Abstract
The mammalian translational initiation machinery is a tightly controlled system that is composed of eukaryotic initiation factors, and which controls the recruitment of ribosomes to mediate cap-dependent translation. Accordingly, the mTORC1 complex functionally controls this cap-dependent translation machinery through the phosphorylation of its downstream substrates 4E-BPs and S6Ks. It is generally accepted that rapamycin, a specific inhibitor of mTORC1, is a potent translational repressor. Here we report the unexpected discovery that rapamycin's ability to regulate cap-dependent translation varies significantly among cell types. We show that this effect is mechanistically caused by rapamycin's differential effect on 4E-BP1 versus S6Ks. While rapamycin potently inhibits S6K activity throughout the duration of treatment, 4E-BP1 recovers in phosphorylation within 6 h despite initial inhibition (1–3 h). This reemerged 4E-BP1 phosphorylation is rapamycin-resistant but still requires mTOR, Raptor, and mTORC1's activity. Therefore, these results explain how cap-dependent translation can be maintained in the presence of rapamycin. In addition, we have also defined the condition by which rapamycin can control cap-dependent translation in various cell types. Finally, we show that mTOR catalytic inhibitors are effective inhibitors of the rapamycin-resistant phenotype.
Keywords: cap-dependent translation, mTORC1, rapamycin resistance
The mammalian translational initiation machinery governs the recruitment of ribosomes to mRNA to commence the production of protein synthesis. This machinery consists of various eukaryotic initiation factors (eIFs) that tightly regulate protein synthesis based on environmental cues. Importantly, initiation is an important step for cellular control because it is the rate-limiting step of translation (1).
Two predominant pathways translate mammalian mRNA through cap-dependent and independent mechanisms. The capping of the 5′ end of mRNA by m7GTP allows the recruitment of the eIF4F complex, eIF3, and the 40S ribosomal subunit to the 5′ mRNA cap. Cap-independent translation is mediated by an internal RNA structure called internal ribosome entry site (IRES), which recruits the ribosome independent of both the cap and the entire eIF4F complex (2).
The initiation of cap-dependent translation is tightly regulated by extracellular conditions including glucose, nutrient, and growth factor levels. These factors control cap-dependent translation by regulating the evolutionarily conserved mTORC1 (mTOR, Raptor, mLST8) pathway (3). Activation of mTORC1 positively stimulates mRNA translation via its downstream substrates S6Ks and 4E-BP1/eIF4E (4–7). Phosphorylation of 4E-BP1 by mTORC1 results in its dissociation from eIF4E, promoting assembly of the eIF4F complex. It is thought that S6K1 can phosphorylate translational regulators such as eIF4B and PDCD4 to enhance the translational efficiency of mRNAs with highly structured 5′ UTRs (8–10).
Therefore, growth factors positively regulate cap-dependent translation via mTORC1-dependent or rapamycin-sensitive phosphorylation of 4E-BP1 and through the regulation of S6Ks.
Based on the described effects of rapamycin and mTORC1 on 4E-BP1 phosphorylation and S6K activity, it is generally accepted that rapamycin is a global inhibitor of cap-dependent translation in most cell types (11). To understand the biological effects of long-term rapamycin treatment on translational control, we used a cap-dependent translational reporter vector to discover unexpectedly that rapamycin exhibits differential regulation of its known downstream substrates S6Ks and 4E-BP1 in a cell-specific manner. In all tested cell types, rapamycin potently inhibited S6K activity throughout the duration of treatment (24+ h). However, despite initial (1–3 h) inhibition of 4E-BP1 phosphorylation on growth factor-sensitive sites by rapamycin, 4E-BP1 phosphorylation recovered on all sites despite continued S6K inhibition. These results suggest a differential regulation of the 2 known mTORC1 substrates; to our knowledge, this has not been previously described. This process required rapamycin-induced de novo protein synthesis in a cell-autonomous manner. Mechanistically, the rapamycin-induced effect on 4E-BP1 required rapamycin-resistant mTORC1 activity, suggesting a substrate-specific gain of rapamycin resistance. Importantly, cap-dependent translation reinitiated despite the presence of rapamycin and S6K inhibition through a 4E-BP1 phosphorylation-dependent manner. Finally, we show that catalytic inhibitors of mTOR prevent rapamycin-resistant rephosphorylation of 4E-BP1 supporting their clinical promise.
Results
Rapamycin Does Not Functionally Mimic 4E-BP1 Hypophosphorylation.
