forces between hydrogen bonding groups in proteins includes all of the coulombic interactions. J Mol Biol. 1995;248:361–373. doi: 10.1016/s0022-2836(95)80056-5. [DOI] [PubMed] [Google Scholar]
- 11.Maccallum PH, Poet R, Milner-White EJ. Coulombic attractions between partially charged main-chain atoms stabilizes the right-handed twist found in most β-strands. J Mol Biol. 1995;248:374–384. doi: 10.1016/s0022-2836(95)80057-3. [DOI] [PubMed] [Google Scholar]
- 12.Deane CM, Allen FH, Taylor R, Blundell TL. Carbonyl-carbonyl interactions stabilize the partially allowed Ramachandran conformations of asparagines and aspartic acid. Protein Eng. 1999;12:1025–1028. doi: 10.1093/protein/12.12.1025. [DOI] [PubMed] [Google Scholar]
- 13.Bergner A, Günther J, Hendlich M, Klebe G, Verdonk M. Use of Relibase for retrieving complex three-dimensional interaction patterns including crystallographic packing effects. Biopolymers. 2002;61:99–110. doi: 10.1002/1097-0282(2001/2002)61:2<99::AID-BIP10075>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Paliwal S, Geib S, Wilcox CS. Molecular torsion balances for weak molecular recognition forces. Effects of “tilted-T” edge-to-face aromatic interactions on conformational selection and solid state structure. J Am Chem Soc. 1994;116:4497–4498. [Google Scholar]
- 15.Kim E-I, Paliwal S, Wilcox CS. Measurement of molecular electrostatic field effects in edge-to-face aromatic interactions and CH–π interactions with implications for protein folding and molecular recognition. J Am Chem Soc. 1998;120:11192–11193. [Google Scholar]
- 16.Bhayana B, Wilcox CS. A minimal protein folding model to measure hydrophobic and CH–π effects on interactions between nonpolar surfaces in water. Angew Chem Int Ed. 2007;46:6833–6836. doi: 10.1002/anie.200700932. [DOI] [PubMed] [Google Scholar]
- 17.Cockroft SL, Hunter CA. Chemical double-mutant cycles: Dissecting non-covalent interactions. Chem Soc Rev. 2007;36:172–188. doi: 10.1039/b603842p. [DOI] [PubMed] [Google Scholar]
- 18.Iancu CV, Borza T, Fromm HJ, Honzatko RB. IMP, GTP, and 6-phosphoryl-IMP complexes of recombinant mouse muscle adenylosuccinate synthetase. J Biol Chem. 2002;277:26779–26787. doi: 10.1074/jbc.M203730200. [DOI] [PubMed] [Google Scholar]
- 19.Castro B, Evin G, Selve C, Seyer R. Peptide coupling reagents: VIII. A high yielding preparation of phenyl esters of amino acids using benzotriazolyloxytris[dimethylamino]phosphonium hexafluorophosphate (BOP reagent) Synthesis. 1977:413–413. [Google Scholar]
- 20.Tröger J. Über einige mittels nascierenden Formaldehyds entstehende Basen (About some bases formed via nascent formaldehyde) J Prakt Chem. 1887;36:225–245. [Google Scholar]
- 21.Fabian WMF. Tautomeric equilibria of heterocyclic molecules. A test of the semiempirical AM1 and MNDO-PM3 methods. J Comput Chem. 1991;12:17–35. [Google Scholar]
- 22.Wood PA, Borwick SJ, Watkin DJ, Motherwell WDS, Allen FH. Dipolar C≡N⋯C≡N interactions in organic crystal structures: Database analysis and calculation of interaction energies. Acta Crystallogr B. 2008;64:393–396. doi: 10.1107/S0108768108010239. [DOI] [PubMed] [Google Scholar]
- 23.Allen FH. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr B. 2002;58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 24.Hayes IC, Stone AJ. An intermolecular perturbation theory for the region of moderate overlap. J Mol Phys. 1984;53:83–105. [Google Scholar]
- 25.Amos RD, et al. CADPAC 6.5. The Cambridge Analytical Derivatives Package: A Suite of Quantum Chemistry Programs. Lensfield Road, Cambridge, England: Chemistry Department, Cambridge University; 1998. [Google Scholar]
- 26.Stone AJ. The theory of intermolecular forces. Oxford: Clarendon Press; 1996. [Google Scholar]
- 27.Lommerse JPM, Stone AJ, Taylor R, Allen FH. The nature and geometry of intermolecular interactions between halogens and oxygen or nitrogen. J Am Chem Soc. 1996;118:3108–3116. [Google Scholar]
- 28.Dunitz JD, Taylor R. Organic fluorine hardly ever accepts hydrogen bonds. Chem Eur J. 1997;3:89–98. [Google Scholar]
- 29.Nobeli I, Yeoh SL, Price SL, Taylor R. On the hydrogen bonding abilities of phenol and anisoles. Chem Phys Lett. 1997;280:196–202. [Google Scholar]
- 30.Stone AJ. Computation of charge-transfer energies by perturbation theory. Chem Phys Lett. 1993;211:101–109. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.