Abstract
Apolipoprotein H (apoH, also named β-2 glycoprotein I) is found on several classes of lipoproteins, and is involved in the activation of lipoprotein lipase in lipid metabolism. We have comprehensively investigated the association of variation in the apoH gene (APOH) with lipid traits in hepatic cholesterol transport, dietary cholesterol transport (DCT), and reverse cholesterol transport (RCT). Our study population consisted of families from the Genetic Epidemiology Network of Arteriopathy multicenter study that include African Americans, Mexican Americans, and European Americans. We individually tested 36 single-nucleotide polymorphisms (SNPs) that span the APOH locus, including nonsynonymous variants that result in known apoH charge isoforms. In addition, we constructed haplotypes from SNPs in the 5′ promoter region that comprise cis-acting regulatory elements, as well as haplotypes for multiple amino acid substitutions. We found point-wise significant associations of APOH variants with various lipid measures in the three racial groups. The strongest associations were found for DCT traits (triglyceride and apoE levels) in Mexican Americans with a nonsynonymous variant (SNP 14917, Cys306Gly) that may alter apoH protein folding in a region involved in phospholipid binding. In conclusion, family-based analyses of APOH variants have identified associations with measures of lipid metabolism in three American racial groups.
Keywords: dietary cholesterol transport, reverse cholesterol transport, hepatic cholesterol transport, apolipoprotein E levels, triglyceride levels, HDL cholesterol, LDL cholesterol, functional polymorphisms, nonsynonymous substitutions, SNP haplotypes, protein folding
Apolipoprotein H (apoH, also named β-2 glycoprotein I) is synthesized mainly in the liver, but the specific functions of apoH remain unknown. ApoH was first thought to be involved mainly in lipid metabolism, because much of the circulating protein is bound to lipoproteins (1, 2), and apoH activates lipoprotein lipase (LPL) in triglyceride (TG) metabolism (3). However, apoH is also thought to be involved in coagulative and atherosclerotic pathways through the immunological response (4–6). Recent studies have shown that apoH binds to phospholipid particles, forming a major epitope for auto-antibodies involved in antiphospholipid syndrome (7–9). The gene for apoH (APOH) on chromosome 17q is 18 kb in length and contains 8 exons (10). Genetic variations in APOH result in five charge isoforms, including H*1, H*2, H*3, H*3B, and H*3W (9, 11, 12). H*2 is the most common isoform, with a relative allele frequency >85% in all studied populations, whereas frequencies for the other isoforms range only as high as 6% (9, 11, 12). A recent effort to resequence APOH in 23 European Americans and 24 African Americans discovered 150 single-nucleotide polymorphisms (SNPs) and characterized linkage disequilibrium (LD) patterns for APOH variants (13).
Previous studies have associated apoH charge isoforms and other amino acid substitutions with various lipid traits (14–17). However, population-based associations of the extensive variation in APOH characterized by Chen and Kamboh (13) have not yet been investigated in relation to lipid traits. In this study, we have comprehensively investigated this extensive APOH variation in the family-based Genetic Epidemiology Network of Arteriopathy (GENOA) study, which includes African Americans (AAs), Mexican Americans (MAs), and European Americans (EAs) (18). We genotyped GENOA subjects for 36 SNPs, including variants that underlie the known apoH charge isoforms. These APOH genotypes were used for family-based statistical analyses to identify associations with cardiovascular disease risk factors involved in hepatic cholesterol transport (HCT), dietary cholesterol transport (DCT), and reverse cholesterol transport (RCT).
MATERIALS AND METHODS
Study subjects
The study subjects participated in the GENOA study, which is composed of AAs from Jackson, MS, MAs from Starr County, TX, and EAs from Rochester MN (18). For AAs and EAs, sibships were recruited that contained at least two hypertensive subjects. For MAs, sibships were recruited based on at least two subjects with type II diabetes. GENOA provided demographic and pedigree information, medical history, anthropometric measures, informed consent, and various plasma measures for study subjects. Genomic DNA samples were available for 4,941 individuals, and pedigree information and phenotypic measurements were available for 5,242 individuals. After exclusion of subjects who were missing data, the current study included a total of 4,748 subjects (1,696 AA subjects from 583 families, 1,643 MAs from 415 families, and 1,409 EAs from 498 families). All GENOA protocols were approved by appropriate institutional review boards for the protection of human subjects, and all GENOA subjects provided written informed consent.
APOH genotyping
We selected 36 SNPs spanning APOH from the SeattleSNPs Program for Genome Application database (http://pga.gs.washington.edu/data/APOH) that were identified by resequencing of 23 EAs and 24 AAs (13). We included SNPs based on minor allele frequencies (MAFs) >5%, and potential functional properties (nonsynonymous SNPs). Although many of these SNPs were correlated, we chose to genotype multiple SNPs rather than a few tag SNPs that rely on LD relationships that differ among the racial groups. The potentially functional SNPs included nucleotide substitutions that underlie the known apoH protein isoforms (9, 11, 12). Genotyping of APOH SNPs in GENOA DNA samples used several high-throughput genotyping platforms, including Taqman assays (Applied Biosystems), the LightTyper platform (Roche Applied Science), and the SNPlex platform (Applied Biosystems). In addition, we resequenced exon 5 of APOH to obtain genotypes for several closely spaced SNPs using the Applied Biosystems 3730xl DNA analyzer. The identity and location of the 36 SNPs are provided in the Results section (Table 2), and the primer and probe sequences are available from the authors upon request.
TABLE 2.