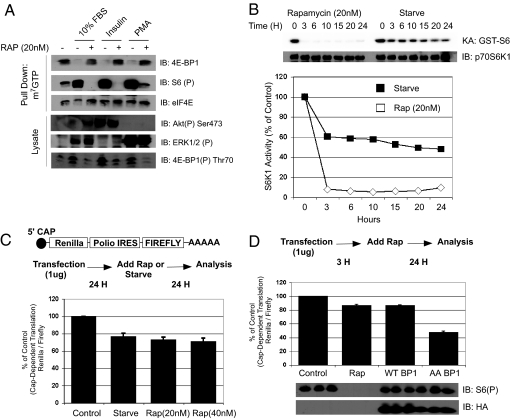
To investigate the dependence of growth factors to regulate the phosphorylation of 4E-BP1 and eIF4E activity, we initially starved HEK293 cells and pretreated the cells with rapamycin before stimulation with 10% FBS, insulin, and PMA. As shown in Fig. 1A, rapamycin completely inhibited the phosphorylation of 4E-BP1 and consequently prevented its dissociation from eIF4E. The binding of 4E-BP1 to eIF4E was measured by m7GTP Sepharose beads, which mimic the 5′ mRNA cap to precipitate cap-interacting proteins. The rapamycin-induced binding of 4E-BP1 with the m7GTP beads is consistent with previous evidence that suggests that multiple growth factors converge onto the TSC complex upstream of mTORC1 (3). Therefore, irrespective of the growth factor, mTORC1 activity is required for 4E-BP1 phosphorylation. We measured the activity of mTOR as a function of S6K1 activity and observed that rapamycin's effect on mTORC1 activity was immediate (Fig. 1B) and complete attenuation was achieved within 5 min (12). This is in contrast to serum starvation, which is a more gradual process. Therefore, we speculate that rapamycin would be much more potent than serum starvation in attenuating cap-dependent translation in vivo.
Fig. 1.
Rapamycin does not functionally mimic hypophosphorylation of 4E-BP1. (A) HEK293 cells were serum-starved for 24 h, pretreated with rapamycin (20 nM) for 30 min, and stimulated with 10% FBS, insulin (100 nM), or PMA (100 ng/mL). 4E-BP1 cap binding activity was measured with m7GTP Sepharose association. (B) S6K1 activity was measured with GST-S6 as a substrate. (C) In vivo cap-dependent translational assays were conducted with a dual Renilla/firefly luciferase assay with the Polio virus IRES driving firefly expression. One microgram of DNA was transfected into HEK293 cells in 6-well plates and 24 h later were either starved or treated with rapamycin for 24 more hours. (D) Dominant negative 4E-BP1 (Thr-37/46 AA), WT 4E-BP1, or control plasmids were cotransfected with the translational vector at a 1:2 ratio, and rapamycin or ethanol was treated in other samples. Luciferase activity was measured and is shown as relative cap-dependent translation. Phospho-S6 and 4E-BP1 expression is also shown.
To measure the effect of rapamycin on cap-dependent translation in cells, we used a dual luciferase reporter vector that distinguishes cap versus cap-independent translation by separating Renilla luciferase from firefly with the Polio IRES (13). Therefore, the Renilla/firefly ratio would determine the cap-dependent translation ratio in cells. Serum starvation attenuated cap-dependent translation by ≈20% when compared with the control (Fig. 1C). Surprisingly, rapamycin only inhibited cap-dependent translation by ≈20% at 2 different concentrations, and this effect was not due to alteration in Polio IRES-driven translation [supporting information (SI) Fig. S1]. This same effect was also observed with HCV IRES-driven translational vectors (data not shown). Furthermore, transfection of a dominant-negative 4E-BP1 with alanine mutations at Thr-37/46 was much more potent than rapamycin treatment in inhibiting cap-dependent translation (≈20% versus ≈65%) (Fig. 1D). Therefore, rapamycin appears to exhibit minimal effects on cap-dependent translation when compared with 4E-BP1 hypophosphorylation in cells.
Rapamycin Differentially Affects S6Ks Versus 4E-BP1.
