Abstract
The thiamin- and flavin-dependent peripheral membrane enzyme pyruvate oxidase from E. coli catalyzes the oxidative decarboxylation of the central metabolite pyruvate to CO2 and acetate. Concomitant reduction of the enzyme-bound flavin triggers membrane binding of the C terminus and shuttling of 2 electrons to ubiquinone 8, a membrane-bound mobile carrier of the electron transport chain. Binding to the membrane in vivo or limited proteolysis in vitro stimulate the catalytic proficiency by 2 orders of magnitude. The molecular mechanisms by which membrane binding and activation are governed have remained enigmatic. Here, we present the X-ray crystal structures of the full-length enzyme and a proteolytically activated truncation variant lacking the last 23 C-terminal residues inferred as important in membrane binding. In conjunction with spectroscopic results, the structural data pinpoint a conformational rearrangement upon activation that exposes the autoinhibitory C terminus, thereby freeing the active site. In the activated enzyme, Phe-465 swings into the active site and wires both cofactors for efficient electron transfer. The isolated C terminus, which has no intrinsic helix propensity, folds into a helical structure in the presence of micelles.
Keywords: electron transfer, membrane protein, X-ray crystallography
Reversible binding of peripheral membrane proteins to the lipid bilayer regulates cell signaling, lipid metabolism and many other cellular events. Proteins that adhere directly to the biological membrane are termed amphitropic proteins and can attach to the bilayer through interaction of amphipathic helices, hydrophobic loops, ions, or covalently attached lipids (1, 2). In many cases studied, these proteins exhibit a very low basal membrane affinity, becoming recruited to the membrane from the cytosol only after a conformational transition or electrostatic switch that not only triggers membrane binding but may also initiate or elevate biological activity (3). Despite many recent advances in understanding how membrane binding and concomitant functional activation of proteins are regulated, there remains a paucity of structural data that allow detailed atomic insights into the nature of reversible protein-membrane interaction and of structural transitions that trigger membrane binding and functionality.
In this regard, the thiamin diphosphate- (ThDP, the functional derivative of vitamin B1) and flavin-dependent pyruvate oxidase from Escherichia coli (EcPOX, EC 1.2.2.2) is a particularly interesting and extensively studied peripheral membrane protein that feeds electrons from the cytosol directly into the respiratory chain at the membrane (4–11). EcPOX supports aerobic growth in E. coli as a backup system to the pyruvate dehydrogenase multienzyme complex and catalyzes the oxidative decarboxylation of the metabolite pyruvate to carbon dioxide and acetate (12). The 2 electrons arising from oxidation of pyruvate at the ThDP site are transferred initially to the neighboring flavin (Eq. 1). Reduction of the flavin is thought to induce a structural rearrangement of the protein that exposes a high affinity lipid binding site at the C terminus (13, 14). After adhering to the biological membrane, the 2 electrons residing on the flavin are shuttled to ubiquinone 8 (Q8) (7), a membrane-bound mobile electron carrier of the electron transport chain (Eq. 2).
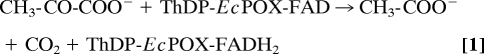 |
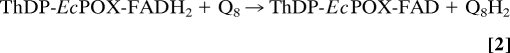 |
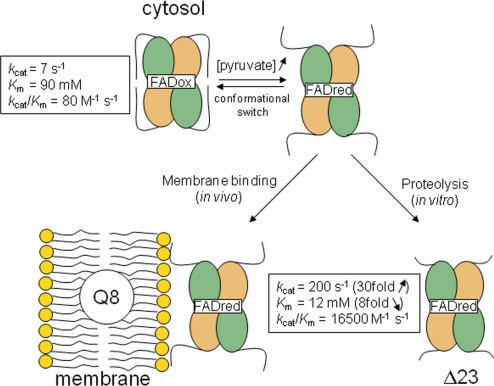
In vitro, the activity of EcPOX can be monitored in reductase assays where artificial electron acceptors such as ferricyanide substitute for the native substrate Q8. The basal enzymatic reductase activity of EcPOX is rather low but is stimulated after binding to lipid amphiphiles (6), or alternatively, under in vitro conditions, by limited proteolytic digestion (Fig. 1) (15, 16). Activation of EcPOX is of a hybrid V- and K-type, affecting both turnover (Vmax is ≈30-fold increased) and affinity for the substrate pyruvate (Kmapp is ≈10-fold decreased) (15–18). Transient kinetic studies have suggested that electron transfer (ET) between the thiamin and flavin, enhanced by several orders of magnitude in the activated enzyme, is the major source of catalytic (Vmax) stimulation (19). In addition, the spectroscopic signature of the FAD in the Vis-region is changed upon activation, suggesting a more open and solvent accessible active site, in line with the observed substrate Km decrease after activation (13, 14).
Fig. 1.
Activation model of EcPOX. At low pyruvate concentrations, the enzyme is located in the cytosol and exhibits a low basal activity and low substrate affinity. High pyruvate concentrations cause reduction of the flavin and a structural rearrangement that exposes the C terminus as a high affinity lipid binding site. Binding of the reduced enzyme to the membrane (in vivo) or mild proteolytic digestion (in vitro) lock in the activated conformation with elevated activity and substrate affinity (17).
