Abstract
Recent analysis of the human and mouse genomes has revealed that highly identical duplicated elements account for >5% of the sequence content. These elements vary in copy number between individuals. Copy number variations (CNVs) contribute significantly to genetic differences among individuals and are increasingly recognized as a causal factor in human diseases with different etiologies. In inbred mouse strains, CNVs have been fixed by inbreeding, but they are highly variable among strains. Within strains, de novo germ-line CNVs can occur, leading to interindividual variation. By analyzing the genome of clonal isolates of mouse ES cells derived from common parental lines, we have uncovered extensive and recurrent CNVs. This variation arises during mitosis and can be cotransmitted into the mouse germ line along with engineered alleles, contributing to genetic variability. The frequency and extent of these genomic changes in ES cells suggests that all somatic tissues in individuals will be mosaics composed of variants of the zygotic genome. Human ES (hES) cells and derived somatic lineages may be similarly affected, challenging the concept of a stable somatic genome.
Keywords: inbred mouse strains, comparative genomic hybridization, BAC arrays
Copy number variation (CNV) of DNA segments in the human genome can involve large segments of DNA (1–5) that occur in phenotypically normal individuals and these can be disease-associated (6). Within an inbred mouse strain, CNVs also occur among individuals (7, 8) and are presumed to arise de novo by meiotic homologous recombination between nearly identical duplicated sequences and through nonhomologous end-joining (9–12). Although the frequency of homologous recombination is several orders of magnitude greater than the single-nucleotide mutation rate, many duplications contain active genes; thus, a CNV arising de novo is expected to contribute more phenotypic variation (on average) than single-nucleotide alterations.
ES cell lines established from several different strains have been the major route through which thousands of new mutations have been established in the mouse germ line over the last 2 decades. To limit the impact of interstrain variation, the programs generating genome-wide resources of knockout alleles (13) use ES cells from a single genetic background, C57BL6/N. The underlying genetic stability of ES cell lines used for these resources is critical, because the modified ES cell clones used to establish new germ-line alleles should be genetically identical to the parental cell lines.
Compared with other cultured cell lines, ES cells are relatively stable. Karyotypic variants that arise, such as trisomies or loss of the Y chromosome (14, 15), preclude germ-line transmission; thus, they do not impact genetic studies. However, very little is known about other types of structural variation, such as CNV, which is likely to be compatible with germ-line transmission.
In humans, CNVs have been shown to have a meiotic origin (16). Recently, the comparison of monozygotic twins has identified CNVs, suggesting these genome alterations also occur during somatic development (17). Because recombination between duplicated sequences occurs at measurable frequencies in mouse ES cells (18), it would be expected that CNVs would be generated de novo in ES cells in culture. Given that ES cells undergo 30–40 mitotic divisions in vitro before resuming their normal developmental route into the germ line, we reasoned that ES cells may accumulate CNVs, which might be transmitted into the mouse germ line along with targeted alleles, contributing phenotypic variability to the analysis of mutant phenotypes.
Results
CNVs in ES Cell Clones.
We examined the genomes of 50 different ES cell clones for evidence of CNV by comparative genomic hybridization (CGH) against BAC arrays. Given that some types of variation may be incompatible with germ-line transmission, and we wished to formulate an unbiased view of the extent of this variation, we analyzed clones with confirmed and compromised germ-line potential. To maximize the chance of discovering independent events, the clones examined in this study were derived from 3 different parental lines: 2 widely used lines, AB2.2, E14, and the JM8 line that is the basis for the EUCOMM and KOMP mutation resource.
Of 26 clones that could not contribute to the mouse germ line, trisomies were detected in 7 which involved chromosomes 1, 6, 8, and 11, [supporting information (SI) Fig. S1 and Tables S1 and S2]. In 5 cases, loss of the Y chromosome was detected. These types of aneuploidy have been observed previously, and they explain the germ-line transmission failure of nearly half of these ES clones. In addition to gains and losses of whole chromosomes, 14 germ-line-compromised clones exhibited subchromosomal changes of 3- to 5-Mb intervals (deletions or duplications) (Fig. S2 and Tables S1 and S2). These smaller genomic alterations may directly explain the germ-line transmission failure of these clones, or they may identify breakpoints of more substantial structural rearrangements, such as inversions or translocations that cannot be directly detected by CGH. In total, CGH analysis identified genetic changes in 19 of these 26 clones.