APOH SNP positions and minor allele frequencies in African Americans, Mexican Americans, and European Americans
| MAF
|
||||||
|---|---|---|---|---|---|---|
| Position | Reference | Allele | Functional Location | AA | MA | EA |
| −1283 | rs8178818 | C/G | Promoter | 0 | 0.020 | 0.002 |
| −1218 | rs8178819 | G/A | Promoter | 0.012 | 0.025 | 0.080 |
| −1054 | rs8178897 | T/G | Promoter | 0.011 | 0 | 0 |
| −758 | rs8178820 | A/G | Promoter | 0.051 | 0.090 | 0.266 |
| −700 | rs3760291 | C/A | Promoter | 0.064 | 0.085 | 0.257 |
| −643 | hcv268405 | T/C | Promoter | 0.123 | 0.234 | 0.133 |
| −627 | rs8178898 | A/C | Promoter | 0.039 | 0.005 | 0 |
| −581 | rs8178899 | A/C | Promoter | 0.051 | 0.008 | 0.001 |
| −32 | rs8178822 | C/A | 5′ UTR | 0.058 | 0.042 | 0.069 |
| 71 | rs8178901 | C/T | Splicing | 0.047 | 0.001 | 0 |
| 3333a | rs8178833 | G/A | Ser88Asn | 0.014 | 0.027 | 0.041 |
| 5247 | rs8178835 | T/C | Intron 3 | 0.439 | 0.452 | 0.227 |
| 5956 | rs8178838 | A/G | Intron 4 | 0.081 | 0.050 | 0.072 |
| 6482 | rs3785617 | A/G | Intron 4 | 0.267 | 0.172 | 0.378 |
| 8627 | reference 14 | A/C | Splicing | 0.080 | 0.049 | 0.070 |
| 8643a | rs1803122 | T/C | Ile122Thr | 0.083 | 0.048 | 0.069 |
| 8682a | rs8178847 | G/A | Arg135His | 0.084 | 0.050 | 0.070 |
| 8698 | rs8178925 | A/G | Ser140Ser | 0.010 | 0 | 0 |
| 8700a | reference 14 | C/A | Ala141Asp | 0.019 | 0 | 0 |
| 8853 | hcv265411 | T/C | Intron 5 | 0.119 | 0.360 | 0.393 |
| 8906 | rs8178926 | G/A | Intron 5 | 0.059 | 0 | 0 |
| 9926 | rs2215415 | T/C | Intron 5 | 0.132 | 0.360 | 0.395 |
| 10099 | rs7212060 | A/C | Intron 5 | 0.347 | 0.486 | 0.703 |
| 13255 | rs4366742 | G/A | Intron 6 | 0.119 | 0.360 | 0.394 |
| 13324 | rs8178857 | C/A | Intron 6 | 0.097 | 0.075 | 0.111 |
| 13524 | rs2873966 | C/T | Intron 6 | 0.187 | 0.125 | 0.306 |
| 14740 | rs3176975 | G/T | Val247Leu | 0.470 | 0.542 | 0.773 |
| 14917 | rs4791077 | T/G | Cys306Gly | 0.007 | 0.049 | 0.026 |
| 14969 | rs1544556 | C/T | Intron 7 | 0.051 | 0.089 | 0.268 |
| 15937 | rs8178943 | T/G | Intron 7 | 0.097 | 0.001 | 0 |
| 15957 | rs4791079 | C/A | Intron 7 | 0.195 | 0.365 | 0.396 |
| 16219 | rs4790914 | C/G | Intron 7 | 0.187 | 0.364 | 0.393 |
| 17212a | rs8178862 | G/C | Thr316Ser | 0.005 | 0.041 | 0.061 |
| 18368 | rs8178818 | G/A | 3′ Flanking region | 0.110 | 0.260 | 0.143 |
| 18774 | rs8178819 | T/G | 3′ Flanking region | 0.114 | 0.263 | 0.144 |
| 18801 | rs8178897 | A/T | 3′ Flanking region | 0.278 | 0.396 | 0.419 |
SNP, single-nucleotide polymorphism; AA, African American; MA, Mexican American; EA, European American; MAF, minor allele frequency; UTR, untranslated region.
SNPs that determine known apoH protein charge isoforms.
Statistical analysis
Correlations among APOH SNPs were estimated using LDSelect (r2 > 0.8) (19). For association studies, we grouped plasma lipoprotein and apolipoprotein measures according to their involvement in HCT [total cholesterol (TC), LDL-cholesterol (LDL-C), and apoB], DCT (TG and apoE), and RCT (HDL-C and apoA-I). Plasma levels of TC, HDL-C, TGs, apoA-I, and apoE were ln-transformed to provide normalized trait distributions. All traits were adjusted for sex, age, height, weight, and waist-hip ratio. We also adjusted for diabetes and hypertension status because GENOA ascertainment resulted in enrichment of diabetes in MAs and hypertension in AAs and EAs. To adjust for covariates, we used the Generalized Estimating Equation (GEE) that accounts for pedigree structures. Heritabilities of the lipid and lipoprotein measures were estimated with SOLAR (Sequential Oligogenic Linkage Analysis Routines) (20, 21). F-tests were used to test for differences in variances of quantitative measures among the three GENOA racial groups. Association analysis used the Family Based Association Test (FBAT) algorithm with an additive model for genotypic effects (22). Haplotypes were statistically estimated using the Expecation Maximization algorithm as part of FBAT, and association tests were performed by haplotype-based association tests. In some cases, GEE was used to confirm the FBAT results. Corrections for multiple testing were based on false discovery rates (FDRs) (23). FDR-adjusted P values were calculated separately for each trait and racial group. Briefly, FBAT P values were ranked, and Q values for each SNP were calculated as qi = kpi/i, where i denotes rank and k gives the total number of tests (k = 34 for AAs, 27 for MAs, and 28 for EAs). The FDR-adjusted P value for SNPi is the minimum q value between qi and qk. We performed conditional linkage analysis using two approaches, including incorporation of associated SNPs as covariates for linkage analysis, and by adjusting apolipoprotein measures for SNP genotypes by regression (GEE) for linkage analysis with residual values.
