Abstract
Peripheral T cell lymphomas (PTCL) account for about 12% of lymphoid tumours worldwide. Almost half show such morphological and molecular variability as to hamper any further classification, and to justify their inclusion in a waste-basket category termed “not otherwise specified (NOS)”. The latter term is used for neoplasms with aggressive presentation, poor response to therapy and dismal prognosis. In contrast to B cell lymphomas, PTCL have been the subject of only a limited number of studies to elucidate their pathobiology and identify novel pharmacological approaches. Herewith, the authors revise the most recent contributions on the subject based on the experience they have gained in the extensive application of microarray technologies. PTCL/NOS are characterised by erratic expression of T cell associated antigens, including CD4 and CD52, which have recently been proposed as targets for ad hoc immunotherapies. PTCL/NOS also show variable Ki-67 marking, with rates >80% heralding a worse prognosis. Gene expression profiling studies have revealed that PTCL/NOS derive from activated T lymphocytes, more often of the CD4+ type, and bear a signature composed of 155 genes and related products that play a pivotal role in cell signalling transduction, proliferation, apoptosis and matrix remodelling. This observation seems to pave the way for the use of innovative drugs such as tyrosine kinase and histone deacetylase inhibitors whose efficacy has been proven in PTCL primary cell cultures. Gene expression profiling also allows better distinction of PTCL/NOS from angioimmunoblastic T cell lymphoma, the latter being characterised by follicular T helper lymphocyte derivation and CXCL13, PD1 and vascular endothelial growth factor expression.
Peripheral T cell lymphomas (PTCL) represent approximately 12% of lymphoid neoplasms.1 Their incidence varies among countries, and it is higher in human T-cell lymphotropic virus-1 endemic areas.1 PTCL are a heterogeneous group of tumours that can be roughly subdivided into: specified and not otherwise specified (NOS) (Box 1).1 2 While specified tumours correspond to distinct but rare entities often occurring at extranodal sites, NOS represent the commonest type of TCL (40–50%), followed by the angioimmunoblastic (AITL) and the anaplastic large cell (ALCL) types.
Box 1: Mature T cell and NK cell neoplasms1
Peripheral T cell lymphoma, not otherwise specified (PTCL/NOS)
Peripheral T cell lymphoma, specified
Leukaemic:
T cell prolymphocytic leukaemia
T cell large granular lymphocytic leukaemia
Aggressive NK cell leukaemia
Systemic Epstein–Barr virus positive T cell lymphoproliferative disease of childhood (associated with chronic active EBV infection)
Hydroa vaccineforme-like lymphoma
Adult T cell leukaemia/lymphoma
Extranodal:
Extranodal NK/T cell lymphoma, nasal type
Enteropathy-associated T cell lymphoma
Hepatosplenic T cell lymphoma
Subcutaneous panniculitis-like T cell lymphoma
Mycosis fungoides
Sézary syndrome
Primary cutaneous anaplastic large-cell lymphoma
Primary cutaneous aggressive epidermotropic CD8+ cytotoxic T cell lymphoma (provisional entity)
Primary cutaneous γδ T cell lymphoma
Primary cutaneous small/medium CD4+ T cell lymphoma (provisional entity)
Prevalently nodal:
Angioimmunoblastic T cell lymphoma
Anaplastic large cell lymphoma (ALCL), anaplastic large cell lymphoma kinase (ALK) positive
ALCL, ALK negative (provisional entity)
PTCL/NOS cannot be further classified based on morphology, phenotype and molecular biology in most instances,3–5 although rare distinctive variants have been reported (ie, follicular and lymphoepithelioid).6–8 Usually, PTCL/NOS occurs in the fifth to sixth decade of life, and there is no evidence of sex predilection.4 9 10 PTCL/NOS more often presents in stage III–IV, with nodal, skin, liver, spleen, bone-marrow or peripheral blood involvement.4 9 10
The tumour is highly variable in terms of cell morphology and may contain prominent reactive components.1 3
Immunohistochemistry usually shows T cell associated molecule expression, although the phenotypic profile is aberrant in about 80% of cases.1 3
Clonal rearrangements of T cell receptor encoding genes are generally detected.11 The karyotype is aberrant in most cases, and is often characterised by complex abnormalities.12 Recently, recurrent chromosomal gains and losses have been documented in PTCL/NOS by comparative genomic hybridisation, and these have been found to differ from those seen in AITL and ALCL.12 13
The molecular pathobiology of PTCL/NOS, as in general in all T cell neoplasms, is poorly understood. In particular, only limited numbers of studies have explored the gene expression profile (GEP).14–22
On clinical grounds, PTCL/NOS are among the most aggressive non-Hodgkin lymphomas. Their response to conventional chemotherapy is indeed poor, with 5-year relapse-free and overall survival rates of 26% and 20%, respectively.4 5 9 23–26 Neither the morphology nor the international prognostic index (IPI) significantly correlates with the outcome. Clinical or clinicobiological scores have been proposed to identify cases with different prognoses.26 27 However, the molecular bases of PTCL/NOS drug resistance and aggressiveness remain elusive.
In the following, the results recently obtained by our group through the extensive application of microarray technologies will be summarised and commented on, with the scope of defining the pathobiological characteristics of PTCL/NOS, tracing the borders between it and AITL on the one hand and anaplastic large cell lymphoma kinase (ALK)-negative ALCL on the other, and drawing attention to potentially novel prognosticators and therapeutic targets.19–22 27
PHENOTYPIC PROFILE OF PTCL/NOS
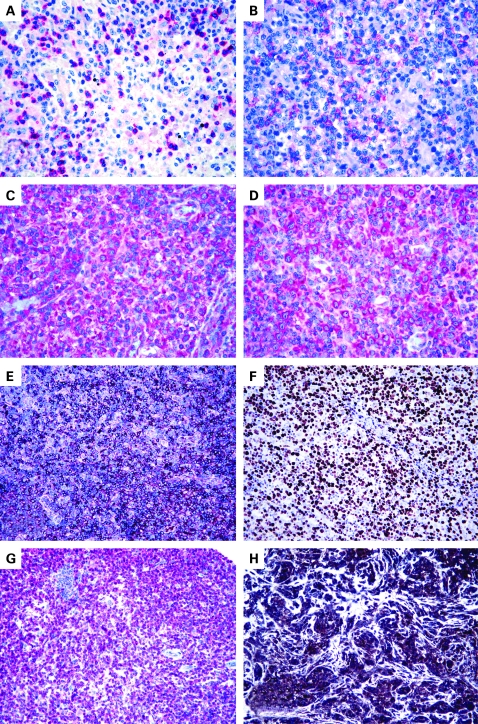
As mentioned above, PTCL/NOS usually carry phenotypic aberrations, the exact prevalence and spectrum of which have remained unresolved.8 11 25 28 In 2006, we reported PTCL from 193 Italian patients (148 NOS and 45 AITL) that had been collected on tissue microarrays and tested by immunohistochemistry and Epstein–Barr virus encoded RNA 1 (EBER1) and EBER2 in situ hybridisation.27 The βF1 antibody (raised against the T cell receptor β chain) reacted with 96% of tumours. NOS and AITL PTCL demonstrated frequent loss of CD5 and CD7, with CD3 being the conventional marker most commonly expressed in NOS types, and CD2 in the AITL types. CD4 was detected in 46% of cases (see fig 1A) and CD8 in 15% of cases; these results are in line with those reported in previous publications.8 11 25 28 29 Interestingly, we found 32% of AITLs to be CD8+; this is in the upper range of reported values.27 30–44 In contrast, the incidence of CD4 positivity (42%) was much lower than expected.27 45 Interestingly, a huge number of PTCL/NOS and AITL (55%) turned out to be either CD4/CD8 double-negative or, more rarely, double-positive. Such profiles, which are usually observed during intrathymic T cell development,1 27 had previously been reported in isolated PTCL cases46 47 and a proportion of cutaneous T cell tumours.27 48 Furthermore, CD10 expression was detected in only 39% of AITL, even when adopting a low cut-off value.27 Such rates did not vary between tissue microarrays and conventional sections.