Remarkably, limited proteolysis and binding to lipid amphiphiles yield activated enzyme with similar kinetic properties, suggesting that both activation methods generate enzyme species with similar structural and functional traits (17). For proteolytic activation (13, 20), the full-length enzyme (EcPOX1–572) is commonly treated with α-chymotrypsin, which cleaves off the last 23 residues from the C terminus—referred to as the “α-peptide” (AP, EcPOX550–572) (21)—to give a fully active Δ23 truncation variant (EcPOXΔ23) that serves as a functional substitute for lipid-activated enzyme. On the basis of the primary structure of the putative membrane-binding AP, it has been postulated that residues G559-D-E-V-I-E-L-A-K-T568 fold into an amphipathic helix that mediates membrane binding (21). In line with this hypothesis, the isolated AP has been shown to bind tightly to phospholipid vesicles in vitro (22). A genetically engineered Δ24 EcPOX1–548 variant was found to be deficient in membrane binding while in steady-state reductase assays it exhibited kinetic constants comparable to the proteolytically generated Δ23 truncation variant (23).
Here, we present the X-ray structures of the full-length EcPOX and a proteolytically activated carboxyl-terminal Δ23 variant. These provide not only detailed insights into structural determinants of lipid association and catalytic activation in response to a conformational transition of the carboxyl terminus in concert with an active center loop, but also shed light on how enzymes may switch on and off ET between 2 spatially proximal redox cofactors.
Results and Discussion
The activity of full-length EcPOX and EcPOXΔ23 was determined in established reductase assays where ferricyanide substitutes for the native substrate Q8. The kinetic steady-state constants are in good agreement with those reported in the literature (17) (see Fig. 1), clearly confirming that proteolytic treatment has a dual activating affect on catalysis as both Vmax and substrate affinity are multifold enhanced. Limited proteolysis studies on EcPOX indicate that activation of EcPOX involves 2 structural transitions, the first of which is caused by the formation of covalent intermediates at the thiamin site and the second by reduction of the flavin [supporting information (SI) Fig. S1]. The latter step is necessary for exposing the lipid binding site, suggesting a redox-sensing mechanism.
Overall Structure of Full-Length EcPOX.
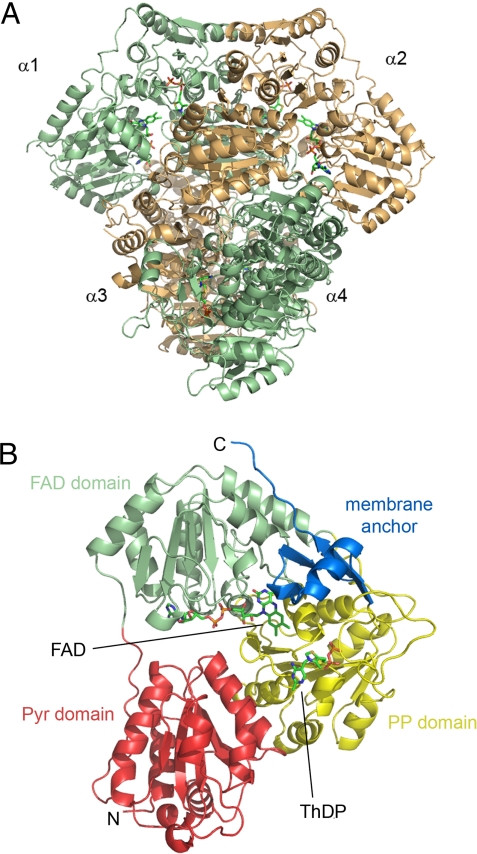
The crystal structure of EcPOX in the full-length form has been determined by molecular replacement-phasing using the structure of the related pyruvate oxidase from Lactobacillus plantarum as a search model (LpPOX; PDB entry 1POW), and refined to an Rcryst/Rfree of 0.181/0.216 against data to 2.9 Å resolution (Table S1). The protein crystallizes in the tetragonal spacegroup P43212 with 2 monomers in the crystallographic asymmetric unit. Two functional dimers with identical active sites at the dimer interface constitute the biologically relevant homotetramer (Fig. 2A). There is no structural non-equivalence of the monomers detectable as observed for pyruvate dehydrogenase E1 component, for which a half-of-sites mechanism has been suggested (Fig. S2) (24, 25). The subunit structure of EcPOX (Fig. 2B) is very similar to that of LpPOX (Fig. S3) (26, 27) and related enzymes of the thiamin enzyme superfamily (28), consisting of 3 main domains:(i) the Pyr domain (1–182) that contacts the pyrimidine moiety of ThDP of the corresponding subunit; (ii) the FAD-binding domain (183–344); and (iii) the PP-binding domain (345–530) that binds the diphosphate (formerly termed pyrophosphate) portion of ThDP. The C-terminal membrane binding region (residues 531–572, blue in Fig. 2B), a unique motif within the thiamin enzyme superfamily, constitutes a separate domain.
Fig. 2.
Overall structure of the tetramer (A) and a monomer (B) of full-length EcPOX in diagram representation. The cofactors ThDP and FAD, marking the positions of the active site, are shown as sticks. (A) The corresponding monomers of the 2 functional dimers (α1-α2 and α3-α4) are colored in orange and green, respectively. (B) The 4 different domains of EcPOX monomer α1 (Pyr domain − FAD domain − PP domain − membrane binding C terminus) are indicated and colored individually. View is rotated ≈90° clockwise from A.
Structure of the Membrane-Binding C Terminus.