RESULTS
Table 1 shows the number of GENOA subjects and families, diabetes and hypertension status, plasma levels of lipoproteins and apolipoproteins, and anthropometric measures by racial group. Higher numbers of diabetics were found in MAs owing to ascertainment by diabetes status. Similarly, higher numbers of hypertensives were found in AAs and EAs owing to ascertainment by hypertensive status. All traits were significantly different among the three racial groups (P < 0.05). We calculated highly significant heritability estimates for the lipoprotein and apolipoprotein measures (all significant at P < 0.0001), indicating a strong genetic determinant of these traits in GENOA subjects. Heritabilities ranged from 0.42 (TG) to 0.71 (HDL-C) in AAs, 0.39 (LDL-C) to 0.54 (HDL-C) in MAs, and 0.26 (TC) to 0.64 (HDL-C) in EAs. We genotyped GENOA subjects for 36 SNPs spanning APOH based on MAFs >0.05 and potential functionality (13). Table 2 shows a list of the 36 SNPs, as well as their gene locations and MAFs in AAs, MAs, and EAs. Among the potentially functional SNPs, we genotyped 8 SNPs that result in amino acid substitutions, including those that underlie known apoH charge isoforms (9, 11, 12).
TABLE 1.
General characteristics of GENOA subjects
| GENOA | European Americans | African Americans | Mexican Americans | F-Test |
|---|---|---|---|---|
| Subjects | 1,409 | 1,696 | 1,643 | |
| Families | 498 | 583 | 415 | |
| Diabetics | 161 | 354 | 1002 | |
| Hypertensives | 1,067 | 1,268 | 815 | |
| Age | 55.92 ± 10.89 | 57.94 ± 10.26 | 55.72 ± 12.08 | <0.0001 |
| Height (cm) | 168.73 ± 9.25 | 168.71 ± 8.81 | 162.94 ± 9.21 | <0.0001 |
| Weight (kg) | 86.67 ± 19.93 | 87.77 ± 18.69 | 81.88 ± 17.30 | <0.0001 |
| BMI (kg/m2) | 30.36 ± 6.30 | 30.90 ± 6.59 | 30.83 ± 6.08 | 0.042 |
| W/H | 0.913 ± 0.09 | 0.910 ± 0.07 | 0.974 ± 0.08 | <0.0001 |
| TC | 210.49 ± 38.79 | 205.10 ± 56.88 | 205.22 ± 46.36 | 0.0008 |
| LDL-C | 121.40 ± 33.90 | 120.70 ± 40.80 | 116.40 ± 35.50 | 0.0017 |
| HDL-C | 51.53 ± 16.35 | 55.52 ± 17.99 | 44.89 ± 12.64 | <0.0001 |
| TG | 193.59 ± 103.58 | 145.45 ± 90.87 | 234.18 ± 176.99 | <0.0001 |
| ApoE | 5.16 ± 1.65 | 5.31 ± 2.20 | 5.43 ± 2.22 | 0.0014 |
| ApoB | 106.65 ± 23.12 | 102.59 ± 25.95 | 108.54 ± 26.30 | <0.0001 |
| ApoA-I | 157.47 ± 31.36 | 158.83 ± 32.26 | 144.91 ± 25.67 | <0.0001 |
| Glucose | 100.11 ± 28.84 | 111.90 ± 49.09 | 152.82 ± 71.90 | <0.0001 |
| Lipid-lowering medication | 21.38% | 7.6% | 19.77% | |
| Anti-hypertensive medication | 68.88% | 66.8% | 49.84% | |
| Anti-diabetic medication | 8.28% | 21.98% | 62.66% |
GENOA, Genetic Epidemiology Network of Arteriopathy study; ApoE, apolipoprotein E; BMI, body mass index; W/H, waist-to-hip ratio; TG, triglycerides, HDL-C, HDL cholesterol; LDL-C, LDL cholesterol; TC, total cholesterol. Plasma levels of lipids, lipoproteins, apolipoproteins, and glucose are measured in mg/dl. Data are given as mean ± SD. Percentages of individuals taking medications were calculated for 991 African Americans, 1,196 Mexican Americans, and 1,038 European Americans.
Single SNP associations
For association studies, we assigned plasma lipoprotein and apolipoprotein measures according to three cholesterol transport pathways including HCT (HCT includes TC, LDL-C, and apoB), DCT (DCT includes TG and apoE), and RCT (RCT includes HDL-C and apoA-I). These groupings reflect involvement in common transport pathways and known correlations among traits (e.g., LDL-C and apoB, HDL-C and apoA-I). However, these assignments are not exact, because some traits are components of overlapping transport pathways. Figure 1 shows FBAT results for association of individual APOH SNPs with apolipoprotein components of the three cholesterol transport groupings. Table 3 shows P values for APOH SNPs that showed point-wise significant associations (unadjusted P < 0.05) for all traits in all races, as well as experiment-wise P values adjusted for multiple testing based on FDRs. Supplementary Table I shows the means of lipoprotein and apolipoprotein levels for APOH SNP genotypes that showed point-wise significance (unadjusted P < 0.05).
Fig. 1.
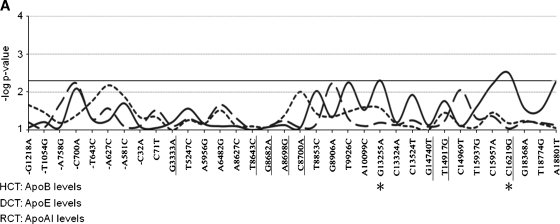
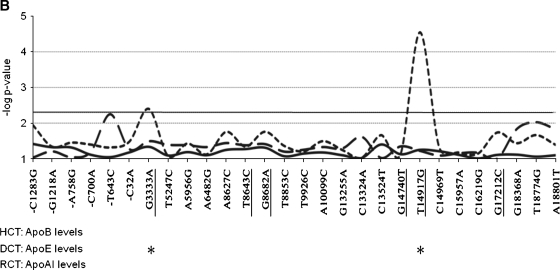
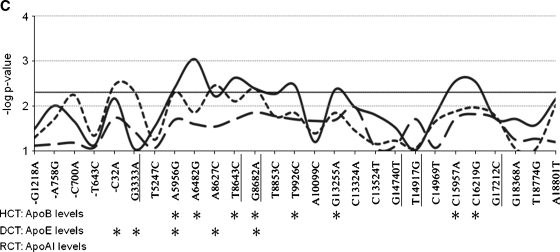
Association of individual APOH single-nucleotide polymorphisms (SNPs) with plasma apolipoprotein levels in GENOA. Each APOH SNP is listed on the x axis, and numbered relative to the translational start site. The underlined SNPs are polymorphisms located in the coding sequence of the gene. The y axis shows the point-wise –log P values (not adjusted for multiple testing) for associations (P = 0.05 corresponds to –log P value of 2.3, solid horizontal line). The solid curved lines show associations with apolipoprotein B (apoB) levels (hepatic cholesterol transport), the short dashed curves show associations with apoE levels (dietary cholesterol transport), and the long dashed curves show associations with apoA-I levels (reverse cholesterol transport). Asterisks below the graph mark APOH SNPs that show point-wise significant associations (unadjusted P > 0.05) with each apolipoprotein measure. A: Associations for African American families. B: Associations for Mexican Americans. C: Associations for European Americans.