Figure 1. (A) Lymphomatous cells do not express CD4; however, CD4 is detected in some reactive small lymphocytes (alkaline phosphatase anti-alkaline phosphatase (APAAP) technique, Gill’s haematoxylin nuclear counterstaining, ×250). (B) Partial CD30 expression; it should be noted that the tumour has no anaplastic morphology (APAAP technique, Gill’s haematoxylin nuclear counterstaining, ×250). (C) Positivity for platelet-derived growth factor receptor α (PDGFRα) (APAAP technique, Gill’s haematoxylin nuclear counterstaining, ×400). (D) PDGFRα is phosphorylated (APAAP technique, Gill’s haematoxylin nuclear counterstaining, ×400). (E) CXCL13 expression by neoplastic elements in angioimmunoblastic T cell (EnVision+ technique, Gill’s haematoxylin nuclear counterstaining, ×100). (F) Ki-67 marking exceeds the 80% value (EnVision+ technique, Gill’s haematoxylin nuclear counterstaining, ×200). (G) CD52 positivity in a peripheral T cell lymphoma, not otherwise specified (APAAP technique, Gill’s haematoxylin nuclear counterstaining, ×100). (H) Strong expression of vascular endothelial growth factor in an angioimmunoblastic T cell lymphoma (EnVision+ technique, Gill’s haematoxylin nuclear counterstaining, ×200).
CD56 was detected in 5% of PTCL/NOS: all cases stained with βF1 and three co-expressed TIA-1. Interestingly, CD56 expression suggests a malignant phenotype: in fact, under physiological conditions it is limited to T lymphocytes with spontaneous non-MHC-restricted cytotoxicity.27 49 CD57 was seen in 10% and 5% of PTCL/NOS and AITL respectively. Although numbers of CD57+ normal T lymphocytes increase with age,49 no correlation was found between patient age and CD57 expression.27 50
CD30 was recorded in 6% of cases (see fig 1B), CD15 in 4%, and CD20 in 1%27; these rates of positivity may undoubtedly cause diagnostic difficulties. In particular, CD20 was detected in only two PTCL/NOS that were negative for CD79a, in keeping with previous observations of CD20 positivity in isolated PTCL/NOS, and CD79a aberrant expression in “specified” PTCL.27 51–53 Co-expression of CD15 and CD30 was found in only 3/183 of cases that were able to be evaluated. This is the first reliable estimate of the random incidence of such a phenomenon in a large cohort of patients with PTCL; in fact, the previous reports of Barry et al54 and Gorczyka et al55 referred to a highly selected series. In spite of its rarity, such a finding raises the question of how to differentiate between PTCL and classic Hodgkin lymphoma (CHL) under these circumstances: the polymorphism of neoplastic elements, the possible lack of Reed-Sternberg cells and B cell specific activator protein negativity favour the diagnosis of PTCL and vice versa. In particular, B cell specific activator protein is a valuable B cell marker that is found in about 90% of cases of CHL,56 but it is exceptional in PTCL/NOS.57
In our hands, the mean percentage of Ki-67+ neoplastic cells was around 50%, with 11% of PTCL/NOS exceeding the 80% value. Finally, EBV integration was found at the neoplastic cell level in 5% and 3% of PTCL/NOS and AITL respectively; this value is definitely lower than the one recorded by Dupuis et al in a French cohort.58
GEP OF PTCL/NOS
PTCL have been the subject of a limited number of GEP studies14–22 59 60 (table 1). In particular, Tracey et al,60 Lamant et al16 and de Leval et al17 focused on mycosis fungoides, ALK-positive and -negative ALCLs, and AITL, respectively. In contrast, Martinez-Delgado et al14 and Ballester et al15 analysed large collections of PTCL of the NOS, AITL and ALCL types. However, their studies suffered limitations that varied from the usage of chips with a restricted number of genes14 15 to the lack of a reliable normal counterpart for comparison.14 Martinez-Delgado et al14 reported that PTCL/NOS corresponded to a heterogeneous group of tumours whose GEP was difficult to interpret due to the amount of infiltrating reactive cells. According to those authors, the only clinically relevant information provided by GEP pertains the NF-κB gene expression level (see below).14 Ballester et al15 reported that GEP could discriminate among PTCL of the NOS, AITL and ALCL types, although NOS did not share a single profile. Using a multiclass predictor, the authors separated their cases into three molecular subgroups: U1, U2 and U3. However, the corresponding signatures might have been, at least in part, influenced by reactive components, as suggested by the fact that, for instance, the U3 subgroup consisted almost entirely of histiocyte-rich tumours.
Table 1. The main studies dealing with gene expression profiling of peripheral T cell lymphomas.
| Reference | Disease(s) explored | Comments |
| Tracey et al60 | FM | The GEP of FM was investigated, and it showed concurrent deregulation of multiple genes involved in the tumour necrosis factor signalling pathway. |
| Martinez-Delgado et al14 | PTCL/NOS | The authors found significant differences between the peripheral and lymphoblastic T cell lymphomas. The differences included a deregulation of the nuclear factor-κB signalling pathway. |
| Martinez-Delgado et al98 | PTCL/NOS | The authors found two different subgroups of PTCL based on the expression of NF-κB related genes. One-third of PTCL clearly showed reduced expression of NF-κB genes, while the other group was characterised by high expression of these genes. Of interest, the expression profile associated with reduced expression of NF-κB genes was significantly associated with shorter survival of patients. |
| Ballester et al15 | PTCL/NOS, AILT, ALCL | According to this study, PTCL/NOS could be divided into three molecular subgroups: U1, U2 and U3. The U1 gene expression signature included genes known to be associated with poor outcome in other tumours, such as CCND2. The U2 subgroup was associated with overexpression of genes involved in T cell activation and apoptosis, including NF-κB1 and BCL-2. The U3 subgroup was mainly defined by overexpression of genes involved in the IFN/JAK/STAT pathway. Notably, such distinction possibly reflected, at least in part, the presence of reactive components in the PTCL samples. |
| de Leval et al17 | AILT | The molecular profile of AILT was characterised by a strong microenvironment and overexpression of several genes characteristic of normal follicular helper T (TFH) cells: CXCL13, BCL6, PDCD1, CD40L and NFATC1. Such a finding was reinforced by gene set enrichment analysis, which demonstrated that the AITL molecular signature was significantly enriched in TFH-specific genes. |
| Piccaluga et al20 | PTCL/NOS | The authors showed that PTCL/NOS are most closely related to activated peripheral T lymphocytes, either CD4+ or CD8+, based on the GEP. In addition, PTCL/NOS displayed deregulation of relevant functional cell programmes. In particular, among others, PDGFRA, a gene encoding for a tyrosine kinase receptor, turned out to be aberrantly expressed by PTCL/NOS. Notably, phosphorylation of PDGFRA and sensitivity of cultured PTCL cells to imatinib were demonstrated. |
| Piccaluga et al21 | PTCL/NOS | The authors found that CD52 is expressed in approximately 40% of PTCL/NOS at the same level as in normal T lymphocytes, being aberrantly downregulated in the remaining cases. Notably, they concluded that the estimation of CD52 expression may provide a rationale for the selection of patients with a higher probability of response to the anti-CD52 antibody alemtuzumab. |
| Piccaluga et al22 | AILT | In this manuscript, the authors reported that AILT and other PTCL have rather similar GEP, possibly sharing common oncogenic pathways. In addition, they found that the molecular signature of follicular T helper cells was significantly overexpressed in AILT. Finally, several genes, such as PDGFRA and VEGF, which are deregulated in AILT and represent potential therapeutic targets, were identified. |
| Lamant et al16 | ALCL | This was the first study to focus on ALCL. Unsupervised analysis classified ALCL in two clusters, corresponding essentially to morphological subgroups and clinical variables. Supervised analysis showed that ALK-positive ALCL and ALK-negative ALCL have different GEP, further confirming that they are different entities. |
| Cuadros et al18 | PTCL/NOS | Five clusters of genes were identified, and their expression varied significantly among the samples. Genes in these clusters were functionally related to different cellular processes such as proliferation, inflammatory response, and T cell or B cell lineages. Notably, overexpression of genes in the proliferation signature was significantly associated with shorter survival of patients. |
AILT, peripheral T cell lymphoma, angioimmunoblastic type; ALCL, anaplastic large cell lymphoma; ALK, anaplastic large cell lymphoma kinase; FM, mycosis fungoides; GEP, gene expression profile; PDGFRA, platelet-derived growth factor receptor α; PTCL/NOS, peripheral T cell lymphoma, not otherwise specified.