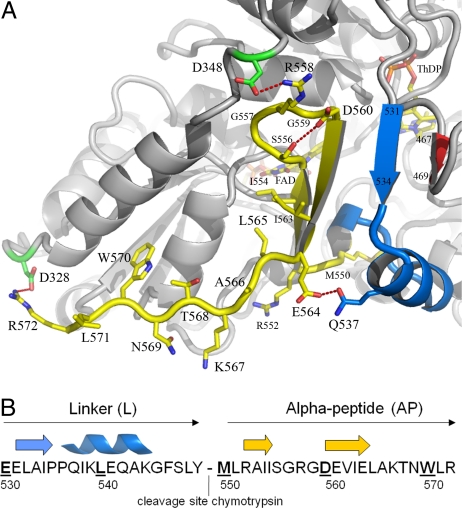
The C-terminal domain of EcPOX exhibits well defined electron density for the main chain and all side chains (Fig. S4). It covers the active center cleft such that only the thiazolium part of ThDP is solvent accessible (Fig. 3). The domain may be further subdivided into a short linker region (residues 531–549) and the alpha-peptide part (AP, residues 550–572), which in the activated state serves as the membrane anchor. The linker region consists of a single stranded ß-sheet (ß531–534) and an alpha-helix (α536–544). The AP forms a 2-stranded antiparallel ß-sheet structure (residues 550–564) that together with 1 strand of an active center loop (ß467–469) and the linker strand constitutes a 4-stranded half barrel motif. The very last 8 C-terminal amino acids (L565-AKTNWL-R572) do not exhibit a defined secondary structure (Fig. 3). The loop connecting the antiparallel strands of AP contains 2 glycine residues and appears to be stabilized by numerous main chain contacts and an intramolecular hydrogen-bonding interaction between S556 and D560. Additionally, E564 of AP and N537 of the neighboring linker helix form a hydrogen bond. The side chain of R558, located in the loop between the antiparallel sheets, contacts the side chain of D348, which belongs to the partially unwound part of the final helix of the FAD domain (α330–349). Finally, the carboxyl-terminal R572 forms an additional salt bridge with D328 of the FAD domain. The structure thus reveals that the C-terminal domain is held firmly in place by the half-barrel super secondary structure and a large number of hydrogen-bonding and electrostatic interactions evenly distributed over the whole sequence. Earlier mutagenesis data had already indicated that several residues of the C terminus (E564, R572) and residues of the active center loop (A467) interacting with the half-barrel motif play critical roles for membrane binding and activity (29, 30). Most remarkably, many of the residues implicated in forming the proposed membrane-binding amphipathic helix (G559-D-E-V-I-E-L-A-K-T568) are located in the sheet structure. To analyze the structure of AP550–572 without the constraints imposed by other domains of the protein, AP550–572 was chemically synthesized and subjected to CD spectroscopic analysis (Fig. S5). In aqueous buffered solution, the CD spectra of AP suggest a random structure with minor amounts of turn and sheet elements. In contrast, AP550–572 clearly forms a helical structure in the presence of micelles as also observed with a positive control in the presence of 50% trifluorethanol. These data support and extend the amphipathic helix model suggested for membrane binding of EcPOX. Release of the C-terminal domain from interactions with the other domains should result in a structurally flexible peptide that can undergo a disorder-order (helix) transition upon contact with the membrane.
Fig. 3.
Structure of the C-terminal membrane-binding domain of EcPOX. (A) Diagram representation of EcPOX in gray with the C terminus highlighted: blue, linker region (residues 531–549); yellow, alpha-peptide part (residues 550–572). The cofactors ThDP and FAD, and selected amino acid side chains are shown in stick representation. Selected hydrogen-bonding and electrostatic interactions are indicated. (B) Primary sequence and secondary structure assignment of the C-terminal domain.
Structure of the Active Site of Full-Length EcPOX.
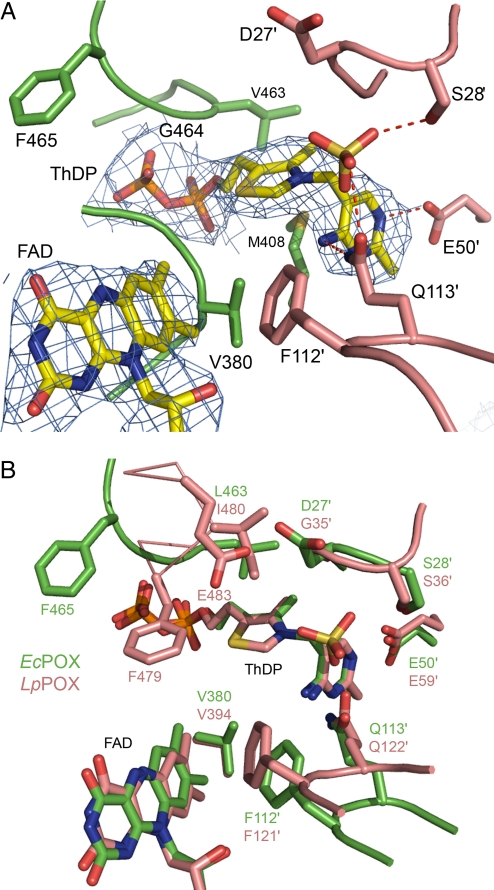
The structurally identical active centers of EcPOX are located at the dimer interface, but are covered by the C-terminal domain. Both cofactors are bound in close proximity (≈7 Å edge-to-edge distance of ThDP-C2 and FAD-C7M), with the dimethylbenzene moiety of the flavin reactive isoalloxazine ring pointing directly toward the thiazolium of ThDP (Fig. 4A). The thiamin cofactor adopts the canonical V conformation juxtaposing the reactive C2 carbon of the thiazolium and the 4′-amino group of the pyrimidine ring, which acts as an intramolecular acid/base catalyst with high effective molarity and delicately balanced protonic equilibria (31–33). The isoalloxazine part of FAD is slightly bent over the N5-N10 axis (15° distortion), a structural feature that is thought to be beneficial for ET as this conformation resembles the reduced state of the flavin (27). The active center is constituted from numerous loops originating from both subunits of the functional dimer. As previously observed for other enzymes of the pyruvate