Next, we analyzed the 24 germ-line-competent clones and, as expected, none of these clones exhibited gains or losses of entire chromosomes. However, 7 of these had 1- to 2-Mb deletions and/or duplications (Table S2). Subclones derived from all 3 parental cell lines exhibited this type of variation, including 2 clonal isolates of the recently derived JM8 cell line. A total of 9 different CNVs were detected in 7 of the 24 germ-line-transmittable ES cell clones. Five of these variants were also detected in germ-line-compromised clones, indicating these specific changes could not explain the germ-line transmission failure of these clones. However, the number and size of the variants were greater in the germ-line-compromised clones.
Germ-Line Transmission of CNVs.
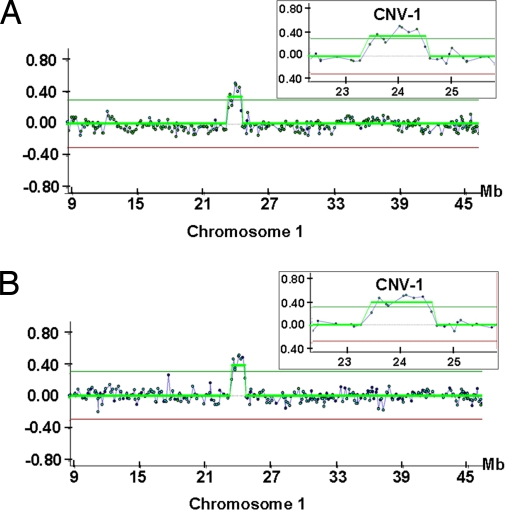
We next examined whether these CNVs were transmitted into the mouse germ line. CNV-1, a 1.5-Mb duplication on chromosome 1 in AB2.2 subclones involving 10 genes, was transmitted into the germ line along with the engineered mutation (Fig. 1).
Fig. 1.
A 1.5-Mb amplification in Chr.1 (CNV-1) detected in a germ-line-competent AB2.2 ES cell clone that was transmitted to its descendents. (A) CGH array analysis of the ES cell clone vs. the AB2.2 parental cells as control. (B) CGH array analysis of F1 generation mice vs. the same control.
CNV Accumulation.
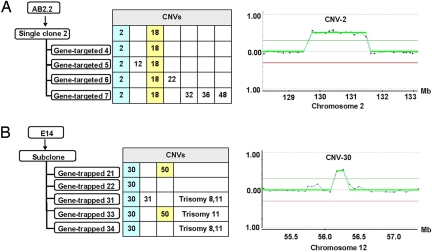
The mitotic mutation rate for CNVs generated through homologous recombination appears to be high enough for these variants to accumulate through successive cell divisions. For instance, a targeted AB2.2 clone (which can achieve a high rate of germ-line transmission) contains CNV-2 and CNV-18 (Fig. 2A, Fig. S2, and Tables S1 and S2). Four double-targeted clones derived from this clone inherited these CNVs, as expected; however, 5 new CNVs arose in 4 subclones (Fig. 2A). Similarly, analysis of the E14 clones from the SIGTR gene-trap library identified 5 clones with the same CNV (CNV-30). In addition, 2 of these 5 clones also share CNV-50 (Fig. 2B, Fig. S2, and Tables S1 and S2), suggesting that CNV-30 arose in the parental cell population first and was preferentially enriched; subsequently, CNV-50 occurred in the subpopulation of the parental cells that already contained CNV-30. The mitotic recombination rate between direct repeats on the same chromosome depends on the length, location, and identity of the duplicated sequences and ranges from 3.8 × 10−3 to 4.3 × 10−6 per cell per generation (18). Given this rate, such a cell will have segregated several daughter cells with a deletion of the sequence between the repeats by the time the cell has undergone 18 doublings.