Recently, we20 published a GEP study based on the analysis of 28 PTCL/NOS, all corresponding to lymph node biopsy samples containing an amount of neoplastic cells exceeding 70% value of the whole examined population. The mRNA extracted from these cases was hybridised on the HG U133 2.0 Plus gene chip. The results obtained were compared with those of six AITL, six ALCL (two ALK-positive and four ALK-negative) and 20 samples of normal T lymphocytes, which were purified from the peripheral blood and tonsil and corresponded to the main T cell subsets (CD4+, CD8+, resting and activated). Such a study significantly differed from most previous reports14 60 in terms of methodology and selection criteria. In addition, for the first time it provided the rationale for possible targeted therapies in PTCL/NOS by offering clear evidence of their ex vivo effectiveness.
In particular, the GEP we detected20 indicated that PTCL/NOS are distinct from normal T and B lymphocytes and are more closely related to activated rather than resting T cells. As in normal mature T lymphocytes, it was possible to identify two main subgroups of PTCL/NOS, with GEPs related to either CD4 or CD8 elements. Notably, this characteristic did not reflect the expression of CD4 and CD8 molecules.
In addition to histogenetic information, our analysis20 provided several insights into the functional alterations of PTCL/NOS. A careful comparison of PTCL/NOS with the closest normal counterparts revealed the systematic deregulation of 155 genes controlling functions that are typically damaged in malignant cells, such as matrix remodelling, cell adhesion, transcription, proliferation and apoptosis. In particular, our findings might explain the dissemination pattern of PTCL/NOS, with frequent extranodal and bone-marrow involvement and spread to peripheral blood,1 by showing the upregulation of FN1, LAMB1, COL1A2, COL3A1, COL4A1, COL4A2, and COL12A1 (ie, genes that promote local invasion and metastasis in different types of human cancer).61–63 In addition, it revealed the deregulation of genes involved in apoptosis (eg, MOAP1, ING3, GADD45A and GADD45B)64–70 and chemoresistance (such as CYR61 and NNMT).61–63,71–82
Immunohistochemistry provided in situ validation of the genomic data by showing correspondence between mRNA and protein expression, as seen, for example, with GEP, PDGFRα (see fig 1C and D) and BCL10. In addition, by comparison with normal tissues, immunohistochemistry allowed the identification of staining patterns corresponding to the synthesis of ectopic or paraphysiological products by neoplastic cells. Finally, the phenotypic test highlighted the possibility that some of the results obtained by GEP may depend on non-neoplastic components present in the analysed sample, as seen for Caldesmon.
In the course of the same study, we found that all ALCLs tended to cluster together – irrespective of their ALK positivity or negativity – showing a signature distinct from those of PTCL/NOS and AITL.20
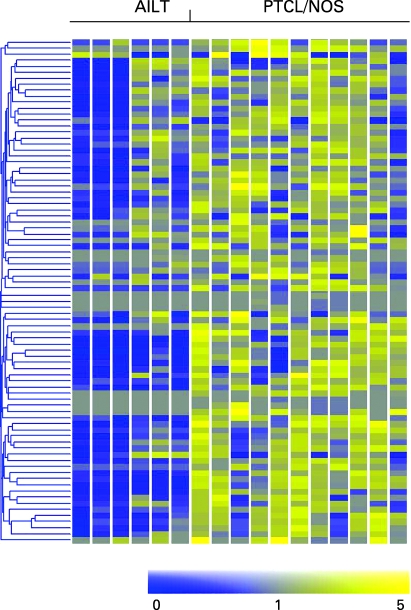
More recently, we succeeded in identifying a gene signature discriminating between PTCL/NOS and AITL (fig 2).22 In addition, the observed AITL global profile strengthened its derivation from the follicular T helper lymphocyte (FTHL), as originally proposed by Rüdiger et al83 and de Leval et al.17 Among upregulated genes, were those encoding for CXC13, PD1 and vascular endothelial growth factor (VEGF).
Figure 2. Peripheral T cell lymphoma, not otherwise specified (PTCL/NOS), and peripheral T cell lymphoma, angioimmunoblastic type (AILT), can be distinguished according to their gene expression profile. Eighty-three differentially expressed genes are plotted in the matrix.
PRACTICAL IMPLICATIONS OF PHENOTYPIC AND MOLECULAR FINDINGS
Diagnosis
Along with clonality studies,11 the phenotype plays a basic role in the distinction of PTCL from reactive conditions—such as paracortex hyperplasia—that can mimic malignant lymphoma. In fact, the lack of one or more T cell associated antigens (see above) is a hallmark of neoplastic cells as opposed to the complete phenotype of normal T lymphocytes.27 Immunohistochemical and molecular findings are also of great value for differential diagnosis among PTCL.
PTCL/NOS versus AITL
Such distinction may be problematic in about 25% of cases, based on conventional criteria.84 Also CD10 staining, proposed as characteristic of AITL,85 86 is actually seen in less than 50% of cases in our experience.27
Notably, the AITL gene signature recently reported by de Leval et al17 and our group22 (see above) provides a rationale to the immunohistochemical observations of Dupuis et al,87 Grogg et al88 and Roncador et al89 who found that most, if not all, AITL stain for typical FTHL-related antigens, such as CXCL13 (see fig 1E) and PD-1. Such molecules can actually represent a powerful tool for the distinction of AITL from PTCL/NOS, due to the exceptional positivity of the latter, a finding also confirmed in our PTCL tissue microarray (unpublished observation).