oxidase family (34, 35), an oxyanion (sulfate or phosphate, an unambigous assignment is impossible as both compounds are in the crystallization mixture) is bound as a placeholder at the presumed binding site of the substrate carboxylate moiety. Structural comparison with the constitutively active, acetylphosphate-producing pyruvate oxidase from Lactobacillus plantarum (LpPOX) (26), which is located exclusively in the cytosol and uses oxygen as the final electron acceptor rather than Q8, reveals a common overall architecture with respect to cofactor binding and active site residues (Fig. 4B). Whereas a Phe-Gln loop (F112-Q113 in EcPOX), which is important for fixing the substrate carbonyl (35), and a valine (V380 in EcPOX) presumably contacting the methyl group of pyruvate (35, 36) are structurally conserved, significant differences in the spatial organisation of other residues are observed. Most remarkably, a conserved Phe (F479 in LpPOX, F465 in EcPOX) occupies different positions in the 2 enzymes. In LpPOX, this residue is held in close proximity to both cofactors and was therefore suggested to function as a relay for ET (26). In contrast, the side chain of F465 in EcPOX points away from the active site with a displacement of ≈6 Å compared with F479 in LpPOX. As the estimated rate constants of ET between the thiamin and flavin cofactor differ so substantially (kobs ≈3 s−1 in EcPOX and ≈400 s−1 in LpPOX) (19, 37, 38), the orientation of this Phe residue might be central to controlling the rate of inter-cofactor ET.
Fig. 4.
Active site of EcPOX. (A) Structure of the active site showing ThDP, FAD, an oxyanion and selected amino acid residues in stick representation. The electron density of the 2 cofactors is contoured at 1.0 σ in a 2Fo − Fc map. Amino acids contributed from the neighboring subunit are shown in a different color code and are labeled with an apostrophe. (B) Superposition of the active centers of EcPOX (green) and the related LpPOX (pink) in stick representation. The 2 active centers are largely conserved, residue F465 (F479 in LpPOX), however, is pointing away from the active site in EcPOX.
Structural Differences of Full-Length and Proteolytically Activated EcPOX.
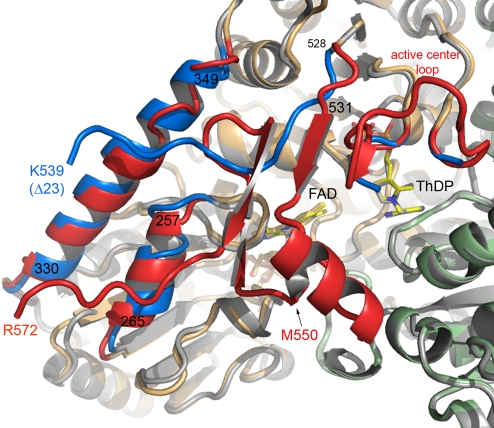
The structure of chymotrypsin-activated EcPOXΔ23, lacking the membrane-anchoring AP550–572 but still containing the C-terminal linker (residues 531–549) (see Fig. 3), was solved by molecular replacement using the structure of the full-length enzyme. EcPOXΔ23 crystallized in the orthorhombic spacegroup P212121 with 12 monomers (3 tetramers) in the asymmetric unit. The overall fold of the monomer is very similar to that of the full-length enzyme with an average Cα displacement of 0.89 Å, but the 2 corresponding dimers of EcPOXΔ23 are more tightly packed, resulting in a Cα-rmsd of 1.57 Å for the full-length and activated tetramers. The major structural differences locate to the C-terminal domain itself, 2 neighboring helices of the FAD domain (α257–265, α330–349) and an active center loop460–480, which interacts with the C-terminal linker sheet (Fig. 5). The C terminus of EcPOXΔ23 exhibits well-defined electron density until K539 (in some monomers until P535), suggesting that the remaining residues of the linker region (540–549) are flexible. The C-terminal linker, which in the full-length enzyme consists of a sheet-helix motif that covers the active site, exhibits no defined secondary structure in EcPOXΔ23 and points away from the active site contacting helix α330–349, which is now fully wound. As a consequence, access to the active site is no longer impaired and both ThDP and the isoalloxazine part of FAD are solvent exposed, providing a rationale for the ≈10-fold decreased KM value of pyruvate in activated EcPOX. Assuming that the same structural transition occurs upon membrane binding and concomitant activation of EcPOX, the quinone part of the final electron acceptor Q8 would have direct access to the active site to facilitate ET from the reduced flavin. The isoalloxazine part of FAD is even more distorted over the N5-N10 axis (19°) than observed in the full-length enzyme (15°).
Fig. 5.
Structural differences between full-length and EcPOXΔ23. Superposition of full-length EcPOX (monomers colored in orange and green) and EcPOXΔ23 (gray) viewed down the substrate channel in diagram representation. Regions with marked structural differences are highlighted: blue, EcPOXΔ23; red, full-length EcPOX. ThDP and FAD are shown in stick representation.