TABLE 3.
P values for significant apoH SNP associations (unadjusted P < 0.05) for all traits in all races, including P values adjusted for multiple testing based on FDR
| AA
|
MA
|
EA
|
|||||
|---|---|---|---|---|---|---|---|
| Position | Trait | P < 0.05 | P (FDR) | P < 0.05 | P (FDR) | P < 0.05 | P (FDR) |
| 5956 | ApoB | 0.039 | 0.146 | ||||
| 6482 | ApoB | 0.009 | 0.146 | ||||
| 8643a | ApoB | 0.023 | 0.146 | ||||
| 8682a | ApoB | 0.04 | 0.146 | ||||
| 9926 | ApoB | 0.035 | 0.146 | ||||
| 13255 | ApoB | 0.049 | 0.423 | 0.043 | 0.146 | ||
| 15957 | ApoB | 0.028 | 0.146 | ||||
| 16219 | ApoB | 0.03 | 0.423 | 0.029 | 0.146 | ||
| −1054 | LDL-C | 0.047 | 0.915 | ||||
| 6482 | LDL-C | 0.013 | 0.224 | ||||
| 5956 | TC | 0.045 | 0.208 | ||||
| 6482 | TC | 0.021 | 0.208 | ||||
| 8643a | TC | 0.031 | 0.208 | ||||
| −32 | ApoE | 0.034 | 0.257 | ||||
| 3333a | ApoE | 0.039 | 0.559 | 0.048 | 0.257 | ||
| 5956 | ApoE | 0.049 | 0.257 | ||||
| 8627 | ApoE | 0.035 | 0.257 | ||||
| 8682a | ApoE | 0.041 | 0.257 | ||||
| 14917 | ApoE | <0.001 | 0.007 | ||||
| 8700a | TG | 0.027 | 0.180 | ||||
| 9926 | TG | 0.04 | 0.217 | ||||
| 13255 | TG | 0.044 | 0.217 | ||||
| 14917 | TG | 0.001 | 0.052 | ||||
| 15937 | TG | 0.012 | 0.143 | ||||
| 15957 | TG | 0.019 | 0.168 | ||||
| 18368 | TG | 0.004 | 0.112 | ||||
| 18774 | TG | 0.006 | 0.112 | ||||
| 14969 | HDL-C | 0.031 | 0.770 | ||||
FDR, false discovery rate.
SNPs that determine known apoH protein charge isoforms.
For HCT in EAs, APOH associations reflected the high correlations among HCT traits (Fig. 1C). For example, intronic SNP 6482 A/G showed point-wise highly significant associations with plasma apoB levels, as well as for TC and LDL-C (Table 3). Four other intronic SNPs (9926 T/C, 13255 G/A, 15957 C/A, 16219 C/G) also showed associations with apoB levels in EAs. These 4 intronic SNPs are strongly correlated (r2 > 0.8). In addition, 2 correlated nonsynonymous SNPs that distinguish the H*3 protein isoform were associated with apoB levels (8643 T/C, 8682 G/A), and TC (8643 T/C). In AAs (Fig. 1A), 2 SNPs showed associations with apoB levels, including 13255 G/A and 16219 C/G, which were also associated with apoB levels in EAs (Table 3). Overall, the less common alleles for these SNPs were associated with increased plasma apoB levels in both EAs and AAs (see supplementary Table I). In addition, SNP −1054 T/G in the promoter region was associated with LDL-C levels in AAs. No associations of APOH variants and HCT traits were observed in MAs at the P < 0.05 level.
For DCT in MAs, a nonsynonymous SNP, 14917 T/G (Cys306Gly), showed point-wise highly significant associations for both apoE levels (Fig. 1B) and TG levels (Table 3). This association remained statistically significant after adjustment for multiple testing using FDRs (P = 0.007; Table 3). In addition, a nonsynonymous SNP that distinguishes the H*1 protein isoform (3333 G/A encoding Ser88Asn) was associated with apoE levels in MAs (and EAs). In EAs, 5 correlated SNPs (r2 > 0.8) showed point-wise significant associations with plasma apoE levels, including promoter SNP −32 C/A, intronic SNPs 5956 A/G and 8627 A/C, and nonsynonymous SNPs 3333 G/A (Ser88Asn) and 8682 G/A (Arg135His) (Fig. 1C). All of these correlated SNPs showed point-wise significant associations with apoE levels at P < 0.05 (Table 3). For all of these SNPs, the less common alleles were associated with reduced apoE levels in EAs (see supplementary Table I). In AAs, the strongest associations for DCT were with TG levels for 2 correlated SNPs in the 3′ flanking region (18368 G/A and 18774 T/G) and 2 correlated intronic SNPs (9926 T/C and 13255 G/A) (Table 3). Two independent intronic SNPs also showed associations with TG levels in AAs (15937 T/G and 15957 C/A). In addition, a nonsynonymous SNP, 8700 C/A, which distinguishes the protein isoform H*3B (Ala141Asp), showed point-wise significant association with TG levels in AAs.