PTCL/NOS versus ALCL
Lamant et al16 reported that ALK-positive and ALK-negative ALCL have different GEPs. In particular, they found that BCL-6, PTPN12, C/EBPβ and serpinA1 genes overexpressed in ALK-positive ALCL, a result also confirmed at the protein level. In contrast, the molecular signature of ALK-negative ALCL included overexpression of CCR7, CNTFR, IL22 and IL21 genes, but did not provide any obvious clues to its molecular pathogenesis. This led to the question of whether ALK-negative ALCL should be included in PTCL/NOS. In the course of our GEP study, we found that all ALCL tended to cluster together irrespective of their ALK status, and this signature was clearly distinct from that of PTCL/NOS.20 In addition to suggesting that ALK-positive and ALK-negative ALCL probably share a set of deregulated pathways, our findings did not support the proposal that ALK-negative ALCL is a subtype of PTCL/NOS. Such a viewpoint is strengthened by the results of a recent clinicopathological trial showing that ALK-negative ALCL—although more aggressive than ALK-positive ALCL—has 5-year failure-free and overall survival rates that are significantly better than PTCL/NOS.84
Prognosis
EBV, CD15 and proliferation
In our series of Italian patients, we found that high Ki-67 expression (see fig 1F), EBV status and CD15 staining were associated with the worst outcome in PTCL/NOS.27 Interestingly, a proliferation signature has recently been reported to correlate with an aggressive clinical course,18 and EBV has repeatedly been proposed as a negative prognosticator in PTCL.58 90 91 No other phenotypic marker alone or in combination was associated with a poor outcome, although patients with tumours expressing a CD57 or CD4+/CD8− profile showed a tendency towards a more favourable outcome, as also observed by others.25 48
Clinicopathological score
Based on our collective results and those published in the literature,26 58 92–96 we developed a new score that integrates patient- and tumour-specific characteristics (age ⩾60 years, performance status, lactate dehydrogenase, and Ki-67 marking >80%) and identifies three clear-cut groups of patients with different prognosis. Such a score seems to be more effective than previous indices, including international prognostic index and prognostic index for peripheral T cell lymphoma, not otherwise specified.26
CYP3A
Recently, Rodríguez-Antona et al97 measured tumour CYP3A mRNA content in 44 T cell lymphomas and found a large variation in its expression that might be due to gains affecting the corresponding gene. To test whether CYP3A could influence PTCL treatment outcome, its expression levels were compared with the patient clinical response and survival, and it was observed that a high CYP3A4 expression was significantly associated with a lower complete remission rate. These results indicate that CYP3A as a potential predictor of tumour chemosensitivity.
NF-κB pathway
Different GEP studies have suggested that PTCL/NOS may show up- or downregulation of NF-κB molecules,14 15 98 with possible prognostic implications (see above).14 98 However, these studies included a limited number of PTCL/NOS14 or cases with prominent non-neoplastic components.15 By contrast, we found that PTCL/NOS mostly consisting of neoplastic cells present with global downregulation of NF-κB genes in comparison with normal T lymphocytes. This observation was corroborated by consistent cytoplasmic localisation of NF-κB molecules, the latter moving to the nucleus in the case of NF-κB pathway activation (unpublished observation).
Therapy
CD4 and CD52 expression
The in vivo administration of monoclonal antibodies targeted to CD4 and CD52 has recently been proposed for the treatment of patients with PTCL.99 However, in our experience this should be regarded with caution when referring to PTCL/NOS. The latter, in fact, characteristically lacks the expression of one or more T cell associated antigens, including those antigens that these antibodies are targeted towards. In particular, we found that CD4 is lacking at the neoplastic cell level in up to 50% of cases.27 CD52 is a molecule expressed by most peripheral blood lymphocytes, macrophages, and monocytes.102 Campath-1H (alemtuzumab) is a humanised antibody against CD52 currently approved for B cell chronic lymphocytic leukaemia therapy,103–106 and it has also shown interesting activity in T prolymphocytic leukaemia and cutaneous TCLs.107 Although other factors can affect its response in vivo, the lack of CD52 expression may play a major role in causing refractoriness to the compound. Few data are available regarding the use alemtuzumab in PTCL/NOS.108 109 We studied the expression of CD52 on tissue microarrays containing 97 PTCL/NOS.21 In addition, in 28 cases for which frozen material was available, GEP were generated and compared with those of 20 samples of normal T lymphocytes.21 We found that 17/28 (60%) PTCL/NOS showed CD52 gene expression level lower than the lowest one recorded in normal T cells.21 In addition, the gene product was detected by immunohistochemistry in 40/97 (41%) PTCL (see fig 1G).21 Interestingly, such data are in keeping with the clinical results obtained by Enblad et al108 who found an overall response rate of 36% in PTCL treated with alemtuzumab. Based on these findings, we think that the estimation of CD52 expression may provide a rationale for the selection of patients with higher probability of responding to alemtuzumab, by avoiding the risk of unwanted toxicity.21 Similar conclusions were achieved by Rodig et al100 and Chang et al,101 who reported immunohistochemical detection of CD52 in 0–40% of PTCL.
PDGFRα
The regular detection of PDGFRα overexpression at the mRNA and protein levels, as well as its frequent phosphorylation (see fig 1D), prompted us20 to design an ex vivo experiment aimed testing the sensitivity of PTCL/NOS cells to imatinib, a well-known PDGFRα inhibitor.110 The results obtained were of interest, with about 50% cytotoxic effect seen at 48 h with a 1 μmol concentration. Such an effect became even higher (75%) with a 10 μmol dose. Notably, imatinib exerted a limited effect on the viability of normal lymphocytes.
Take-home messages
Peripheral T cell lymphomas (PTCL) represent about 12% of all lymphoid tumours worldwide. Around half belong to the not otherwise specified (NOS) type.
Conventional morphological and molecular criteria do not assist in the subclassification of PTCL/NOS, as anthracycline-based therapies fail to cure it, and most patients die of their disease within 5 years.
Novel microarray technologies allow the identification of peculiar features that may in turn be useful for the diagnosis, prognosis and treatment of PTCL/NOS.
PTCL/NOS is characterised by frequent defective expression of T associated antigens, including CD4 and CD52, which have recently been proposed as targets for humanised monoclonal antibodies.
The growth fraction >80% has a prognostic impact.
Gene expression profiling studies show derivation from activated peripheral T lymphocytes and systematic deregulation of 155 genes and related products that may provide the rationale for the unprecedented usage of drugs such as tyrosine kinase and histone deacetylase inhibitors.
The gene expression profile also contributes to the better definition of the boundaries between PTCL/NOS and angioimmunoblastic T cell lymphoma, the latter deriving from follicular T helper lymphocytes and characteristically expressing CXCL13 and PD1 along with vascular endothelial growth factor.
Histone deacetylation
Since silencing of certain genes (such as GADD45A and GADD45B) can be regulated by epigenetic mechanisms including acetylation, we tested a histone deacetylase inhibitor (HDACi) (ITF2357) against PTCL/NOS primary cells. Notably, the compound induced dramatic G0–G1 cell cycle arrest and apoptosis at therapeutic concentrations, suggesting a possible role for this class of drugs in PTCL/NOS therapy, as also supported by preliminary clinical observations.111 Interestingly, the combination of ITF2357 and daunorubicin apparently had a slight additive effect, as already observed with other HDACi.112
VEGF
Recently, we observed upregulation of the VEGF gene in AITL.22 The same finding had previously been reported by de Leval et al17 who had attributed it to the rich vascular component of the tumour. However, by immunohistochemistry on tissue microarrays, we showed that neoplastic cells strongly express both VEGF (see fig 1H) and its receptor KDR.22 This fact suggests possible AITL sensitivity to anti-angiogenetic drugs, such as thalidomide and bevacizumab.113
CONCLUSIONS
For a long time, PTCL have represented an orphan pathology. This can be explained by their relatively low prevalence (which is in any case higher than that of a “common” tumour, such as CHL), diagnostic difficulties and dismal prognosis. Based on recent advances in the genomic and translational fields, a new scenario can now be envisaged leading the way to more successful therapeutic strategies. This may be the right time to live a dream, never forgetting however that “the truth is not always pure and never simple” (Oscar Wilde).