Fig. 2.
Accumulation of CNVs in ES cells during rounds of single-cell cloning. (A) CNVs detected in AB2.2 ES cell clones. (B) CNVs in E14 ES cell clones. The clone ID corresponds to the ID in Tables S1 and S2. The CNV numbers detected in each clone are indicated. The filled colored squares indicate recurrent CNVs detected in different clones. CNV-2 and -30 are examples of CNVs shared between these clones.
Recurrent CNVs.
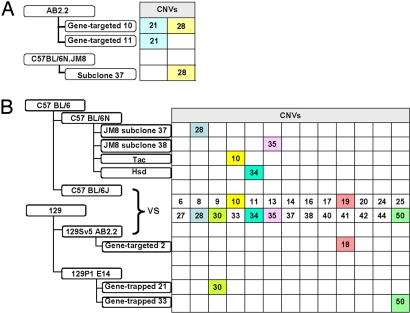
Analysis of the E14 and AB2.2 clones revealed that several contained the same CNVs. These may represent subclones of daughter cells, or they could have arisen independently. To resolve this, we looked for recurrent CNVs in different cell lines. CNV-28 was found in both AB2.2 and C57BL6/N JM8 ES cell clones (Fig. 3A, Fig. S2, and Tables S1 and S2); thus, this CNV arose independently. We also compared the CNVs detected in ES clones with those revealed by comparisons between the 129S5 and C57BL6/N mouse strains (Fig. 3B; Table S1). Several CNVs were identified (CNV-28, -30, -35, and -50) that have arisen independently in ES cells in culture (Fig. 3B and Tables S1 and S2).
Fig. 3.
Examples of recurrent CNVs in ES cells and mice. (A) CNV-28 that arose independently in subclones of 2 different parental ES cell lines. (B) CNVs detected in ES cell clones or fixed in different mouse strains. CNV-18 and -19 are adjacent and have the same breakpoints.
Studies in humans have revealed that many CNVs are generated by nonallelic homologous recombination between low-copy repeats (19). To investigate whether a similar mechanism was responsible for the CNVs observed in this study, we compared the nucleotide sequence at the breakpoints of the recurrent CNVs. Because of the resolution of the BAC arrays, the CNV breakpoints are only approximately determined; therefore, we aligned 1 Mb of sequence (repeat masked) from each breakpoint. This analysis identified low-copy repeats ranging from 1 to 5 kb that were 97–99% identical (Table 1).
Table 1.
Number of low-copy repeats indetified by aligning sequences around the breakpoints of recurrent CNVs
| Copy number variants | Chromosome | Coordinates(Ensembl m37) |
>1 kb |
>5 kb |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Region 1* | Region 2* | 97% | 98% | 99% | 97% | 98% | 99% | ||
| CNV-10 | 4 | 121356475 | 121656475 | 11 | 11 | 11 | 9 | 4 | 4 |
| CNV-18 | 7 | 17550532 | 17850532 | 0 | 0 | 0 | 0 | 0 | 0 |
| CNV-19 | 7 | 20612611 | 20912611 | 34 | 18 | 6 | 9 | 5 | 2 |
| CNV-21 | 7 | 38032340 | 38332340 | 43 | 43 | 43 | 0 | 0 | 0 |
| CNV-28 | 10 | 79225351 | 79525351 | 0 | 0 | 0 | 0 | 0 | 0 |
| CNV-30 | 12 | 56061731 | 56361731 | 45 | 43 | 39 | 0 | 0 | 0 |
| CNV-34 | 13 | 65475239 | 65775239 | 35 | 21 | 2 | 1 | 1 | 0 |
| CNV-35 | 14 | 3080383 | 3380383 | 97 | 74 | 49 | 2 | 2 | 1 |
| CNV-50 | X | 3084190 | 3384190 | 32 | 26 | 20 | 12 | 7 | 3 |
The sequences of region 1 and region 2 used for comparison are 1 Mb around the breakpoints (the breakpoints ± 0.5 Mb).