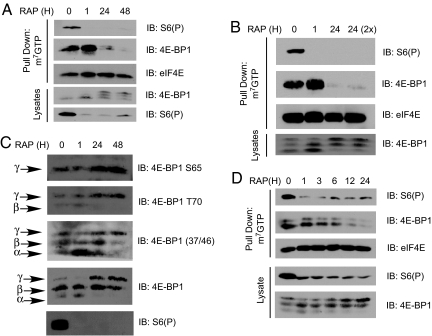
The minimal effect of rapamycin on cap-dependent translation in cells was perplexing because rapamycin effectively attenuated 4E-BP1 phosphorylation in cells (Fig. 1A) and the addition of recombinant 4E-BP1 almost completely inhibited in vitro translation of capped mRNAs [Fig. 1E and Fig. S2]. However, overexpression of a dominant negative 4E-BP1 (37/46AA) was much more potent than rapamycin in inhibiting cap-dependent translation (Fig. 1D). In contrast, rapamycin was similar to WT 4E-BP1, which is effectively phosphorylated in vivo (Fig. 1D). Therefore, we treated rapamycin in asynchronously growing cells for 24 h and analyzed the status of known mTORC1 substrates to recapitulate the conditions of the translational assays. While a 1-h treatment with rapamycin induced 4E-BP1 dephosphorylation and association with eIF4E, prolonged treatment for 24 and 48 h led to hyperphosphorylation of 4E-BP1 and dissociation from eIF4E (Fig. 2A). This reemerging phosphorylation occurred despite continued mTORC1 inhibition as measured by S6 phosphorylation. Additionally, the activities of both S6K1 and S6K2 were continuously inhibited 24 h after rapamycin treatment, as well as phosphorylation of T389 on S6K1 (Fig. S3 A and B). This effect was not due to the differential kinetics in recovery from rapamycin treatment between S6Ks and 4E-BP1, because neither increasing rapamycin concentration nor readdition of rapamycin to 24-h treatment groups failed to induce the dephosphorylation of 4E-BP1 (Fig. 2B and Fig. S4). The phosphorylation-induced gel shifts of 4E-BP1 were also concomitant with increases in known phosphorylation sites on 4E-BP1 including Thr-37/46, Thr-70, and Ser-65 (Fig. 2C). These results were also observed in HeLa, TSC2−/− p53−/− MEFs, and DU145 but were less apparent in p53−/− MEFs (Fig. S5). However, this effect was not observed in all cells including PC3, MCF7, and U2OS (see Fig. 4E). Therefore, depending on the cell type, rapamycin differentially inhibits the phosphorylation of its downstream targets 4E-BP1 and the S6Ks.
Fig. 2.
Rapamycin exhibits differential effects toward S6Ks versus 4E-BP1. (A) HEK293 cells were treated with rapamycin for 1, 24, and 48 h or ethanol for 48 h and were analyzed for the binding of 4E-BP1 to the cap complex. (B) Rapamycin or ethanol was readded to the 24-h-treated sample 1 h before lysis, and it was analyzed for 4E-BP1 phosphorylation. (C) Lysates from rapamycin-treated samples were blotted for the phosphorylation status of 4E-BP1. The sites analyzed were Ser-65, Thr-70, Thr-37/46, and total 4E-BP1. Gel shifts can be observed in samples treated with rapamycin for 24 or 48 h. The α-β-γ isoforms represent the phosphorylation status of 4E-BP1 with α being hypophosphorylated and γ being hyperphosphorylated. (D) The kinetics of rapamycin-induced 4E-BP1 was measured in HEK293 and treated according to the time listed. 4E-BP1 binding to the m7GTP Sepharose and gel shifts on lysates are shown.
Fig. 4.
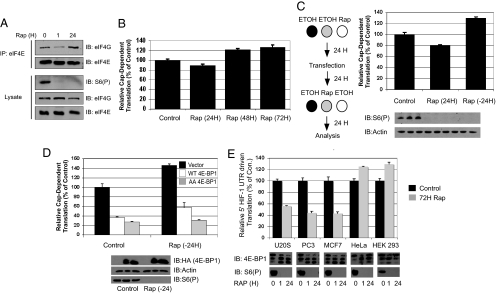
Rapamycin-induced 4E-BP1 phosphorylation stimulates cap-dependent translation and regulates cell-specific inhibition of translation. (A) The interaction between eIF4E and eIF4G was measured in coimmunoprecipitation experiments in HEK293 cells treated with rapamycin for 1 or 24 h. (B) In vivo cap-dependent translational assays were performed except rapamycin was added 3 h after transfection for 24, 48, or 72 h. (C) Phospho-S6 and actin were blotted for normalization. (D) The experiments were completed in the same way as in C, except WT 4E-BP1, AA 4E-BP1, or a control vector was cotransfected. Phospho-S6, actin, and HA (4E-BP1) blots are shown. Black, control vector; white, WT 4E-BP1; gray, AA 4E-BP1. (E) Different cell lines that differentially exhibit rapamycin-induced 4E-BP1 hyperphosphorylation were transfected with the HIF-1α 5′ UTR-driven translational vector and were treated with rapamycin or ethanol 3 h after transfection for 72 h. Black, control; gray, rapamycin (20 nM). The 4E-BP1 and S6(P) Western blots are shown in conjunction with the translational data.