The active center loop460–480, fully traceable for the full-length enzyme, is more flexible in EcPOXΔ23 and exhibits no defined density for residues 467–478. As a consequence of the different conformation of the defined parts of the loop, the side chain of F465 now approaches the thiazolium of ThDP and the isoalloxazine moiety of the flavin, whereas the conformation of all other active site residues is virtually unaltered (Fig. 6A). In conjunction with the transient kinetic studies on EcPOX, which gave evidence for a dramatic rate enhancement of inter-cofactor ET upon activation from ≈3 s−1 in the full-length form up to ≈200 s−1 (proteolysis) or 400 s−1 (lipid binding) (19), this finding supports a crucial role of F465 for ET. Remarkably, in the constitutively activated LpPOX where the rate of ET amounts for ≈400 s−1 (as in activated EcPOX) F479 occupies a position akin to F465 in activated EcPOXΔ23. An equivalent to F465 is not present in other enzymes of the pyruvate oxidase family with a similar cofactor set (glyoxylate carboligase, GCL; acetohydroxyacid synthase, AHAS) (34, 39) that catalyze reactions that rely on carboligation rather than ET. In these enzymes, ET side reactions occur either very slowly (2 s−1 in AHAS) or not at all (GCL) (40). We thus hypothesize that the catalytic stimulation (Vmax) in response to activation of EcPOX results from the structural rearrangement of F465 facilitating transfer of 2 reducing equivalents from the thiamin to the flavin cofactor. In the spatial orientation observed for the activated enzyme, F465 would cause a strong repulsive interaction with the side chain of Y549 (edge-to-edge distance <1.7 Å) of the C-terminal linker region of the full-length enzyme (Fig. 6B). This invites speculation that F465 might have a key role for not only facilitating ET between the thiamin and flavin cofactors but also for expelling the C terminus from the active site, allowing membrane localization and unrestrained access for substrates.
Fig. 6.
Changes of the active site structure upon proteolytic activation of EcPOX. (A) Superposition of the active sites of full-length EcPOX (green) and EcPOXΔ23 (yellow). Amino acids contributed from the neighboring subunit are labeled with an apostrophe. Note the position of F465 in EcPOXΔ23, which adopts a conformation identical to that of F479 in LpPOX. (B) Illustration of the structural transition of the C-terminal linker region (residues 531–549) and an active center loop harboring F465 upon proteolytic activation. The full-length enzyme is shown in green and EcPOXΔ23 is shown in yellow. Residues Y549 and F465 are highlighted.
Conclusions
The current structural study reveals new molecular insights into membrane binding and catalytic activation of EcPOX. In the cytosol, the enzyme adopts a structure in which the membrane-anchoring C-terminal domain folds over the active site as a half-barrel/helix motif, impairing access for the substrate pyruvate. The active centers are even more deeply buried at the bottom of the funnel-shaped substrate channel than reported for the related pyruvate dehydrogenase multienzyme E1 component explaining the poor affinity of EcPOX for the substrate pyruvate (Fig. S6). Upon activation (flavin reduction and membrane binding in vivo, proteolysis in vitro), a conformational transition takes place that leads to the exposure of the C terminus, making the active site fully accessible for binding pyruvate and the final electron acceptor Q8 (see Movie S1 and SI Text). When freed from its interactions with the rest of the protein, the membrane anchor becomes structurally flexible, folding into an amphipathic helix upon binding to the membrane. This reversible order (half-barrel) ↔ disorder ↔ order (helix) transition driven by the redox state of the flavin cofactor appears to be a unique regulation mechanism for peripheral membrane association. In many peripheral membrane proteins studied, membrane association/dissociation involves a change of the net charge of basic membrane binding patches by phosphorylation/dephosphorylation (protein kinase C) or charge neutralization by calcium binding (C2 domains of phospholipase A2 and 5-lipoxygenase) (41). There are also instances where basic patches are occluded becoming uncovered upon binding/dissociation of effectors (Gßγ/Gα complex) (41). Although all basic residues (R552, R558, K567, R572) of the AP are exposed to the solvent, full-length EcPOX in the oxidized state does not bind to membranes (see Fig. 3), hence nonspecific electrostatic interactions may not be the sole driving force for membrane association, but rather both nonpolar and electrostatic contributions after formation of an amphipathic helix.
Intriguingly, the side chain of F465 swings into the active site upon activation and leads to a rate enhancement of ET between ThDP and FAD. There are different viable mechanisms that could account for this observation (42, 43). At first, because electron tunneling is by far more efficient through bonded orbitals than through space, a mechanism can be envisioned in which the electrons are transferred in a combined through-space/through-bond mechanism with F465 as a way-station that efficiently promotes coupling of the electron donor and acceptor wavefunctions. The side chain of F465 in EcPOXΔ23 would be positioned close to van der Waals distance (≈3 Å) both to the methyl group of the redox active hydroxyethyl-ThDP enamine intermediate and the C7 methyl group of FAD. Second, the spatial orientation of F465 could affect the reorganization energy λ, and/or the driving force ΔG by influencing the redox potentials of the cofactors. These different modes notwithstanding, it is surprising that ET in pyruvate oxidases proceeds with relatively slow rates, especially in view of the short edge-to-edge distance between the 2 cofactors (≈7 Å) for which higher rates are expected with no obvious necessity for a molecular wire (42). However, in a precedent, kinetic and thermodynamic studies on ET in the related LpPOX revealed that a large reorganization energy and small intrinsic driving force translate into slow ET rates with the reaction being in the low driving force regime (−ΔG ≪ λ) (38). It remains to be studied in which discrete way F465 facilitates the redox reaction between the thiamin and flavin cofactor in EcPOX and why in related enzymes such as AHAS or GCL ET between likewise proximal cofactors occurs that slowly or not at all.