*Regions 1 and 2 represent the chromosomal coordinates of the 2 breakpoints for each CNV.
CNVs in Inbred Mouse Strains.
Given that CNVs are generated mitotically during ES cell culture, we looked for evidence that these also arise during meiosis. We compared several isolated breeding colonies of the C57BL6/N line. From this analysis, a 1-Mb duplication in chromosome 4 was identified (CNV-10, Figs. S2 and S3 and Tables S1 and S2) in the mice from C57BL/6N Tac and a 1.6-Mb duplication (CNV-34, Figs. S2 and S3 and Tables S1 and S2) was observed in the C57BL/6N Hsd mice on chromosome 13. Thus, carefully maintained inbred strains have fixed significant genetic differences.
Discussion
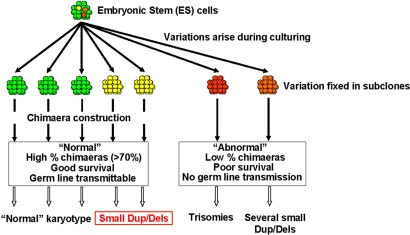
We have demonstrated that CNV involving gains or losses of millions of base pairs occurs frequently during mitotic divisions of mouse ES cells during routine culture involving relatively few cellular divisions. These variants do not interfere with germ-line transmission and can be cotransmitted into the mouse germ line along with the engineered allele(s) contributing to genetic and phenotypic variability (Fig. 4).
Fig. 4.
Genomic variation arising during ES cell culture. The parental cells (green) segregate variants (yellow, orange, and red). Clones with major chromosomal changes (red) cannot be transmitted into the mouse germ line and typically exhibit trisomies or multiple deletions or duplications. Some clones have a few small (1- to 2-Mb) CNVs that do not affect germ-line transmission. These CNVs may be transmitted into the mouse germ line along with an engineered allele.
Among 50 different ES cell clones from 3 different genetic backgrounds, we detected 28 different CNVs involving a total of 1,218 genes. In this study, we used a BAC array; thus, the resolution of our analysis is limited to 100 kb. Undoubtedly, smaller CNVs remain undetected in these ES cell clones and mouse lines. One-third of the CNVs identified in this study were detected in multiple ES cell clones. Although some of these may be daughter cells descended from a cell with a CNV that arose early during the expansion of a population, more than half of these CNVs appear to have arisen independently, because they were observed in subclones isolated from different parental ES cell lines (Fig. 3B and Table S2). It is unlikely these variants come from the mice used to generate these parental ES cell lines, because ES cell lines are usually derived from single cell clones; thus, any underlying variation from the mouse would be fixed during the process of isolating an ES cell line. The incidence of CNVs detected in different ES cell lines varied in this study. This is likely to reflect the history of the cell lines selected for analysis; the AB2.2 subclones have a high passage number (40+), because they are mostly double targeted clones, whereas the E14 clones have only been subject to a single round of gene trapping. The JM8 clones are single-cell subclones isolated without a targeted or gene-trap alteration from an early passage of the parental cell line.
Seven CNVs (CNV-10, -11, -19, -30, -31, -34, and -40) have been described previously (12, 20). Recurrent CNVs may have a higher mutation rate than the other CNVs identified in this study, although this conclusion will require more detailed analysis. Alignment of the sequences at the breakpoints of the recurrent CNVs indentified in this study identified several highly related sequences extending from 1 to 5 kb, suggesting these recurrent CNVs may be generated through nonallelic homologous recombination.