Rapamycin Requires de Novo Protein Synthesis to Stimulate 4E-BP1 Phosphorylation in a Cell-Autonomous Manner.
Next, we measured the kinetics of rapamycin-induced 4E-BP1 hyperphosphorylation. Despite an obvious attenuation of 4E-BP1 phosphorylation at 1–3 h after rapamycin treatment, by 6 h 4E-BP1 phosphorylation reemerged and was almost completely rephosphorylated by 12 h (Fig. 2D). More importantly, cotreatment of HEK293 cells with rapamycin and either cycloheximide or actinomycin D prevented the hyperphosphorylation of 4E-BP1 (Fig. S6). Therefore, de novo protein synthesis in the presence of rapamycin is required for the 4E-BP1 hyperphosphorylation to occur. This result also argues against 4E-BP1 turnover as the basis for free eIF4E, because cycloheximide increased 4E-BP1 interaction with the 5′ cap when treated in the presence of rapamycin. This effect was also cell-autonomous as transfer of media from long-term rapamycin-treated cells did not confer rapamycin-resistant 4E-BP1 phosphorylation in nontreated cells.
Cyclin-Dependent Kinases (CDKs) Are Not Involved in the Phosphorylation of 4E-BP1.
It was previously reported that various kinases, including CDKs, could phosphorylate 4E-BP1 in a rapamycin-independent fashion (14). To determine whether CDKs could be responsible for the rapamycin-stimulated rephosphorylation of 4EBP1, we treated cells with saturating concentrations of roscovitine (50 μM), a CDK inhibitor. Rapamycin-induced 4E-BP1 hyperphosphorylation was not affected by roscovitine (data not shown, A.Y.C. and J.B.). This effect is also distinct from previous reports suggesting that mitotic arrest or microtubule stabilizing agents stimulated 4E-BP1 phosphorylation (14, 15). As shown in Fig. S7A, the microtubule stabilizer nocodazole robustly stimulated 4E-BP1 phosphorylation, and this effect could be reversed with purvalanol A (PurA), a nonspecific CDK inhibitor, but not by rapamycin. However, PurA failed to inhibit the hyperphosphorylated status of rapamycin-induced 4E-BP1, suggesting that the mitotic-arrest-mediated regulation of 4E-BP1 is distinct from the gain of rapamycin resistance that we observed (Fig. S7B). When taken together, our data suggest that the rapamycin-induced 4E-BP1 phosphorylation is not CDK-mediated.
Rapamycin-Induced 4E-BP1 Phosphorylation Is Independent of the PI3K and MEK-ERK Pathways.
In addition to rapamycin, inhibition of the PI3 kinase (PI3K) and MEK-ERK pathways also inhibits the activation of mTORC1 (3). Therefore, we examined the ability of wortmannin, a PI3K-selective inhibitor at 50 nM, and UO126, a MEK1/2/5 inhibitor, to antagonize the rapamycin-mediated 4E-BP1 phosphorylation. A 1-h treatment with rapamycin inhibited both 4E-BP1 and S6 phosphorylation (Fig. S8). However, neither wortmannin nor UO126 significantly affected the stimulation of 4E-BP1 phosphorylation induced by rapamycin (Fig. S8), suggesting that the PI3K and MEK-ERK pathways are not involved in rapamycin-stimulated 4E-BP1 phosphorylation. Accordingly, such mTORC1-induced hyperphosphorylation renders the cell insensitive to subsequent rapamycin retreatment and PI3K or MEK-ERK inhibition (Fig. S8).
Rapamycin-Resistant mTORC1 Activity Is Necessary for Rapamycin-Induced 4E-BP1 Phosphorylation.