Methods
X-Ray Crystallography.
Crystals of full-length and EcPOXΔ23 were grown as detailed in ref. 44. A redundant dataset of a single EcPOX crystal was collected in-house in a 100 K nitrogen cryostream (XSTREAM2000; Rigaku/MSC) after gradually transferring the crystal into a cryoprotectant containing mother liquor supplemented with 5%, 15%, and 30% (vol/vol) glycerol. The crystal diffracted up to 2.90 Å with Cu Kα radiation (λ = 1.5418 Å), using a rotating-anode source (RA Micro 007, RigakuMSC) and image plate detector (R-AXIS IV++, Rigaku/MSC). Oscillation photographs were integrated, merged and scaled using the XDS program package (45).
The structure of full-length EcPOX was determined by molecular replacement (MR), using data from 30 to 2.9 Å, and using a monomer of LpPOX (PDB code 1pow) as search model with PHASER (46). The asymmetric unit comprises 2 monomers, forming half of the biological tetramer. The structure was manually rebuilt and verified against simulated annealing (SA) omit maps and sigmaa weighted difference Fourier maps using programs O and Coot (47, 48). The refinement was carried out with CNS (49) against a maximum likelihood target and was based on slow cooling SA (both Torsion Angle Dynamics and Cartesian Dynamics) combined with standard minimization and individually restrained B-factor refinement. Both overall anisotropic B-factor and bulk solvent corrections were applied. Initially, strong NCS restraints between equivalent residues in the 2 monomers were applied but these were later relaxed for regions showing different conformations in each monomer. The model was refined to R and Rfree of 0.2129 and 0.2412. The analysis of atomic displacements parameters of the refined structure revealed that the B-factors generally increase with distance from the center of the molecule, implying a rigid body libration of the 3 main domains of the monomer (Pyr domain, FAD domain, PP domain). To model anisotropic displacements of these domains, the final refinement was carried out with PHENIX (50) implementing 6 TLS groups (3 groups per monomer). Both CNS and PHENIX used the same Rfree set. The final model consists of residues 2 to 572, 2 sulfate or phosphate ions, 2 FAD, 2 ThDP and 2 Mg2+, and has been refined to R and Rfree factors of 0.1832 and 0.2160 against data up to 2.9 Å resolution. No water molecules have been included to the model. The stereochemistry of the structure was assessed with PROCHECK (51).
A dataset of EcPOXΔ23 was collected at BESSY beam line BL 14.1 equipped with a fast scanning 225 mm CCD-mosaic detector from MARRESEACH. The data were processed using MOSFLM (52) and subsequently reduced and scaled with SCALA (46). The structure of EcPOXΔ23 was determined by MR with data from 30 to 2.5 Å, using a monomer of full-length EcPOX as a starting model. The asymmetric unit comprises 12 monomers (3 tetramers). The structure was refined and manually rebuilt in a similar way as described for full-length EcPOX. The structure was refined using CNS to R and Rfree factors of 0.2248 and 0.2577, respectively. The final model consists of residues 1 to 539 (in some monomers 1–535) with 467–479 missing, 1329 water molecules, 36 phosphate ions, 12 FAD, 12 ThDP and 12 Mg2+, and was refined at 2.50 Å resolution to an R-factor of 0.1834 and Rfree of 0.1977. The stereochemistry of the structure was assessed with PROCHECK (51).
Supplementary Material
Acknowledgments.
We thank John Cronan, Jr. for providing plasmid pYYC102. We thank the Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY) for access to their synchrotron radiation beamtime. This work was supported by the Deutsche Forschungsgemeinschaft-Graduiertenkolleg 1026 “Conformational Transitions in Macromolecular Interactions” and the European Community–Research Infrastructure Action under the Sixth EU Framework Programme “Structuring the European Research Area” Program through the Integrated Infrastructure Initiative“ Integrating Activity on Synchrotron and Free Electron Laser Science Contract R II 3-CT-2004-506008.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.J.M. is a guest editor invited by the Editorial Board.
Data deposition: The model and structure factors have been deposited in the Research Collaboratory for Structural Biology database, www.rcsb.org [accession nos. 3EY9 (full-length EcPOX) and 3EYA (EcPOXΔ23)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0805027105/DCSupplemental.