Footnotes
Funding: Associazione Italiana per la Ricerca sul Cancro (AIRC, Milan, Italy), Ministero dell’Università e della Ricerca Scientifica e Tecnologica (PRIN/COFIN and FIRB, Rome, Italy), BolognAIL (Bologna, Italy) and Fondazione Cassa di Risparmio in Bologna (Bologna, Italy).
Competing interests: None.
REFERENCES
- 1.Jaffe E S, Harris N L, Stein H, Vardiman J W, eds. Pathology and genetics: tumours of haematopoietic and lymphoid tissues (World Health Organization Classification of Tumours). Lyon: IARC Press, 2001 [Google Scholar]
- 2.Zucca E, Zinzani PL. Understanding the group of peripheral T-cell lymphomas, unspecified. Curr Hematol Rep 2005;4:23–30 [PubMed] [Google Scholar]
- 3.Harris NL, Jaffe ES, Stein H, et al. A revised European–American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994;84:1361–92 [PubMed] [Google Scholar]
- 4.Evens AM, Gartenhaus RB. Treatment of T-cell non-Hodgkin’s lymphoma. Curr Treat Options Oncol 2004;5:289–303 [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Guillermo A, Cid J, Salar A, et al. Peripheral T-cell lymphomas: initial features, natural history, and prognostic factors in a series of 174 patients diagnosed according to the REAL Classification. Ann Oncol 1998;9:849–55 [DOI] [PubMed] [Google Scholar]
- 6.de Leval L, Savilo E, Longtine J, et al. Peripheral T-cell lymphoma with follicular involvement and a CD4+/bcl-6+ phenotype. Am J Surg Pathol 2001;25:395–400 [DOI] [PubMed] [Google Scholar]
- 7.Rudiger T, Ichinohasama R, Ott MM, et al. Peripheral T-cell lymphoma with distinct perifollicular growth pattern: a distinct subtype of T-cell lymphoma? Am J Surg Pathol 2000;24:117–22 [DOI] [PubMed] [Google Scholar]
- 8.Geissinger E, Odenwald T, Seung-Souk L, et al. Nodal peripheral T-cell lymphomas and, in particular, their lymphoepithelioid (Lennert’s) variant are often derived from CD8+ cytotoxic cells. Virchows Arch 2004;445:334–43 [DOI] [PubMed] [Google Scholar]
- 9.Gisselbrecht C, Gaulard P, Lepage E, et al. Prognostic significance of T-cell phenotype in aggressive non-Hodgkin’s lymphomas. Groupe d’Etudes des Lymphomes de l’Adulte (GELA). Blood 1998;92:76–82 [PubMed] [Google Scholar]
- 10.The Non-Hodgkin’s Lymphoma Classification Project Effect of age on the characteristics and clinical behavior of non-Hodgkin’s lymphoma patients. Ann Oncol 1997;8:973–8 [PubMed] [Google Scholar]
- 11.Rudiger T, Weisenburger DD, Anderson JR, et al. Peripheral T-cell lymphoma (excluding anaplastic large-cell lymphoma): results from the Non-Hodgkin’s Lymphoma Classification Project. Ann Oncol 2002;13:140–9 [DOI] [PubMed] [Google Scholar]
- 12.Zettl A, Rudiger T, Konrad MA, et al. Genomic profiling of peripheral T-cell lymphoma, unspecified, and anaplastic large T-cell lymphoma delineates novel recurrent chromosomal alterations. Am J Pathol 2004;164:1837–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oshiro A, Tagawa H, Ohshima K, et al. Identification of subtype-specific genomic alterations in aggressive adult T-cell leukemia/lymphoma. Blood 2006;107:4500–7 [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Delgado B, Melendez B, Cuadros M, et al. Expression profiling of T-cell lymphomas differentiates peripheral and lymphoblastic lymphomas and defines survival related genes. Clin Cancer Res 2004;10:4971–82 [DOI] [PubMed] [Google Scholar]
- 15.Ballester B, Ramuz O, Gisselbrecht C, et al. Gene expression profiling identifies molecular subgroups among nodal peripheral T-cell lymphomas. Oncogene 2006;25:1560–70 [DOI] [PubMed] [Google Scholar]
- 16.Lamant L, De Reynies A, Duplantier MM, et al. Gene expression profiling of systemic anaplastic large cell lymphoma reveals differences depending on ALK status and two distinct morphological ALK+ subtypes. Blood 2007;109:2156–64 [DOI] [PubMed] [Google Scholar]
- 17.de Leval L, Rickman DS, Thielen C, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood 2007;109:4952–63 [DOI] [PubMed] [Google Scholar]
- 18.Cuadros M, Dave SS, Jaffe ES, et al. Identification of a proliferation signature related to survival in nodal peripheral T-cell lymphomas. J Clin Oncol 2007;25:3321–9 [DOI] [PubMed] [Google Scholar]
- 19.Piccaluga PP, Agostinelli C, Zinzani PL, et al. Expression of platelet-derived growth factor receptor α in peripheral T-cell lymphoma not otherwise specified. Lancet Oncol 2005;6:440. [DOI] [PubMed] [Google Scholar]
- 20.Piccaluga PP, Agostinelli C, Califano A, et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest 2007;117:823–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piccaluga PP, Agostinelli C, Righi S, et al. Expression of CD52 in peripheral T-cell lymphoma. Haematologica 2007;92:566–7 [DOI] [PubMed] [Google Scholar]
- 22.Piccaluga PP, Agostinelli C, Califano A, et al. Gene expression analysis of angioimmunoblastic lymphoma indicates derivation from T follicular helper cells and vascular endothelial growth factor deregulation. Cancer Res 2007;67:10703–10 [DOI] [PubMed] [Google Scholar]
- 23.Jantunen E, Wiklund T, Juvonen E, et al. Autologous stem cell transplantation in adult patients with peripheral T-cell lymphoma: a nation-wide survey. Bone Marrow Transplant 2004;33:405–10 [DOI] [PubMed] [Google Scholar]
- 24.Kahl C, Leithauser M, Wolff D, et al. Treatment of peripheral T-cell lymphomas (PTCL) with high-dose chemotherapy and autologous or allogeneic hematopoietic transplantation. Ann Hematol 2002;81:646–50 [DOI] [PubMed] [Google Scholar]
- 25.Kojima H, Hasegawa Y, Suzukawa K, et al. Clinicopathological features and prognostic factors of Japanese patients with “peripheral T-cell lymphoma, unspecified” diagnosed according to the WHO classification. Leuk Res 2004;28:1287–92 [DOI] [PubMed] [Google Scholar]
- 26.Gallamini A, Stelitano C, Calvi R, et al. Peripheral T-cell lymphoma unspecified (PTCL-U): a new prognostic model from a retrospective multicentric clinical study. Blood 2004;103:2474–9 [DOI] [PubMed] [Google Scholar]
- 27.Went P, Agostinelli C, Gallamini A, et al. Marker expression in peripheral T-cell lymphoma: a proposed clinical-pathologic prognostic score. J Clin Oncol 2006;24:2472–9 [DOI] [PubMed] [Google Scholar]