For RCT in AAs, intronic SNP 14969 C/T showed point-wise significant associations with HDL-C levels (P = 0.031; Table 3). In MAs and EAs, no associations were found for the RCT pathway at P < 0.05.
Promoter haplotype associations
Because SNP combinations in regulatory elements may comprise cis-acting functional units that control gene transcription, we constructed haplotypes for SNPs in the APOH promoter region. We genotyped 8 SNPs in the APOH proximal promoter region (positions −1283 to −581) to construct haplotypes. Table 4 shows the nine common haplotypes we found in the three GENOA racial groups, as well as their relative frequencies. We used these haplotypes for statistical analyses that identified significant associations in AAs for a relatively rare haplotype (haplotype 8, MAF = 0.021) with reductions of plasma measures in HCT (TC, LDL-C, apoB) and DCT (apoE). In addition, we found point-wise significant associations of haplotype 9 in AAs with increased LDL-C levels (P = 0.022). We did not detect any significant associations of APOH promoter haplotypes with lipoprotein and apolipoprotein measures in MAs or EAs.
TABLE 4.
Identity and frequencies of SNP haplotypes for the apoH promoter region and results of HBAT association analysis
| Association Results African Americans
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Promoter SNPs
|
GENOA Frequencies
|
HCT
|
DCT
|
||||||||||||
| Haplotype | -1283 | -1218 | -1054 | -758 | -700 | -643 | -627 | -581 | EA | AA | MA | TC | LDL-C | ApoB | ApoE |
| Hap1 | C | G | T | A | C | T | A | A | 0.539 | 0.608 | 0.583 | ns | ns | ns | ns |
| Hap2 | C | G | T | G | A | T | A | A | 0.209 | 0.045 | 0.072 | ns | ns | ns | ns |
| Hap3 | C | G | T | A | C | C | A | A | 0.163 | 0.157 | 0.273 | ns | ns | ns | ns |
| Hap4 | C | A | T | G | A | T | A | A | 0.078 | 0.013 | 0.027 | ns | ns | ns | ns |
| Hap5 | G | G | T | A | C | T | A | A | 0.022 | na | na | na | na | ||
| Hap6 | C | G | T | A | C | T | A | C | 0.068 | ns | ns | ns | ns | ||
| Hap7 | C | G | T | A | C | T | C | A | 0.058 | ns | ns | ns | ns | ||
| Hap8 | C | G | T | A | A | T | A | A | 0.021 | 0.006 | 0.030 | 0.001 | 0.006 | ||
| Hap9 | C | G | G | A | C | T | A | A | 0.013 | ns | 0.022 | ns | ns | ||
HBAT, haplotype-based association test; HCT, hepatic cholesterol transport; DCT, dietary cholesterol transport; ns, not significant (P > 0.05); na, not available; number of informative families <10.
Amino acid haplotype associations
We used 7 nonsynonymous SNPs to construct amino acid haplotypes that define allelic apoH proteins in GENOA subjects. Five of the nonsynonymous SNPs determine known charge isoforms, including 3333 G/A (Ser88Asn, isoform H*1), 8643 T/C (Ile122Thr, H*3), 8682 G/A (Arg135His, H*3), 8700 C/A (Ala141Asp, H*3B), and 17212 G/C (Trp316Ser, H*3W) (9, 11, 12). Table 5 shows the seven most common amino acid haplotypes grouped according to charge isoforms and their relative frequencies in AAs, MAs, and EAs. In EAs, we found associations of H*1 with RCT traits (increased HDL-C and apoA-I levels), and associations of H*3 with HCT traits (reduced TC, LDL-C, and apoB levels). In MAs, we found significant associations of a relatively rare form of H*2 (14917 T/G, Cys306Gly) with DCT traits (reduced TG and apoE levels) and reduced TC levels. This nonsynonymous SNP, 14917 T/G (Cys306Gly), also showed point-wise significant associations for DCT traits in single SNP analysis in MAs (Table 3; Fig. 1B). In addition, we found an association of a rare form of H*3 distinguished by Trp316Ser (H*3W) with apoA-I levels in EAs, and TC and HDL-C levels in MAs. No significant associations with apoH amino acid haplotypes were found for AAs.
TABLE 5.
Identity and frequencies of haplotypes for apoH amino acid substitutions and results of HBAT association analysis
| Association Results European Americans
|
Association Results Mexican Americans
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino Acid Substitutions
|
GENOA Frequencies
|
HCT
|
RCT
|
HCT
|
DCT
|
RCT
|
|||||||||||||
| Protein Isoforms | Ser88Asn | Ile122Thr | Arg135His | Ala141Asp | Val247Leu | Cys306Gly | Trp316Ser | EA | AA | MA | TC | LDLC | ApoB | HDL | ApoA1 | TC | TG | ApoE | HDL |
| H*1 | N | I | R | A | L | C | W | 0.051 | 0.025 | ns | ns | ns | 0.024 | 0.026 | ns | ns | ns | ns | |
| H*2 | S | I | R | A | L | C | W | 0.571 | 0.385 | 0.431 | ns | ns | ns | ns | ns | ns | ns | ns | ns |
| S | I | R | A | V | C | W | 0.255 | 0.558 | 0.431 | ns | ns | ns | ns | ns | ns | ns | ns | ns | |
| S | I | R | A | L | G | W | 0.033 | 0.056 | ns | ns | ns | ns | ns | 0.018 | 0.002 | 0.0007 | ns | ||
| H*3 | S | T | H | A | L | C | W | 0.015 | 0.031 | 0.010 | 0.010 | 0.033 | 0.009 | ns | ns | ns | ns | ns | ns |
| H*3W | S | T | H | A | L | C | S | 0.069 | 0.042 | ns | ns | ns | ns | 0.015 | 0.014 | ns | ns | 0.013 | |
| H*3B | S | T | H | N | L | C | W | na | na | na | na | na | na | na | na | na | |||
RCT, reverse cholesterol transport. ns, not significant (P > 0.05); number of informative families <10.