Although this study was limited to the analysis of mouse ES cells in vitro, CNVs will undoubtedly occur in the somatic descendents of ES cells in humans and mice and are also likely to arise in other stem cell pools whose descendents populate somatic lineages. Currently, the de novo rate with which CNVs are generated in somatic lineages is unknown. This may be lower in vivo, because cells in culture continuously divide compared with the situation in vivo. Thus, human and mouse somatic tissues are mosaics composed of variants of the mitotic genome. Recent analysis of an inversion in humans has demonstrated that mitotic structural variation occurs at a measurable rate (21). It is anticipated that hES cells will also incur this type of genetic change during culture, irrespective of whether they are embryo-derived or induced (22, 23). The occurrence and affects of culture-derived CNVs will need to be considered in the development of hES-cell-based transplantation therapies, especially because therapeutic applications will involve substantial expansion from a limited number of founder cells, increasing the chance of generating and fixing CNVs.
Materials and Methods
ES Cell Lines.
All ES cell clones were selected from the microinjection database of the Wellcome Trust Sanger Institute, which contains the data for all of the clones, including the percentage of chimerism and the germ-line transmission rates. The early-passage cell lines of AB2.2 (129S5), E14 (129P2),and C57 BL6/N JM8 were used as the hybridization controls for the array-CGH experiments. Genomic DNA samples were prepared from ES cells or tail samples by using standard methods. Of the 20 targeted AB2.2 (129S5) clones, 3 were germ-line-competent. Ten of the 16 E14 (129P2) ES cell clones from the SIGTR gene-trap library (24) had germ-line potential. The JM8 (C57Bl6/N) clones were nontargeted subclones from very early-passage JM8 cells of which 11 of 14 had germ-line colonization potential.
Mouse Strains.
We collected C57BL6/N substrains from different breeders: The Jackson Laboratory (C57BL6/N Jax), Taconic (C57BL6/N Tac), Harlan Sprague–Dawley (C57BL6/N Hsd), and Charles River Laboratories (C57BL6/N Crl). The individuals were ordered in 2 different batches to ensure they were not from the same litters; all mice were males. CGH analysis was performed on 3 individual C57BL6/N mice held by different breeders. For each mouse, independent comparisons were conducted against mice from the other breeders and the C57BL/6N JM8 ES cells.
Tiling-Array CGH.
Tiling-array experiments were carried out on a microarray manufactured by the Wellcome Trust Sanger Institute containing 18,294 BACs from the RPCI-23 and -24 libraries. The position of clones on the array was verified by end sequencing or fingerprinting. Each array (1 slide) contained 2 copies of each clone. Each BAC is 150–250 kb in length, and cumulatively they cover the entire mouse genome at tiling-path resolution. Clone information can be obtained from www.ensembl.org/Mus_musculus/cytoview. Test and control DNA samples labeling, dye incorporation, hybridization, and data processing were carried out as described (4).
Fluorescence intensities of Cy5 and Cy3 on each array image and the log2 ratio values were extracted by using Bluefuse software (Bluegnome). Spots with inconsistent fluorescence patterns (“confidence” < 0.3 or “quality” = 0) were excluded before normalizing all log2 ratio values.
Array Data Analysis and CNV Determination.
All experiments were performed in a fluorochrome-reversed pairs of 2-color (Cy3 and Cy5) hybridizations. Fusion of dye-swap results and subsequent analyses were performed by using custom Perl scripts. The median of all ratio values was calculated globally for the whole genome for each individual hybridization. Each ratio was then normalized by the genomic median. The ratios of each clone in the 2 dye-swap hybridizations were then averaged if replicate ratios differed by <50% (i.e., less than a difference of 0.585 on the log2 scale). The 68.2th percentile of the absolute values for all combined ratios was then calculated as an estimation of the standard deviation (StdDev). Dye-swap experiments were accepted only if the following criteria were fulfilled: (i) StdDev between replicate clones was <0.4; (ii) StdDev between dye-swap replicates was <0.3; (iii) clone exclusion rates of the whole genome and the clone exclusion rate for each individual chromosome (except chromosome Y) were <10%.