References
- 1.Johnson JE, Cornell RB. Amphitropic proteins: Regulation by reversible membrane interactions. Mol Membr Biol. 1999;16:217–235. doi: 10.1080/096876899294544. [DOI] [PubMed] [Google Scholar]
- 2.Cornell RB, Taneva SG. Amphipathic helices as mediators of the membrane interaction of amphitropic proteins, and as modulators of bilayer physical properties. Curr Protein Pept Sc. 2006;7:539–552. doi: 10.2174/138920306779025675. [DOI] [PubMed] [Google Scholar]
- 3.Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 4.Williams FR, Hager LP. A crystalline flavin pyruvate oxidase. J Biol Chem. 1961;236:PC36–37. [PubMed] [Google Scholar]
- 5.Williams FR, Hager LP. Crystalline flavin pyruvate oxidase from Escherichia coli. I. Isolation and properties of the flavoprotein. Arch Biochem Biophys. 1966;116:168–176. doi: 10.1016/0003-9861(66)90025-7. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham CC, Hager LP. Crystalline pyruvate oxidase from Escherichia coli. II. Activation by phospholipids. J Biol Chem. 1971;246:1575–1582. [PubMed] [Google Scholar]
- 7.Cunningham CC, Hager LP. Reactivation of the lipid-depleted pyruvate oxidase system from Escherichia coli with cell envelope neutral lipids. J Biol Chem. 1975;250:7139–7146. [PubMed] [Google Scholar]
- 8.Grabau C, Cronan JE., Jr Molecular cloning of the gene (poxB) encoding the pyruvate oxidase of Escherichia coli, a lipid-activated enzyme. J Bacteriol. 1984;160:1088–1092. doi: 10.1128/jb.160.3.1088-1092.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koland JG, Miller MJ, Gennis RB. Reconstitution of the membrane-bound, ubiquinone-dependent pyruvate oxidase respiratory chain of Escherichia coli with the cytochrome d terminal oxidase. Biochemistry. 1984;23:445–453. doi: 10.1021/bi00298a008. [DOI] [PubMed] [Google Scholar]
- 10.Carter K, Gennis RB. Reconstitution of the Ubiquinone-dependent pyruvate oxidase system of Escherichia coli with the cytochrome o terminal oxidase complex. J Biol Chem. 1985;260:10986–10990. [PubMed] [Google Scholar]
- 11.Marchal D, Pantigny J, Laval JM, Moiroux J, Bourdillon C. Rate constants in two dimensions of electron transfer between pyruvate oxidase, a membrane enzyme, and ubiquinone (coenzyme Q8), its water-insoluble electron carrier. Biochemistry. 2001;40:1248–1256. doi: 10.1021/bi002325y. [DOI] [PubMed] [Google Scholar]
- 12.Abdel-Hamid AM, Attwood MM, Guest JR. Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiol. 2001;147:1483–1498. doi: 10.1099/00221287-147-6-1483. [DOI] [PubMed] [Google Scholar]
- 13.Recny MA, Hager LP. Isolation and characterization of the protease-activated form of pyruvate oxidase. Evidence for a conformational change in the environment of the flavin prosthetic group. J Biol Chem. 1983;258:5189–5195. [PubMed] [Google Scholar]
- 14.Mather MW, Gennis RB. Spectroscopic studies of pyruvate oxidase flavoprotein from Escherichia coli trapped in the lipid-activated form by cross-linking. J Biol Chem. 1985;260:10395–10397. [PubMed] [Google Scholar]
- 15.Russell P, Hager LP, Gennis RB. Characterization of the proteolytic activation of pyruvate oxidase. Control by specific ligands and by the flavin oxidation-reduction state. J Biol Chem. 1977;252:7877–7882. [PubMed] [Google Scholar]
- 16.Russell P, Schrock HL, Gennis RB. Lipid activation and protease activation of pyruvate oxidase. Evidence suggesting a common site of interaction on the protein. J Biol Chem. 1977;252:7883–7887. [PubMed] [Google Scholar]
- 17.Hager LP. Studies on the mechanism of activation of pyruvate oxidase by lipids and by limited proteolysis. In: Bisswanger H, Ullrich J, editors. Biochemistry and Physiology of Thiamin Diphosphate Enzymes. Switzerland: Wiley-VCH, Basel; 1991. pp. 277–285. [Google Scholar]
- 18.Mather MW, Gennis RB. Kinetic studies of the lipid-activated pyruvate oxidase flavoprotein of Escherichia coli. J Biol Chem. 1985;260:16148–16155. [PubMed] [Google Scholar]
- 19.Bertagnolli BL, Hager LP. Activation of Escherichia coli pyruvate oxidase enhances the oxidation of hydroxyethylthiamin pyrophosphate. J Biol Chem. 1991;266:10168–10173. [PubMed] [Google Scholar]
- 20.Bertagnolli BL, Hager LP. Minimum requirements for protease activation of flavin pyruvate oxidase. Biochemistry. 1991;30:8131–8137. doi: 10.1021/bi00247a006. [DOI] [PubMed] [Google Scholar]
- 21.Recny MA, Grabau C, Cronan JE, Jr, Hager LP. Characterization of the alpha-peptide released upon protease activation of pyruvate oxidase. J Biol Chem. 1985;260:14287–14291. [PubMed] [Google Scholar]
- 22.Zhang TF, Hager LP. Binding of pyruvate oxidase alpha-peptide to phospholipid vesicles. Arch Biochem Biophys. 1987;255:201–204. doi: 10.1016/0003-9861(87)90311-0. [DOI] [PubMed] [Google Scholar]
- 23.Grabau C, Cronan JE., Jr In vivo function of Escherichia coli pyruvate oxidase specifically requires a functional lipid binding site. Biochemistry. 1986;25:3748–3751. doi: 10.1021/bi00361a003. [DOI] [PubMed] [Google Scholar]
- 24.Frank RA, Titman CM, Pratap JV, Luisi BF, Perham RN. A molecular switch and proton wire synchronize the active sites in thiamine enzymes. Science. 2004;306:872–876. doi: 10.1126/science.1101030. [DOI] [PubMed] [Google Scholar]