DISCUSSION
We have performed a comprehensive investigation of the effects of APOH variation on lipid traits related to HCT, DCT, and RCT in the GENOA study, which includes three racial groups (AA, MA, EA). Single SNP analysis (36 SNPs) identified multiple associations with lipid traits, some of which were consistent across multiple traits and racial groups. For example, intronic SNP 6482 A/G was associated with levels of all three HCT traits (apoB, TC, LDL-C) in EAs, and 16219 C/G was associated with apoB levels in both EAs and AAs.
We examined effects of variants in regions involved in APOH gene expression. In single SNP analysis, we found association of −32 C/A at the transcriptional start site with plasma apoE levels in EAs. Three SNPs that are correlated with −32 C/A (r2 > 0.8) also showed point-wise significant associations (P < 0.05) with apoE levels in EAs (Fig. 1C). The potential functional impact of −32 C/A on apoE levels is supported by a previous study that showed −32 C/A reduced APOH transcriptional activities in human hepatic tissues and transfected COS-1 cells, as well as population-based association with apoH levels in 232 lupus women (24). In addition to single SNP analysis, we constructed haplotypes from SNPs located in the APOH promoter region, because they may comprise cis-acting regulatory elements. In AAs, haplotype 8 showed consistent associations across all three HCT traits for reduced plasma levels of TC, LDL-C, and apoB (Table 4). The −700 C/A allele that distinguishes haplotype 8 from the common haplotype 1 (also in haplotypes 2 and 4) is of particular interest, because this single SNP shows suggestive associations (P < 0.10) with several traits in AAs (apoA-I, HDL-C, apoB), as well as EAs (TC and apoE) (Fig. 1). Although these associations support the possibility that promoter region variation may alter apoH levels and affect lipid pathways, we did not identify any specific sequence binding motifs for transcription factors that were altered by these SNPs.
We were particularly interested in potential functional variants that altered apoH amino acid sequences. We tested for associations with nonsynonymous SNPs individually and as amino acid haplotypes that included the apoH charge isoforms. Previous studies have reported associations of apoH isoforms with lipid measures as well as plasma levels of apoH. For example, the protein isoform H*3 showed associations with TG levels in Italian males (16) and in African females (14). Although the protein isoform H*3 is defined by Ile122Thr and Arg135His, these studies did not differentiate between H*3W (Trp316Ser) and H*3B (Ala141Asp). In our studies of EAs, we found association of H*3 (Ile122Thr and Arg135His) with HCT measures (apoB, TC, LDL-C), but not with TG levels. However, we found associations of Ala141Asp that distinguishes H*3B with TG levels in AAs, although insufficient numbers of H*3B families were available for haplotype analysis among GENOA subjects (Table 5). In addition, H*3W (Trp316Ser) showed associations with HDL-C levels in MAs, as well as apoA-I levels in EAs (Table 5). Analysis of amino acid haplotypes found association of a relatively rare H*2 allele (Cys306Gly) with TG and apoE levels in MAs (Table 5). Single SNP analysis in MAs also identified a strong association of TG and apoE levels with nonsynonymous SNP 14917 (Cys306Gly) that remained significant after adjustment for multiple testing (FDR-adjusted P value = 0.007) (Fig. 1B; Table 3). Previous studies have found associations of Cys306Gly and Trp316Ser with plasma apoH levels (25, 26).
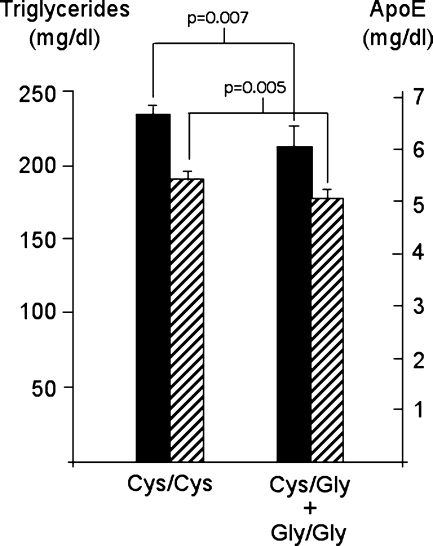
Because the strongest associations were found for SNP 14917 (Cys306Gly), we investigated the effects of this APOH variant on DCT measures in MAs. Figure 2 shows the mean values of TG and apoE levels among Cys306Gly genotypes in MAs. GEE showed significant differences among Cys306Gly genotypes for plasma levels of TG (P = 0.007) and apoE (P = 0.005). Genotypes containing the less common Gly allele were associated with a reduction in TG and apoE levels. We also compared Tg/ApoE ratios for Cys306Gly genotypes, but did not find any significant differences (42% in Cys/Cys, 40% in Cys/Gly, 38% in Gly/Gly) (P > 0.1). Recent studies have shown that Cys306Gly is probably a functional polymorphism that alters binding of apoH to phospholipid particles (9, 27). ApoH contains five tandem domains of 60 amino acids (sushi domains) that contain Cys residues that form disulfide bonds for apoH folding (28). Recent in vitro studies have shown that a hydrophobic region of four amino acids in the fifth sushi domain is involved in phospholipid binding (27). Therefore, the replacement of Cys at position 306 by Gly may affect apoH function by disruption of normal folding in the fifth sushi domain, resulting in alteration of phospholipid binding (9).
Fig. 2.
Plasma levels of triglycerides (TGs) and apoE among Cys306Gly genotypes in MAs. The mean values (±SE) for TG levels for Cys306Gly genotypes in MAs (1,463 Cys/Cys subjects, 151 Cys/Gly, and 3 Gly/Gly) are shown by solid bars (left y axis), and for apoE levels by hatched bars (right y axis).