CNVs were called by using a CGH smoothing algorithm (25). The log2 ratio of each probe from Cy5/Cy3 hybridization represent 1 of 3 states: “up” for duplicated (Dup), “base line” for equivalent and “down” for deleted (Del). The number of clones in region showing a duplication/deletion was ≥2. The log2 ratio threshold for “Duplication” was >0.29999. The log2 ratio threshold for “Deletion” was <0.29999. Each experiment included dye swap, so that each experiment consisted of 1 replicate hybridization done with reversal of the Cy3 and Cy5 dyes relative to each DNA sample and thus included 4 comparative measurements per BAC. Additionally, when a CNV was detected, the experiment was repeated at least once. The hybridization results with consistent breakpoints were confirmed as CNVs.
Supplementary Material
Acknowledgments.
We thank O. Dovey for help with technical assistance and the microarray team for data generation and array analysis. We also thank P. Liu and everybody in Team 82 of the Wellcome Trust Sanger Institute for sharing their ES cell clones and C. S. Smith for comments on this manuscript. This work was supported by the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805638105/DCSupplemental.
References
- 1.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 2.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 3.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 4.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 6.Lee JA, Lupski JR. Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron. 2006;52:103–121. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 7.Li J, et al. Genomic segmental polymorphisms in inbred mouse strains. Nat Genet. 2004;36:952–954. doi: 10.1038/ng1417. [DOI] [PubMed] [Google Scholar]
- 8.Snijders AM, et al. Mapping segmental and sequence variations among laboratory mice using BAC array CGH. Genome Res. 2005;15:302–311. doi: 10.1101/gr.2902505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 10.Han LL, Keller MP, Navidi W, Chance PF, Arnheim N. Unequal exchange at the Charcot–Marie–Tooth disease type 1A recombination hot-spot is not elevated above the genome average rate. Hum Mol Genet. 2000;9:1881–1889. doi: 10.1093/hmg/9.12.1881. [DOI] [PubMed] [Google Scholar]
- 11.Tusie-Luna MT, White PC. Gene conversions and unequal crossovers between CYP21 (steroid 21-hydroxylase gene) and CYP21P involve different mechanisms. Proc Natl Acad Sci USA. 1995;92:10796–10800. doi: 10.1073/pnas.92.23.10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egan CM, Sridhar S, Wigler M, Hall IM. Recurrent DNA copy number variation in the laboratory mouse. Nat Genet. 2007;39:1384–1389. doi: 10.1038/ng.2007.19. [DOI] [PubMed] [Google Scholar]
- 13.Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 14.Caisander G, et al. Chromosomal integrity maintained in 5 human embryonic stem cell lines after prolonged in vitro culture. Chromosome Res. 2006;14:131–137. doi: 10.1007/s10577-006-1019-8. [DOI] [PubMed] [Google Scholar]
- 15.Sugawara A, Goto K, Sotomaru Y, Sofuni T, Ito T. Current status of chromosomal abnormalities in mouse embryonic stem cell lines used in Japan. Comp Med. 2006;56:31–34. [PubMed] [Google Scholar]
- 16.Turner DJ, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruder CE, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasty P, Ramirez-Solis R, Krumlauf R, Bradley A. Introduction of a subtle mutation into the Hox-2.6 locus in embryonic stem cells. Nature. 1991;350:243–246. doi: 10.1038/350243a0. [DOI] [PubMed] [Google Scholar]
- 19.Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39:S43–47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 20.She X, et al. Recurrent DNA inversion rearrangements in the human genome. Proc Natl Acad Sci USA. 2007;104:6099–6106. doi: 10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Zaehres H, Scholer HR. Induction of pluripotency: From mouse to human. Cell. 2007;131:834–835. doi: 10.1016/j.cell.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 24.Skarnes WC, et al. A public gene trap resource for mouse functional genomics. Nat Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jong K, Marchiori E, Meijer G, Vaart AV, Ylstra B. Breakpoint identification and smoothing of array comparative genomic hybridization data. Bioinformatics. 2004;20:3636–3637. doi: 10.1093/bioinformatics/bth355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.