- 25.Seifert F, et al. Direct kinetic evidence for half-of-the-sites reactivity in the E1 component of the human pyruvate dehydrogenase multienzyme complex through alternating sites cofactor activation. Biochemistry. 2006;45:12775–12785. doi: 10.1021/bi061582l. [DOI] [PubMed] [Google Scholar]
- 26.Muller YA, Schulz GE. Structure of the thiamine- and flavin-dependent enzyme pyruvate oxidase. Science. 1993;259:965–967. doi: 10.1126/science.8438155. [DOI] [PubMed] [Google Scholar]
- 27.Muller YA, Schumacher G, Rudolph R, Schulz GE. The refined structures of a stabilized mutant and of wild-type pyruvate oxidase from Lactobacillus plantarum. J Mol Biol. 1994;237:315–335. doi: 10.1006/jmbi.1994.1233. [DOI] [PubMed] [Google Scholar]
- 28.Muller YA, et al. A thiamin diphosphate binding fold revealed by comparison of the crystal structures of transketolase, pyruvate oxidase and pyruvate decarboxylase. Structure. 1993;1:95–103. doi: 10.1016/0969-2126(93)90025-c. [DOI] [PubMed] [Google Scholar]
- 29.Grabau C, Chang YY, Cronan JE., Jr Lipid binding by Escherichia coli pyruvate oxidase is disrupted by small alterations of the carboxyl-terminal region. J Biol Chem. 1989;264:12510–12519. [PubMed] [Google Scholar]
- 30.Chang YY, Cronan JE., Jr Molecular cloning, DNA sequencing, and enzymatic analyses of two Escherichia coli pyruvate oxidase mutants defective in activation by lipids. J Bacteriol. 1986;167:312–318. doi: 10.1128/jb.167.1.312-318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tittmann K, et al. NMR analysis of covalent intermediates in thiamin diphosphate enzymes. Biochemistry. 2003;42:7885–7891. doi: 10.1021/bi034465o. [DOI] [PubMed] [Google Scholar]
- 32.Nemeria N, et al. The 1′,4′-iminopyrimidine tautomer of thiamin diphosphate is poised for catalysis in asymmetric active centers on enzymes. Proc Natl Acad Sci USA. 2007;104:78–82. doi: 10.1073/pnas.0609973104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemeria N, et al. Elucidation of the chemistry of enzyme-bound thiamin diphosphate prior to substrate binding: Defining internal equilibria among tautomeric and ionization states. Biochemistry. 2007;46:10739–10744. doi: 10.1021/bi700838q. [DOI] [PubMed] [Google Scholar]
- 34.Pang SS, Duggleby RG, Guddat LW. Crystal structure of yeast acetohydroxyacid synthase: A target for herbicidal inhibitors. J Mol Biol. 2002;317:249–262. doi: 10.1006/jmbi.2001.5419. [DOI] [PubMed] [Google Scholar]
- 35.Wille G, et al. The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography. Nat Chem Biol. 2006;2:324–328. doi: 10.1038/nchembio788. [DOI] [PubMed] [Google Scholar]
- 36.Chang YY, Cronan JE., Jr Conversion of Escherichia coli pyruvate oxidase to an ‘alpha-ketobutyrate oxidase’. Biochem J. 2000;352(Pt 3):717–724. [PMC free article] [PubMed] [Google Scholar]
- 37.Tittmann K, Golbik R, Ghisla S, Hubner G. Mechanism of elementary catalytic steps of pyruvate oxidase from Lactobacillus plantarum. Biochemistry. 2000;39:10747–10754. doi: 10.1021/bi0004089. [DOI] [PubMed] [Google Scholar]
- 38.Tittmann K, et al. Radical phosphate transfer mechanism for the thiamin diphosphate- and FAD-dependent pyruvate oxidase from Lactobacillus plantarum. Kinetic coupling of intercofactor electron transfer with phosphate transfer to acetyl-thiamin diphosphate via a transient FAD semiquinone/hydroxyethyl-ThDP radical pair. Biochemistry. 2005;44:13291–13303. doi: 10.1021/bi051058z. [DOI] [PubMed] [Google Scholar]
- 39.Kaplun A, et al. Glyoxylate carboligase lacks the canonical active site glutamate of thiamine-dependent enzymes. Nat Chem Biol. 2008;4:113–118. doi: 10.1038/nchembio.62. [DOI] [PubMed] [Google Scholar]
- 40.Tittmann K, et al. Electron transfer in acetohydroxy acid synthase as a side reaction of catalysis. Implications for the reactivity and partitioning of the carbanion/enamine form of (alpha-hydroxyethyl)thiamin diphosphate in a “nonredox” flavoenzyme. Biochemistry. 2004;43:8652–8661. doi: 10.1021/bi049897t. [DOI] [PubMed] [Google Scholar]
- 41.Mulgrew-Nesbitt A, Diraviyam K, Wang J, Singh S, Murray P, Li Z, Rogers L, Mirkovic N, Murray D. The role of electrostatics in protein-membrane interactions. Biochim Biophys Acta. 2006;1761:812–826. doi: 10.1016/j.bbalip.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 42.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 43.Onuchic JN, Beratan DN, Winkler JR, Gray HB. Pathway analysis of protein electron-transfer reactions. Annu Rev Biophy Biomol Struct. 1992;21:349–377. doi: 10.1146/annurev.bb.21.060192.002025. [DOI] [PubMed] [Google Scholar]
- 44.Weidner A, Neumann P, Wille G, Stubbs MT, Tittmann K. Crystallization and preliminary X-ray diffraction analysis of full-length and proteolytically activated pyruvate oxidase from Escherichia coli. Acta Crystallogr F. 2008;64:179–181. doi: 10.1107/S1744309108003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 46.Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. COOT: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 51.Laskowski RA, MacArthur M, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 52.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newslett Protein Crystallogr. 1992 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.