- 28.Au WY, Ma SY, Chim CS, et al. Clinicopathologic features and treatment outcome of mature T-cell and natural killer-cell lymphomas diagnosed according to the World Health Organization classification scheme: a single center experience of 10 years. Ann Oncol 2005;16:206–14 [DOI] [PubMed] [Google Scholar]
- 29.Ascani S, Zinzani PL, Gherlinzoni F, et al. Peripheral T-cell lymphomas. Clinico-pathologic study of 168 cases diagnosed according to the REAL Classification. Ann Oncol 1997;8:583–92 [DOI] [PubMed] [Google Scholar]
- 30.Boulland ML, Kanavaros P, Wechsler J, et al. Cytotoxic protein expression in natural killer cell lymphomas and in αβ and γδ peripheral T cell lymphomas. J Pathol 1997;183:432–9 [DOI] [PubMed] [Google Scholar]
- 31.Chott A, Augustin I, Wrba F, et al. Peripheral T-cell lymphomas: a clinicopathologic study of 75 cases. Hum Pathol 1990;21:1117–25 [DOI] [PubMed] [Google Scholar]
- 32.Doi S, Nasu K, Arita Y, et al. Immunohistochemical analysis of peripheral T-cell lymphoma in Japanese patients. Am J Clin Pathol 1989;91:152–8 [DOI] [PubMed] [Google Scholar]
- 33.Feller AC, Griesser H, Schilling CV, et al. Clonal gene rearrangement patterns correlate with immunophenotype and clinical parameters in patients with angioimmunoblastic lymphadenopathy. Am J Pathol 1988;133:549–56 [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneko Y, Maseki N, Sakurai M, et al. Characteristic karyotypic pattern in T-cell lymphoproliferative disorders with reactive “angioimmunoblastic lymphadenopathy with dysproteinemia-type” features. Blood 1988;72:413–21 [PubMed] [Google Scholar]
- 35.Knecht H, Odermatt BF, Maurer R, et al. Diagnostic and prognostic value of monoclonal antibodies in immunophenotyping of angioimmunoblastic lymphadenopathy/lymphogranulomatosis X. Br J Haematol 1987;67:19–24 [DOI] [PubMed] [Google Scholar]
- 36.Nakamura S, Suchi T. A clinicopathologic study of node-based, low-grade, peripheral T-cell lymphoma. Angioimmunoblastic lymphoma, T-zone lymphoma, and lymphoepithelioid lymphoma. Cancer 1991;67:2566–78 [DOI] [PubMed] [Google Scholar]
- 37.Namikawa R, Suchi T, Ueda R, et al. Phenotyping of proliferating lymphocytes in angioimmunoblastic lymphadenopathy and related lesions by the double immunoenzymatic staining technique. Am J Pathol 1987;127:279–87 [PMC free article] [PubMed] [Google Scholar]
- 38.Ohsaka A, Saito K, Sakai T, et al. Clinicopathologic and therapeutic aspects of angioimmunoblastic lymphadenopathy-related lesions. Cancer 1992;69:1259–67 [DOI] [PubMed] [Google Scholar]
- 39.Ohshima K, Kikuchi M, Hashimoto M, et al. Genetic changes in atypical hyperplasia and lymphoma with angioimmunoblastic lymphadenopathy and dysproteinaemia in the same patients. Virchows Arch 1994;425:25–32 [DOI] [PubMed] [Google Scholar]
- 40.Ree HJ, Kadin ME, Kikuchi M, et al. Angioimmunoblastic lymphoma (AILD-type T-cell lymphoma) with hyperplastic germinal centers. Am J Surg Pathol 1998;22:643–55 [DOI] [PubMed] [Google Scholar]
- 41.Richel DJ, Lepoutre JM, Kapsenberg JG, et al. Epstein–Barr virus in a CD8-positive T cell lymphoma. Am J Pathol 1990;136:1093–9 [PMC free article] [PubMed] [Google Scholar]
- 42.Takagi N, Nakamura S, Ueda R, et al. A phenotypic and genotypic study of three node-based, low-grade peripheral T-cell lymphomas: angioimmunoblastic lymphoma, T-zone lymphoma, and lymphoepithelioid lymphoma. Cancer 1992;69:2571–82 [DOI] [PubMed] [Google Scholar]
- 43.Tobinai K, Minato K, Ohtsu T, et al. Clinicopathologic, immunophenotypic, and immunogenotypic analyses of immunoblastic lymphadenopathy-like T-cell lymphoma. Blood 1988;72:1000–6 [PubMed] [Google Scholar]
- 44.Weiss LM, Strickler JG, Dorfman DM, et al. Clonal T cell populations in angioimmunoblastic lymphadenopathy and angioimmunoblastic lymphadenopathy-like lymphoma. Am J Pathol 1986;122:392–7 [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SS, Rudiger T, Odenwald T, et al. Angioimmunoblastic T cell lymphoma is derived from mature T-helper cells with varying expression and loss of detectable CD4. Int J Cancer 2003;103:12–20 [DOI] [PubMed] [Google Scholar]
- 46.Barth TF, Leithauser F, Dohner H, et al. Primary gastric apoptosis-rich T-cell lymphoma co-expressing CD4, CD8, and cytotoxic molecules. Virchows Arch 2000;436:357–64 [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto Y, Kitajima H, Sakihana H, et al. CD3+CD4−CD8−TCR−alphabeta+ T-cell lymphoma with clinical features of primary effusion lymphoma: an autopsy case. Int J Hematol 2001;74:442–6 [DOI] [PubMed] [Google Scholar]
- 48.Bekkenk MW, Vermeer MH, Jansen PM, et al. Peripheral T-cell lymphomas unspecified presenting in the skin: analysis of prognostic factors in a group of 82 patients. Blood 2003;102:2213–9 [DOI] [PubMed] [Google Scholar]
- 49.Lanier LL, Le AM, Civin CI, et al. The relationship of CD16 (Leu-11) and Leu-19 (NKH-1) antigen expression on human peripheral blood NK cells and cytotoxic T lymphocytes. J Immunol 1986;136:4480–6 [PubMed] [Google Scholar]
- 50.Knowles DM. Immunohistochemical markers useful in the diagnosis and classification of hematopoietic neoplasms. Knowles D M, ed. Neoplastic hematopathology. 2nd edn. Philadelphia: Lippincott Williams & Wilkins, 2001:93–226 [Google Scholar]
- 51.Blakolmer K, Vesely M, Kummer JA, et al. Immunoreactivity of B-cell markers (CD79a, L26) in rare cases of extranodal cytotoxic peripheral T- (NK/T-)cell lymphomas. Mod Pathol 2000;13:766–72 [DOI] [PubMed] [Google Scholar]
- 52.Quintanilla-Martinez L, Preffer F, Rubin D, et al. CD20+ T-cell lymphoma. Neoplastic transformation of a normal T-cell subset. Am J Clin Pathol 1994;102:483–9 [DOI] [PubMed] [Google Scholar]
- 53.Yao X, Teruya-Feldstein J, Raffeld M, et al. Peripheral T-cell lymphoma with aberrant expression of CD79a and CD20: a diagnostic pitfall. Mod Pathol 2001;14:105–10 [DOI] [PubMed] [Google Scholar]
- 54.Barry TS, Jaffe ES, Sorbara L, et al. Peripheral T-cell lymphomas expressing CD30 and CD15. Am J Surg Pathol 2003;27:1513–22 [DOI] [PubMed] [Google Scholar]
- 55.Gorczyca W, Tsang P, Liu Z, et al. CD30-positive T-cell lymphomas co-expressing CD15: an immunohistochemical analysis. Int J Oncol 2003;22:319–24 [PubMed] [Google Scholar]