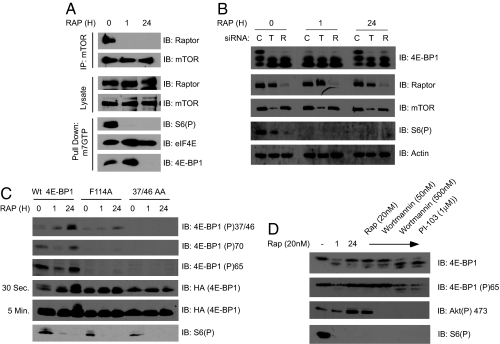
We next analyzed the molecular requirements for the mTORC1 complex to mediate this effect. Accordingly, the mTORC1 complex after either 1 or 24 h of rapamycin treatment existed in a “less active” conformation as determined by mTOR and Raptor coimmunoprecipitation experiments (Fig. 3A). This conformation is measured by evaluating the interaction between Raptor and mTOR after purification with detergent-containing buffers. Although mTOR and Raptor still interact in vivo while in this “less active” conformation, they dissociate during purification because the altered conformation cannot withstand biochemical purification (16). Nonetheless, this reiterates previous results that suggest that rapamycin continues to inhibit mTORC1 at 1 or 24 h after treatment by altering the conformation of this complex (Fig. 2) (16) and is consistent with the sustained inhibition of S6Ks (Fig. 1B and Fig. S3). Surprisingly, mTORC1 components still appear to be required because siRNA knockdown of either mTOR or Raptor abrogated rapamycin-induced 4E-BP1 hyperphosphorylation (Fig. 3B). Furthermore, mutations to either the 4E-BP1 priming phosphorylation sites (Thr-37/46), which are required for both Thr-70 and Ser-65 phosphorylations, or the TOS motif (F114A), which mediates 4E-BP1/Raptor interaction, abrogated the rapamycin-induced 4E-BP1 hyperphosphorylation (Fig. 3C) (17). Last, increasing the concentrations of wortmannin to 500 nM, which catalytically inhibits mTOR (18), or treatment with the dual PI3K and mTOR catalytic inhibitor PI-103 (19) also repressed 4E-BP1 phosphorylation induced by rapamycin treatment (Fig. 3D). The wortmannin/PI-103 sensitivity was observed in both 293 (Fig. S9) and TSC2−/− MEF cells (Fig. 3D), which activates mTORC1 independently of PI3K (18). The requirement of mTOR's catalytic activity is associated with mTORC1 and not mTORC2, because loss of Rictor did not affect the rapamycin-induced 4E-BP1 phosphorylation (Fig. S10). When taken together, the components that positively stimulate the rephosphorylation of 4E-BP1 by rapamycin require the TOS motif, employ a priming phosphorylation pattern similar to that of mTORC1, and structurally and catalytically require mTORC1 components. Consistent with the requirement for mTORC1, the rapamycin-induced effect was insensitive to staurosporine, a potent nonspecific kinase inhibitor that fails to affect mTOR's catalytic activity, even at a saturating concentration of 500 nM (Fig. S11).
Fig. 3.
Rapamycin-induced 4E-BP1 phosphorylation requires rap-resistant mTORC1 but is insensitive to inhibitors of the PI3K and MEK-ERK pathways. (A) The conformation of the mTORC1 complex was determined with mTOR coimmunoprecipitation experiments. The association of Raptor to mTOR is shown. (B) siRNAs against mTOR and Raptor were transfected into HEK293 cells and incubated for 24 h. Thereafter, the cells were treated with rapamycin for 24 more hours or 1 h before lysis and were analyzed. C, Scrambled control siRNA; R, siRNA against Raptor; T, siRNA against mTOR. (C) WT, TOS motif mutant (F114A), and 37/46 AA 4E-BP1s were transfected (3 μg) into a 10-cm plate of HEK293 cells, treated with rapamycin for 24 h, and analyzed for 4E-BP1 phosphorylation. (D) TSC2−/− MEFs were treated with control (DMSO) or rapamycin (20 nM) for either 1 or 24 h. In the 24 h rapamycin-treated samples, either control, rapamycin (20 nM), Wortmannin (50 and 500 nM), or PI-103 (1 μM) was added for 1 h before lysis. The samples were analyzed for 4E-BP1 phosphorylation.
Rapamycin-Mediated 4E-BP1 Phosphorylation Stimulates Cap-Dependent Translation.