Interestingly, a previous GENOA study found that the common apoE isoforms (E2, E3, E4) had much smaller effects in MAs compared with AAs and EAs, accounting for only 1.0% and 0.5% of the variation in apoE levels in males and females, respectively (29). In contrast to AAs and EAs, linkage analysis in MAs detected only a modest linkage signal for apoE levels on chromosome 19 [logarithm of the odds ratio (LOD) = 0.97] that was not reduced after adjustment for genotypic effects of the apoE isoforms (29). Instead, chromosome 17 showed evidence for linkage with apoE levels (LOD = 3.32) in MAs with low TC levels. APOH is located within the linked region on chromosome 17, providing further evidence for the physiological relevance of our observed associations of Cys306Gly on apoE and TG levels in MAs. In MAs with low TC levels, we found that Cys306Gly accounted for 2.8% of the variation in apoE levels (P = 0.0002). However, linkage analysis conditioned for the Cys306Gly genotype only lowered the LOD score on chromosome 17 from 2.8 to 2.5 (11% reduction), indicating that other genes in this region must contribute to the apoE linkage signal in MAs. Interestingly, Bosse and coworkers (30) examined APOH to follow up a major quantitative trait locus for LDL particle size on chromosome 17 that was identified in a genome linkage scan in French Canadians. Although APOH haplotypes showed associations with increased LDL particle size in their study, the relationship of APOH variation to the linkage signal was not determined.
These comprehensive studies of APOH variation have yielded several point-wise significant associations with lipid traits in HCT, DCT, and RCT (unadjusted P < 0.05). However, only one of the point-wise significant associations reached experiment-wise significance after correction for multiple testing using FDR (Cys306Gly with apoE levels in MAs). In part, this may be owing to our relatively conservative application of FDR that assumed independence for all tests, although many of the SNPs are in LD and not truly independent. Yet, the importance of our point-wise results is supported by consistency of associations of multiple correlated SNPs (r2 > 0.8) with particular cholesterol transport measures, as well as consistency of SNP associations with multiple measures in cholesterol transport groups. Biological interpretations of these associations are limited by our incomplete understanding of the precise roles of apoH in lipid metabolism. A large fraction (35%) of circulating apoH is found on lipoprotein particles, including chylomicrons and VLDL (16%), LDL (2%), and HDL (17%) (31). Previous in vivo and in vitro studies have shown that apoH is involved in TG metabolism by activation of LPL (3). Perhaps the best candidate polymorphism for further experimental studies is Cys306Gly, given the strong associations identified with plasma levels of TG and apoE in MAs. Future functional studies would examine how Cys306Gly disruption of protein folding might alter apoH distribution among lipoprotein classes or activation of LPL in TG metabolism.
Published, JLR Papers in Press, August 1, 2008.
Footnotes
This work was supported in part by a graduate student fellowship from the Harry S. and Isabel C. Cameron Foundation (M.S.L.), and National Institutes of Health/National Heart Lung and Blood Institute Grant HL-072810 (K.L.E.K., E.B., J.E.H.). GENOA sample collection and measurements were supported by National Institutes of Health/National Heart Lung and Blood Institute Grants HL-54504, HL-039107, HL-054457, and HL-051021.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of a table.
References
- 1.Polz E., and G. M. Kostner. 1979. The binding of beta 2-glycoprotein-I to human serum lipoproteins: distribution among density fractions. FEBS Lett. 102 183–186. [DOI] [PubMed] [Google Scholar]
- 2.Polz E., G. M. Kostner, and A. Holasek. 1979. Studies on the protein composition of human serum very low density lipoproteins: demonstration of the beta 2-glycoprotein-I. Hoppe Seylers Z. Physiol. Chem. 360 1061–1067. [DOI] [PubMed] [Google Scholar]
- 3.Nakaya Y., E. J. Schaefer, and H. B. Brewer, Jr. 1980. Activation of human post heparin lipoprotein lipase by apolipoprotein H (beta 2-glycoprotein I). Biochem. Biophys. Res. Commun. 95 1168–1172. [DOI] [PubMed] [Google Scholar]
- 4.Schousboe I. 1980. Binding of beta 2-glycoprotein I to platelets: effect of adenylate cyclase activity. Thromb. Res. 19 225–237. [DOI] [PubMed] [Google Scholar]
- 5.Schousboe I. 1985. Beta 2-glycoprotein I: a plasma inhibitor of the contact activation of the intrinsic blood coagulation pathway. Blood. 66 1086–1091. [PubMed] [Google Scholar]
- 6.Nimpf J., E. M. Bevers, P. H. Bomans, U. Till, H. Wurm, G. M. Kostner, and R. F. Zwaal. 1986. Prothrombinase activity of human platelets is inhibited by beta 2-glycoprotein-I. Biochim. Biophys. Acta. 884 142–149. [DOI] [PubMed] [Google Scholar]
- 7.McNeil H. P., R. J. Simpson, C. N. Chesterman, and S. A. Krilis. 1990. Anti-phospholipid antibodies are directed against a complex antigen that includes a lipid-binding inhibitor of coagulation: beta 2-glycoprotein I (apolipoprotein H). Proc. Natl. Acad. Sci. USA. 87 4120–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones J. V., H. James, M. H. Tan, and M. Mansour. 1992. Antiphospholipid antibodies require beta 2-glycoprotein I (apolipoprotein H) as cofactor. J. Rheumatol. 19 1397–1402. [PubMed] [Google Scholar]
- 9.Sanghera D. K., D. R. Wagenknecht, J. A. McIntyre, and M. I. Kamboh. 1997. Identification of structural mutations in the fifth domain of apolipoprotein H (beta 2-glycoprotein I) which affect phospholipid binding. Hum. Mol. Genet. 6 311–316. [DOI] [PubMed] [Google Scholar]