- 56.Browne P, Petrosyan K, Hernandez A, et al. The B-cell transcription factors BSAP, Oct-2, and BOB.1 and the pan-B-cell markers CD20, CD22, and CD79a are useful in the differential diagnosis of classic Hodgkin lymphoma. Am J Clin Pathol 2003;120:767–77 [DOI] [PubMed] [Google Scholar]
- 57.Tzankov AS, Went PT, Munst S, et al. Rare expression of BSAP (PAX-5) in mature T-cell lymphomas. Mod Pathol 2007;20:632–7 [DOI] [PubMed] [Google Scholar]
- 58.Dupuis J, Emile JF, Mounier N, et al. Prognostic significance of Epstein–Barr virus in nodal peripheral T-cell lymphoma, unspecified: A Groupe d’Etude des Lymphomes de l’Adulte (GELA) study. Blood 2006;108:4163–9 [DOI] [PubMed] [Google Scholar]
- 59.Mahadevan D, Spier C, Della Croce K, et al. Transcript profiling in peripheral T-cell lymphoma, not otherwise specified, and diffuse large B-cell lymphoma identifies distinct tumor profile signatures. Mol Cancer Ther 2005;4:1867–79 [DOI] [PubMed] [Google Scholar]
- 60.Tracey L, Villuendas R, Dotor AM, et al. Mycosis fungoides shows concurrent deregulation of multiple genes involved in the TNF signaling pathway: an expression profile study. Blood 2003;102:1042–50 [DOI] [PubMed] [Google Scholar]
- 61.Tapper J, Kettunen E, El-Rifai W, et al. Changes in gene expression during progression of ovarian carcinoma. Cancer Genet Cytogenet 2001;128:1–6 [DOI] [PubMed] [Google Scholar]
- 62.Sado Y, Kagawa M, Naito I, et al. Organization and expression of basement membrane collagen IV genes and their roles in human disorders. J Biochem (Tokyo) 1998;123:767–76 [DOI] [PubMed] [Google Scholar]
- 63.van den Boom J, Wolter M, Kuick R, et al. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol 2003;163:1033–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin S, Tong T, Fan W, et al. GADD45-induced cell cycle G2-M arrest associates with altered subcellular distribution of cyclin B1 and is independent of p38 kinase activity. Oncogene 2002;21:8696–704 [DOI] [PubMed] [Google Scholar]
- 65.Papa S, Zazzeroni F, Bubici C, et al. Gadd45 beta mediates the NF-κ B suppression of JNK signalling by targeting MKK7/JNKK2. Nat Cell Biol 2004;6:146–53 [DOI] [PubMed] [Google Scholar]
- 66.Chen F, Lu Y, Zhang Z, et al. Opposite effect of NF-κB and c-Jun N-terminal kinase on p53-independent GADD45 induction by arsenite. J Biol Chem 2001;276:11414–9 [DOI] [PubMed] [Google Scholar]
- 67.Hirose T, Sowa Y, Takahashi S, et al. p53-independent induction of Gadd45 by histone deacetylase inhibitor: coordinate regulation by transcription factors Oct-1 and NF-Y. Oncogene 2003;22:7762–73 [DOI] [PubMed] [Google Scholar]
- 68.Tan KO, Tan KM, Chan SL, et al. MAP-1, a novel proapoptotic protein containing a BH3-like motif that associates with Bax through its Bcl-2 homology domains. J Biol Chem 2001;276:2802–7 [DOI] [PubMed] [Google Scholar]
- 69.Nagashima M, Shiseki M, Pedeux RM, et al. A novel PHD-finger motif protein, p47ING3, modulates p53-mediated transcription, cell cycle control, and apoptosis. Oncogene 2003;22:343–50 [DOI] [PubMed] [Google Scholar]
- 70.Gunduz M, Ouchida M, Fukushima K, et al. Allelic loss and reduced expression of the ING3, a candidate tumor suppressor gene at 7q31, in human head and neck cancers. Oncogene 2002;21:4462–70 [DOI] [PubMed] [Google Scholar]
- 71.Lee MS, Hanspers K, Barker CS, et al. Gene expression profiles during human CD4+ T cell differentiation. Int Immunol 2004;16:1109–24 [DOI] [PubMed] [Google Scholar]
- 72.Chtanova T, Newton R, Liu SM, et al. Identification of T cell-restricted genes, and signatures for different T cell responses, using a comprehensive collection of microarray datasets. J Immunol 2005;175:7837–47 [DOI] [PubMed] [Google Scholar]
- 73.Chtanova T, Tangye SG, Newton R, et al. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol 2004;173:68–78 [DOI] [PubMed] [Google Scholar]
- 74.Hosack DA, Dennis G, Jr, Sherman BT, et al. Identifying biological themes within lists of genes with EASE. Genome Biol 2003;4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han JS, Macarak E, Rosenbloom J, et al. Regulation of Cyr61/CCN1 gene expression through RhoA GTPase and p38MAPK signaling pathways. Eur J Biochem 2003;270:3408–21 [DOI] [PubMed] [Google Scholar]
- 76.Leu SJ, Liu Y, Chen N, et al. Identification of a novel integrin alpha 6 beta 1 binding site in the angiogenic inducer CCN1 (CYR61). J Biol Chem 2003;278:33801–8 [DOI] [PubMed] [Google Scholar]
- 77.Schober JM, Lau LF, Ugarova TP, et al. Identification of a novel integrin alphaMbeta2 binding site in CCN1 (CYR61), a matricellular protein expressed in healing wounds and atherosclerotic lesions. J Biol Chem 2003;278:25808–15 [DOI] [PubMed] [Google Scholar]
- 78.Tsai MS, Bogart DF, Castaneda JM, et al. Cyr61 promotes breast tumorigenesis and cancer progression. Oncogene 2002;21:8178–85 [DOI] [PubMed] [Google Scholar]
- 79.Tsai MS, Hornby AE, Lakins J, et al. Expression and function of CYR61, an angiogenic factor, in breast cancer cell lines and tumor biopsies. Cancer Res 2000;60:5603–7 [PubMed] [Google Scholar]
- 80.Lin MT, Chang CC, Chen ST, et al. Cyr61 expression confers resistance to apoptosis in breast cancer MCF-7 cells by a mechanism of NF-kappaB-dependent XIAP up-regulation. J Biol Chem 2004;279:24015–23 [DOI] [PubMed] [Google Scholar]
- 81.Kassem H, Sangar V, Cowan R, et al. A potential role of heat shock proteins and nicotinamide N-methyl transferase in predicting response to radiation in bladder cancer. Int J Cancer 2002;101:454–60 [DOI] [PubMed] [Google Scholar]
- 82.Xu J, Capezzone M, Xu X, et al. Activation of nicotinamide N-methyltransferase gene promoter by hepatocyte nuclear factor-1beta in human papillary thyroid cancer cells. Mol Endocrinol 2005;19:527–39 [DOI] [PubMed] [Google Scholar]
- 83.Rudiger T, Geissinger E, Muller-Hermelink HK. “Normal counterparts” of nodal peripheral T-cell lymphoma. Hematol Oncol 2006;24:175–80 [DOI] [PubMed] [Google Scholar]
- 84.Savage KJ, Harris NL, Vose MJ, et al. ALK-negative anaplastic large-cell lymphoma (ALCL) is clinically and immunophenotypically different from both ALK+ ALCL and peripheral T-cell lymphoma, not otherwise specified: Report from International Peripheral T-cell Lymphoma Project. Blood 2008;111:5496–504 [DOI] [PubMed] [Google Scholar]