To test the functional consequences of the rapamycin-induced 4E-BP1 phosphorylation, we investigated the formation of the eIF4F translational initiation complex. Under growth factor deprivation or rapamycin treatment, 4E-BP1 normally binds to eIF4E and prevents the association of eIF4G with eIF4E. As shown in Fig. 4A, a 1-h rapamycin treatment in asynchronously growing cells decreased the eIF4E-4G interaction. However, consistent with the dissociation of 4E-BP1 with eIF4E, 24 h of rapamycin treatment restimulated the association of eIF4G from eIF4E. This effect also coincided with an increase in eIF4E phosphorylation at Ser-209, which occurs as a result of eIF4G interaction with eIF4E to scaffold the kinases Mnk1/2 (Fig. S12A) (20). In addition to the reformation of the eIF4F initiation complex, we also observed an increase in cap-dependent translation with 48–72 h of rapamycin treatment (Fig. 4B) when compared with treatment for just 24 h (Fig. 1). The rapamycin-induced increase in cap-dependent translation was also not a function of decreased IRES-driven translation, but rather an increase in cap-dependent translation (Fig. S12B). Likewise, when we pretreated HEK293 cells with rapamycin for 24 h before transfection of reporter constructs to “prehyperphosphorylate” 4E-BP1, we obtained an ≈30% increase in total cap-dependent translation (Fig. 4C). Consistent with the idea that the increase in cap-dependent translation with rapamycin is a result of 4E-BP1 rephosphorylation, expression of a dominant negative 4E-BP1 almost completely attenuated the rapamycin-mediated increase in cap-dependent translation (Fig. 4D). Overexpression of WT 4E-BP1 also decreased cap-dependent translation in both control and rapamycin-treated groups. This inhibition is likely due to a dramatic increase in the total 4E-BP1 protein level, a significant percentage of which is not phosphorylated (Fig. 4D). Nevertheless, more potent inhibition was observed with the AA mutant, suggesting that phosphorylation of 4E-BP1 was important for the increase in cap-dependent translation. Therefore, rapamycin-induced 4E-BP1 phosphorylation is necessary for the increase in total cap-dependent translation.
In addition to an increase in total cap-dependent translation, an increase in eIF4E availability results in preferential translation of mRNAs with highly structured 5′ UTRs (21). It has been demonstrated that translational inhibition of HIF-1α mRNA by rapamycin is specifically regulated by its 5′ UTR region (22). Therefore, the HIF-1α 5′ UTR was inserted in front of the Renilla luciferase, and IRES-driven firefly luciferase was used as an internal normalizing factor. As shown in Fig. S14A, prolonged (72-h) rapamycin treatment increased cap-dependent translation by ≈70% when compared with control. However, insertion of the 5′ UTR region of HIF-1α increased the cap-dependent translation by only ≈30% (Fig. S14A). This difference between with or without 5′ UTR-driven translation may reflect the requirement for eIF4B phosphorylation by S6K1. Kinases such as RSK could also compensate during S6K1 inhibition, which may explain the ≈30% increase in 5′ UTR-containing translation (9). Nonetheless, chronic rapamycin treatment increased translation driven by the 5′ UTR region of HIF-1α mRNA. This rapamycin-induced increase required the hyperphosphorylation of 4E-BP1, because coexpression with the 4E-BP1 AA mutant completely inhibited its effect (Fig. S14B). Moreover, there was a direct correlation with rapamycin-induced 4E-BP1 hyperphosphorylation and its ability to increase translation driven by the 5′ UTR region of HIF-1α. As shown in Fig. 4G, the inability of prolonged rapamycin treatment to stimulate 4E-BP1 hyperphosphorylation in U2OS, PC3, and MCF7 cells correlated with the inability of rapamycin to increase translation driven by the 5′ UTR region of HIF-1α. Conversely, HeLa and HEK293 cells, which do exhibit rapamycin-induced hyperphosphorylation, all increased translation driven by the HIF-1α 5′ UTR region with chronic rapamycin treatment (Fig. 4E). This effect was independent of the cell's p53 status, because PC3 cells, which are p53-null, and U2OS cells, which have WT p53, both failed to induce 4E-BP1 phosphorylation. When taken together, the rapamycin-induced hyperphosphorylation of 4E-BP1 determines the sensitivity of a specific cell to translational inhibition.
Discussion
The eukaryotic translational initiation machinery is a tightly controlled system that regulates protein synthesis based on many factors including the availability of growth factors, nutrients, and glucose (3). Accordingly, when there are shortages of these factors, protein synthesis stalls. The molecular pathway responsible for linking the environmental cues and translational control is the mTORC1 signaling pathway. mTORC1 is part of an evolutionarily conserved signaling pathway that links environmental status with a host of other cellular processes such as cellular growth, proliferation, metabolism, autophagy, and translational control (3). This pathway is of immense interest because rapamycin (also known as sirolimus), an inhibitor of mTORC1, is already clinically approved as an immunosuppressant for kidney transplantation and for postangioplasty restenosis. Temsirolimus, also a rapamycin analogue, was approved last year for treatment against renal cell carcinoma (23). In addition, temsirolimus, as well as other rapamycin analogues, is currently in clinical trials against other cancers because of its antiangiogenic and cytostatic properties (11).
The implications of our findings suggest that although rapamycin treatment inhibits the function of S6Ks and 4E-BP1 in acute treatment experiments, this may not be the case in all cell types during prolonged treatments. As a consequence, general cap-dependent translation and translation of genes with highly structured 5′ UTRs are reinitiated despite continuous mTORC1 inhibition. These results explain how cap-dependent translation could be differentially controlled despite rapamycin treatment.