- 10.Mehdi H., M. Nunn, D. M. Steel, A. S. Whitehead, M. Perez, L. Walker, and M. E. Peeples. 1991. Nucleotide sequence and expression of the human gene encoding apolipoprotein H (beta 2-glycoprotein I). Gene. 108 293–298. [DOI] [PubMed] [Google Scholar]
- 11.Kamboh M. I., R. E. Ferrell, and B. Sepehrnia. 1988. Genetic studies of human apolipoproteins. IV. Structural heterogeneity of apolipoprotein H (beta 2-glycoprotein I). Am. J. Hum. Genet. 42 452–457. [PMC free article] [PubMed] [Google Scholar]
- 12.Sanghera D. K., T. Kristensen, R. F. Hamman, and M. I. Kamboh. 1997. Molecular basis of the apolipoprotein H (beta 2-glycoprotein I) protein polymorphism. Hum. Genet. 100 57–62. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q., and M. I. Kamboh. 2006. Complete DNA sequence variation in the apolipoprotein H (beta-glycoprotein I) gene and identification of informative SNPs. Ann. Hum. Genet. 70 1–11. [DOI] [PubMed] [Google Scholar]
- 14.Sepehrnia B., M. I. Kamboh, L. L. Adams-Campbell, C. H. Bunker, M. Nwankwo, P. P. Majumder, and R. E. Ferrell. 1989. Genetic studies of human apolipoproteins. VIII. Role of the apolipoprotein H polymorphism in relation to serum lipoprotein concentrations. Hum. Genet. 82 118–122. [DOI] [PubMed] [Google Scholar]
- 15.Kaprio J., R. E. Ferrell, B. A. Kottke, M. I. Kamboh, and C. F. Sing. 1991. Effects of polymorphisms in apolipoproteins E, A-IV, and H on quantitative traits related to risk for cardiovascular disease. Arterioscler. Thromb. 11 1330–1348. [DOI] [PubMed] [Google Scholar]
- 16.Cassader M., G. Ruiu, R. Gambino, F. Guzzon, A. Pagano, F. Veglia, R. Pagni, and G. Pagano. 1994. Influence of apolipoprotein H polymorphism on levels of triglycerides. Atherosclerosis. 110 45–51. [DOI] [PubMed] [Google Scholar]
- 17.Takada D., Y. Ezura, S. Ono, Y. Iino, Y. Katayama, Y. Xin, L. L. Wu, S. Larringa-Shum, S. H. Stephenson, S. C. Hunt, et al. 2003. Apolipoprotein H variant modifies plasma triglyceride phenotype in familial hypercholesterolemia: a molecular study in an eight-generation hyperlipidemic family. J. Atheroscler. Thromb. 10 79–84. [DOI] [PubMed] [Google Scholar]
- 18.Daniels P. R., S. L. Kardia, C. L. Hanis, C. A. Brown, R. Hutchinson, E. Boerwinkle, and S. T. Turner. 2004. Genetic Epidemiology Network of Arteriopathy Study. The familial aggregation of hypertension treatment and control in the GENOA study. Am. J. Med. 116 676–681. [DOI] [PubMed] [Google Scholar]
- 19.Carlson C. S., M. A. Eberle, M. J. Rieder, Q. Yi, L. Kruglyak, and D. A. Nickerson. 2004. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 74 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almasy L., and J. Blangero. 1998. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blangero J., J. T. Williams, and L. Almasy. 2000. Quantitative trait locus mapping using human pedigrees. Hum. Biol. 72 35–62. [PubMed] [Google Scholar]
- 22.Laird N. M., S. Horvath, and X. Xu. 2000. Implementing a unified approach to family-based tests of association. Genet. Epidemiol. 19 (Suppl.): 36–42. [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y., D. Drai, G. Elmer, N. Kafkafi, and I. Golani. 2001. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125 279–284. [DOI] [PubMed] [Google Scholar]
- 24.Mehdi H., S. Manzi, P. Desai, Q. Chen, C. Nestlerode, F. Bontempo, S. C. Strom, R. Zarnegar, and M. I. Kamboh. 2003. A functional polymorphism at the transcriptional initiation site in beta2-glycoprotein I (apolipoprotein H) associated with reduced gene expression and lower plasma levels of beta2-glycoprotein I. Eur. J. Biochem. 270 230–238. [DOI] [PubMed] [Google Scholar]
- 25.Mehdi H., C. E. Aston, D. K. Sanghera, R. F. Hamman, and M. I. Kamboh. 1999. Genetic variation in the apolipoprotein H (beta2-glycoprotein I) gene affects plasma apolipoprotein H concentrations. Hum. Genet. 105 63–71. [DOI] [PubMed] [Google Scholar]
- 26.Kamboh M. I., S. Manzi, H. Mehdi, S. Fitzgerald, D. K. Sanghera, L. H. Kuller, and C. E. Atson. 1999. Genetic variation in apolipoprotein H (beta2-glycoprotein I) affects the occurrence of antiphospholipid antibodies and apolipoprotein H concentrations in systemic lupus erythematosus. Lupus. 8 742–750. [DOI] [PubMed] [Google Scholar]
- 27.Mehdi H., A. Naqvi, and M. I. Kamboh. 2000. A hydrophobic sequence at position 313–316 (Leu-Ala-Phe-Trp) in the fifth domain of apolipoprotein H (beta2-glycoprotein I) is crucial for cardiolipin binding. Eur. J. Biochem. 267 1770–1776. [DOI] [PubMed] [Google Scholar]
- 28.Schwarzenbacher R., K. Zeth, K. Diederichs, A. Gries, G. M. Kostner, P. Laggner, and R. Prassl. 1999. Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome. EMBO J. 18 6228–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klos K. L., S. L. Kardia, J. E. Hixson, S. T. Turner, C. Hanis, E. Boerwinkle, and C. F. Sing. 2005. Linkage analysis of plasma apoE in three racial groups: multiple genes with context-dependent effects. Ann. Hum. Genet. 69 157–167. [DOI] [PubMed] [Google Scholar]
- 30.Bosse Y., M. F. Feitosa, J-P. Despres, B. Lamarche, T. Rice, D. C. Rao, C. Bouchard, L. Perusse, and M-C. Vohl. 2005. Detection of a major gene effect for LDL peak particle diameter and association with apolipoprotein H gene haplotype. Atherosclerosis. 182 231–239. [DOI] [PubMed] [Google Scholar]
- 31.Polz E., H. Wurm, and G. M. Kostner. 1980. Investigations on beta 2-glycoprotein-I in the rat: isolation from serum and demonstration in lipoprotein density fractions. Int. J. Biochem. 11 265–270. [DOI] [PubMed] [Google Scholar]