- 85.Attygalle A, Al-Jehani R, Diss TC, et al. Neoplastic T cells in angioimmunoblastic T-cell lymphoma express CD10. Blood 2002;99:627–33 [DOI] [PubMed] [Google Scholar]
- 86.Attygalle AD, Diss TC, Munson P, et al. CD10 expression in extranodal dissemination of angioimmunoblastic T-cell lymphoma. Am J Surg Pathol 2004;28:54–61 [DOI] [PubMed] [Google Scholar]
- 87.Dupuis J, Boye K, Martin N, et al. Expression of CXCL13 by neoplastic cells in angioimmunoblastic T-cell lymphoma (AITL): a new diagnostic marker providing evidence that AITL derives from follicular helper T cells. Am J Surg Pathol 2006;30:490–4 [DOI] [PubMed] [Google Scholar]
- 88.Grogg KL, Attygale AD, Macon WR, et al. Expression of CXCL13, a chemokine highly upregulated in germinal center T-helper cells, distinguishes angioimmunoblastic T-cell lymphoma from peripheral T-cell lymphoma, unspecified. Mod Pathol 2006;19:1101–7 [DOI] [PubMed] [Google Scholar]
- 89.Roncador G, Garcia Verdes-Montenegro JF, Tedoldi S, et al. Expression of two markers of germinal center T cells (SAP and PD-1) in angioimmunoblastic T-cell lymphoma. Haematologica 2007;92:1059–66 [DOI] [PubMed] [Google Scholar]
- 90.Cheng AL, Su IJ, Chen YC, et al. Characteristic clinicopathologic features of Epstein–Barr virus-associated peripheral T-cell lymphoma. Cancer 1993;72:909–16 [DOI] [PubMed] [Google Scholar]
- 91.Kluin PM, Feller A, Gaulard P, et al. Peripheral T/NK-cell lymphoma: a report of the IXth Workshop of the European Association for Haematopathology. Histopathology 2001;38:250–70 [DOI] [PubMed] [Google Scholar]
- 92.Caulet-Maugendre S, Patey M, Granier E, et al. Quantitative analysis of cellular proliferative activity in 35 T-cell non-Hodgkin’s lymphomas. Use of proliferating cell nuclear antigen and Ki-67 (MIB-1) antibodies and nucleolar organizer regions. Anal Quant Cytol Histol 1996;18:337–44 [PubMed] [Google Scholar]
- 93.Miller TP, Grogan TM, Dahlberg S, et al. Prognostic significance of the Ki-67-associated proliferative antigen in aggressive non-Hodgkin’s lymphomas: a prospective Southwest Oncology Group trial. Blood 1994;83:1460–6 [PubMed] [Google Scholar]
- 94.Mochen C, Giardini R, Costa A, et al. MIB-1 and S-phase cell fraction predict survival in non-Hodgkin’s lymphomas. Cell Prolif 1997;30:37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Montalban C, Obeso G, Gallego A, et al. Peripheral T-cell lymphoma: a clinicopathological study of 41 cases and evaluation of the prognostic significance of the updated Kiel classification. Histopathology 1993;22:303–10 [DOI] [PubMed] [Google Scholar]
- 96.Sheval EV, Churakova JV, Dudnik OA, et al. Examination of the proliferative activity of tumor cells in human lymphoid neoplasms using a morphometric approach. Cancer 2004;102:174–85 [DOI] [PubMed] [Google Scholar]
- 97.Rodríguez-Antona C, Leskela S, Zajac M, et al. Expression of CYP3A4 as a predictor of response to chemotherapy in peripheral T-cell lymphomas. Blood 2007;110:3345–51 [DOI] [PubMed] [Google Scholar]
- 98.Martinez-Delgado B, Cuadros M, Honrado E, et al. Differential expression of NF-kappaB pathway genes among peripheral T-cell lymphomas. Leukemia 2005;19:2254–63 [DOI] [PubMed] [Google Scholar]
- 99.Rider DA, Havenith CE, de Ridder R, et al. A human CD4 monoclonal antibody for the treatment of T-cell lymphoma combines inhibition of T-cell signaling by a dual mechanism with potent Fc-dependent effector activity. Cancer Res 2007;67:9945–53 [DOI] [PubMed] [Google Scholar]
- 100.Rodig SJ, Abramson JS, Pinkus GS, et al. Heterogeneous CD52 expression among hematologic neoplasms: implications for the use of alemtuzumab (CAMPATH-1H). Clin Cancer Res 2006;12:7174–9 [DOI] [PubMed] [Google Scholar]
- 101.Chang ST, Lu CL, Chuang SS. CD52 expression in non-mycotic T- and NK/T-cell lymphomas. Leuk Lymphoma 2007;48:117–21 [DOI] [PubMed] [Google Scholar]
- 102.Gilleece MH, Dexter TM. Effect of Campath-1H antibody on human hematopoietic progenitors in vitro. Blood 1993;82:807–12 [PubMed] [Google Scholar]
- 103.Hale G, Dyer MJ, Clark MR, et al. Remission induction in non-Hodgkin lymphoma with reshaped human monoclonal antibody CAMPATH-1H. Lancet 1988;2:1394–9 [DOI] [PubMed] [Google Scholar]
- 104.Hale G, Waldmann H. CAMPATH-1 monoclonal antibodies in bone marrow transplantation. J Hematother 1994;3:15–31 [DOI] [PubMed] [Google Scholar]
- 105.Keating MJ, Flinn I, Jain V, et al. Therapeutic role of alemtuzumab (Campath-1H) in patients who have failed fludarabine: results of a large international study. Blood 2002;99:3554–61 [DOI] [PubMed] [Google Scholar]
- 106.Rai KR, Freter CE, Mercier RJ, et al. Alemtuzumab in previously treated chronic lymphocytic leukemia patients who also had received fludarabine. J Clin Oncol 2002;20:3891–7 [DOI] [PubMed] [Google Scholar]
- 107.Dearden C. The role of alemtuzumab in the management of T-cell malignancies. Semin Oncol 2006;332 Suppl 5:S44–52 [DOI] [PubMed] [Google Scholar]
- 108.Enblad G, Hagberg H, Erlanson M, et al. A pilot study of alemtuzumab (anti-CD52 monoclonal antibody) therapy for patients with relapsed or chemotherapy-refractory peripheral T-cell lymphomas. Blood 2004;103:2920–4 [DOI] [PubMed] [Google Scholar]
- 109.Gallamini A, Zaja F, Patti C, et al. Alemtuzumab (Campath-1H) and CHOP chemotherapy as first-line treatment of peripheral T-cell lymphoma: results of a GITIL (Gruppo Italiano Terapie Innovative nei Linfomi) prospective multicenter trial. Blood 2007;110:2316–23 [DOI] [PubMed] [Google Scholar]
- 110.Peng B, Hayes M, Resta D, et al. Pharmacokinetics and pharmacodynamics of imatinib in a phase I trial with chronic myeloid leukemia patients. J Clin Oncol 2004;22:935–42 [DOI] [PubMed] [Google Scholar]
- 111.Duvic M, Vu J. Vorinostat: a new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opin Investig Drugs 2007;16:1111–20 [DOI] [PubMed] [Google Scholar]
- 112.Sanchez-Gonzalez B, Yang H, Bueso-Ramos C, et al. Antileukemia activity of the combination of an anthracycline with a histone deacetylase inhibitor. Blood 2006;108:1174–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aguiar Bujanda D. Complete response of relapsed angioimmunoblastic T-cell lymphoma following therapy with bevacizumab. Ann Oncol 2008;19:396–7 [DOI] [PubMed] [Google Scholar]