However, it appears that components of mTORC1 are still required for rephosphorylation of 4E-BP1. We showed through multiple methods including siRNA against mTOR/Raptor, mTOR catalytic inhibitors, and 4E-BP1 mutations that mTORC1 is in fact required for the rapamycin-induced 4E-BP1 phosphorylation. So how could an mTORC1 inhibitor like rapamycin stimulate 4E-BP1 phosphorylation but still require mTORC1 components? The molecular nature of rapamycin as an mTORC1 inhibitor differs from the more traditional catalytic kinase inhibitors by binding to the complex with FKBP12 and inhibiting mTORC1's ability to signal to its substrates through an unknown mechanism (24). Therefore, alterations in the molecular interaction among 4E-BP1, mTOR, and Raptor induced by prolonged rapamycin may yield scenarios where the activity of mTORC1 is maintained but the nature of the substrate binding may differ. Indeed, such effects would render mTORC1 rapamycin resistant against selective downstream targets, such as 4E-BP1. However, in vitro kinase assays with mTORC1 complexes immunoprecipitated from short-term (1H) and long-term (24H) rapamycin-treated cells toward GST-4E-BP1 suggest that both are sensitive to FKBP12-rapamycin inhibition in vitro (Fig. S13). It is difficult to determine whether this lack of in vitro inhibition is due to the loss of essential factors during biochemical purification, which is often observed with the mTORC1 complex (25). In addition, the assay was conducted with recombinant GST-4E-BP1 as an in vitro substrate; hence, any modifications on 4E-BP1 in vivo would not be accounted for. Conversely, it is unlikely that inhibition of a phosphatase specific for 4E-BP1 is involved, because calyculin A and okadaic acid-induced 4E-BP1 phosphorylation was sensitive to rapamycin (26). Furthermore, the ability of wortmannin and PI-103, which, like rapamycin, inhibits mTORC1, to attenuate this reemerging phosphorylation suggests that phosphatase inhibition would be insufficient (Fig. 3D). Therefore, it appears that prolonged rapamycin treatment renders mTORC1 to be rapamycin-resistant specifically toward 4E-BP1, which controls global cap-dependent translation.
Our results also suggest that rapamycin can autonomously control cap-dependent translation through de novo protein synthesis. This effect is independent of known proline-directed kinases that could ideally phosphorylate 4E-BP1 including ERKs and CDKs. Rather, the up-regulation appears to be a feedback-like mechanism to maintain 4E-BP1 phosphorylation despite rapamycin treatment. The activation of Akt induced by rapamycin in certain cells is also not involved because neither stauroporine, which catalytically inhibits Akt, nor wortmannin at 50 nM, which specifically inhibits PI3K, affected the phosphorylation of 4E-BP1 (Fig. S11 and Fig. 3D) (18). Whether the de novo protein synthesized during rapamycin treatment (Fig. S6) specifically up-regulates a factor that provokes this rapamycin-resistant 4E-BP1 phosphorylation remains to be determined.
On the contrary, several observations suggest the existence of rapamycin-resistant 4E-BP1 phosphorylations. First, the kinase Pim2 provides a LY294002- and rapamycin-resistant mechanism for phosphorylating 4E-BP1 in hematopoietic cells (27). Accordingly, we were unable to observe Pim2 expression in HEK293/HeLa cells with or without rapamycin treatment, and shRNA directed against PIM2 did not affect rapamycin-induced 4E-BP1 phosphorylation (data not shown). Rapamycin-insensitive regulation of 4E-BP1 was also observed in the livers of mice recovering from partial hepatectomy (28). However, the induction of 4E-BP1 phosphorylation and S6K1 activation in the liver was not rapamycin-induced, indicating that these cells undergoing hepatectomy may have been predisposed to rapamycin-insensitive 4E-BP1 phosphorylations. More interestingly, it is possible that PIM2 and livers undergoing hepatectomy may employ the same mechanism as rapamycin does.
Our findings also illuminate some important questions regarding rapamycin and cancer therapy. First, our results suggest that inhibiting mTORC1 with a catalytic inhibitor may yield better results when compared with rapamycin. Although our results implicate only 4E-BP1, other currently unknown mTORC1 substrates may also be differentially regulated. In addition, recent reports have suggested that 4E-BP1 phosphorylation is directly correlated with the malignancy and severity of various tumors (29). Therefore, using mTOR catalytic inhibitors rather than rapamycin would likely be more effective in dephosphorylating 4E-BP1. Finally, the differential regulation of mTORC1's substrates by rapamycin suggests a reevaluation of phospho-S6 as a biomarker for mTORC1 inhibition, because loss of S6 phosphorylation does not always translate to inhibition of all mTORC1 substrates.
In conclusion, we propose that in certain cells rapamycin autonomously controls cap-dependent translation by differentially regulating its substrates 4E-BP1 versus S6Ks in a cell-type-specific manner. Contrary to the current understanding, this effect maintains global cap-dependent and structured 5′ UTR-mediated translation despite apparent mTORC1 inhibition by rapamycin as monitored by rpS6 phosphorylation. Therefore, we have uncovered an unexpected consequence of long-term rapamycin treatment, which determines the condition that is required for rapamycin to affect cap-dependent translation.
Materials and Methods
The Polio IRES luciferase translation vector was a generous gift from Peter Bitterman (University of Minnesota). HIF-1α 5′ UTR vector and its control vector were from Charles Sawyers and George V. Thomas (Sloan Kettering) (37). The protocol for the translational assay and cap binding assay are described elsewhere (17). In brief, cells were lysed in lysis buffer (10 mM KPO4/1 mM EDTA/10 mM MgCl2/50 mM β-glycerolphosphate/5 mM EGTA/0.5% Nonidet P-40/0.1% Brij/1 mM sodium orthovanadate/appropriate protease inhibitors), incubated with m7GTP-Sepharose for 2 h, and washed 3 times with lysis buffer. A detailed description of all of the other methods appears in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank the J.B. laboratory for critical discussion and technical assistance. We are also grateful to Drs. Peter Bitterman, Nahum Sonenberg, Diane Fingar, Brendan Manning, David Kwiatkowski, and Charles Sawyers for providing reagents. A.Y.C. and J.B. also thank Drs. Lewis Cantley, Joan Brugge, Lee Gehrke, and George V. Thomas for critical discussions. This work was funded by National Institutes of Health Grant GM51405 (to J.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809136105/DCSupplemental.
References
- 1.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 2.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: Mystery of their existence. J Biol Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 3.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signaling controls tumour cell growth. Nature. 2006;441:424–30. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 4.Beretta L, et al. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 5.Brunn GJ, et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 6.Gingras AC, et al. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 8.Raught B, et al. Phosphorylation of eukaryotic translation initation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23:1761–1769. doi: 10.1038/sj.emboj.7600193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shahbazian D, et al. The mTOR/PI3K and Mapk pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25:2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorrello NV, et al. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 11.Fingar DC, Blenis J. Target of rapamycin (TOR): An integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 12.Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd s6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 13.Li S, et al. Translational control of cell fate: Availability of phosphorylation sites on translational repressor 4E-BP1 governs its proapoptotic potency. Mol Cell Biol. 2002;22:2853–2861. doi: 10.1128/MCB.22.8.2853-2861.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heesom KJ, et al. Cell cycle-dependent phosphorylation of the translational repressor eIF4E binding protein-1 (4E-BP1) Curr Biol. 2001;11:1374–1379. doi: 10.1016/s0960-9822(01)00422-5. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg VL, Zimmer SG. Paclitaxel induces the phosphorylation of the eukaryotic translation initiation factor 4E-binding protein 1 through a CDK1-dependent mechanism. Oncogene. 2005;24:4851–4860. doi: 10.1038/sj.onc.1208624. [DOI] [PubMed] [Google Scholar]
- 16.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 17.Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOSmotif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 18.Zhang HH, et al. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol Cell. 2006;24:185–197. doi: 10.1016/j.molcel.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan QW, et al. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyronnet S, et al. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas GV, et al. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12:122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- 23.Costa LJ, Drabkin HA. Renal cell carcinoma: New developments in molecular biology and potential for targeted therapies. 2007;12:1404–1415. doi: 10.1634/theoncologist.12-12-1404. [DOI] [PubMed] [Google Scholar]
- 24.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Sancak Y, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Lin TA, Lawrence JC., Jr Control of PHAS-I phosphorylation in 3T3–L1 adipocytes: Effects of inhibiting protein phosphatases and p70S6K signaling pathway. Diabetologia. 1997;40:S18–S24. doi: 10.1007/s001250051391. [DOI] [PubMed] [Google Scholar]
- 27.Fox CJ, et al. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang YP, Ballou LM, Lin RZ. Rapamycin-insensitive regulation of 4E-BP1 in regenerating rat liver. J Biol Chem. 2001;276:10943–10951. doi: 10.1074/jbc.M007758200. [DOI] [PubMed] [Google Scholar]
- 29.Armengol G, et al. 4E-binding protein 1: A key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 2007;67:7551–7555. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.