Summary
Mutations in pfcrt K76T are associated with chloroquine resistance in Plasmodium falciparum. Previous studies of K76T mutations in Senegal reported the association of T76 with in vitro-resistant isolates, but this mutation was also prevalent in chloroquine-sensitive isolates. This suggests involvement of additional genetic loci in modulating chloroquine resistance. Additional pfcrt polymorphisms at codons A220S, Q271E, N326S and R371I have been found in chloroquine-resistant isolates. We wanted to test if sequential acquisition of mutations at these codons leads to in vitro chloroquine resistance. Stepwise accumulation of mutations was not detected, rather there was almost complete linkage between the pfcrt K76T mutation and polymorphisms in these codons. Therefore these additional polymorphisms do not enhance the correlation between pfcrt T76 and chloroquine resistance in Senegal. These data suggest that in vitro chloroquine resistance requires the genetic background of the pfcrt K76T mutation and additional mutations in genetic loci outside the pfcrt gene.
Keywords: malaria, chloroquine resistance, linkage disequilibrium, Senegal
Introduction
Chloroquine resistance in Plasmodium falciparum is established in many parts of Africa and is becoming more prevalent in areas of West Africa such as Senegal. Resistance to chloroquine was first reported in Dakar in 1988 and more recent treatment failure rates of 13% have been reported in Senegal (Gaye, Soumare & Sambou, 1999). Identifying reliable markers of chloroquine resistance is important to inform both individual malaria treatment and local drug policy.
There has been recent progress in the identification of genetic markers of chloroquine resistance with the discovery of mutations in the P. falciparum pfcrt gene (Djimde et al. 2001a). Genetic mutations in pfcrt are strongly associated with both in vitro and in vivo drug resistance. The correlation of in vivo chloroquine resistance with the pfcrt K76T mutation has been observed at various geographically distinct sites including Indonesia, Laos, Papua New Guinea (PNG), Cameroon, Mozambique, Uganda, Sudan and Brazil (Basco & Ringwald, 2001; Dorsey et al. 2001; Maguire et al. 2001; Pillai et al. 2001; Vieira et al. 2001). Resistance defined as in vivo may be confounded by factors such as host immunity. In contrast, in vitro methods can provide a direct measurement of parasite survival in response to drug treatment. Thus, resistance of different parasite strains can also be defined by the 50% inhibitory concentration or IC50. As measured by this in vitro standard, the mutation in pfcrt K76T is still highly correlated with resistance. Specifically, the presence of the K76T mutation is found in the overwhelming majority of isolates that display in vitro resistance in studies reported from PNG, Thailand, Indonesia, Cameroon and Senegal (Basco et al. 2001; Chen et al. 2001; Maguire et al. 2001).
However, it has been also noted that some parasites that were cleared after administration of chloroquine also had the K76T mutation. This observation has been reported in several studies, including in the original report in which 41% of all isolates contained the K76T mutation, though only 14% of these isolates exhibited in vivo resistance to chloroquine (Djimde et al. 2001a). This clearance of apparently genetically resistant strains has been ascribed to age-related immunity (Djimde et al. 2001b). Yet data from Indonesia showed there was no relation between the history of previous infections and the clearance of genetically resistant parasites in the 12 of 25 parasites that were in vivo sensitive that contained the K76T mutation (Maguire et al. 2001). In addition, a study conducted in Mozambique found that the K76T mutation was present in isolates from 18 of 23 subjects in whom the infection resolved after chloroquine treatment. There was no correlation between age and parasitological outcome after treatment (Mayor, Gomez-Olive & Aponte, 2001). In some areas this marker does not correlate to outcome due to its high penetrance in the population, for example, pfcrt T76 is 100% prevalent in Uganda and no longer predictive of chloroquine treatment failure (Kyosiimire-Lugemwa et al. 2002). A study done in Cameroon (Basco et al. 2001) using in vitro methods for testing chloroquine susceptibility reported 9 isolates with the K76T mutation that had chloroquine IC50s in the sensitive range. Both the in vivo and in vitro data suggest that pfcrt K76T mutation is necessary for chloroquine resistance, but a subset of isolates from various field studies did not manifest this correlation which may suggest that for these isolates, mutations in addition to the pfcrt K76T mutation are needed to confer chloroquine resistance.
Pfcrt has additional polymorphisms at codons A220S, Q271E, N326S and R371I associated with in vivo chloroquine resistance. Mutations at these codons result in amino acid change, for example at amino acid 220, there is a substitution of thymine for guanine, which changes the amino acid from alanine to serine. This study set out to determine whether pfcrt has acquired sequential mutations leading to greater in vitro chloroquine resistance. This analysis was carried out on previously isolated parasite DNA from an area in Senegal. This site was ideal to study the correlation of genetic polymorphisms and chloroquine resistance secondary to the prevalence of single clone infections typical of a hypoendemic transmission region. The pfcrt T76 allele was found in >90% of in vitro-resistant isolates, while isolates with wild-type K76 allele were almost completely chloroquine sensitive. However, a number of isolates harbouring the T76 allele were also in vitro-chloroquine sensitive and therefore the K76T polymorphism did not significantly correlate with resistance in this study (P=0.18Mann–Whitney U test) (Thomas et al. 2002). These isolates were further analysed for the presence of additional pfcrt polymorphisms and correlation with chloroquine resistance.
Materials and Methods
The details of this study population have been previously reported (Thomas et al. 2002). To summarize, mildly symptomatic patients, age 5 years or greater, with peripheral blood smears positive for P. falciparum were invited to participate in the study. Exclusion criteria included severe or complicated malarial disease, pregnancy or recent history of antimalarial treatment. In vitro chloroquine susceptibility testing was done in duplicate using the DELI assay. The in vitro test utilized was the DELI assay, which has shown excellent correlation to the standard isotopic-microtest (Moreno et al. 2001). Parasite DNA was extracted with standard methods using phenol chloroform (Djimde et al. 2001a).
In total 44 patients were enrolled in the study and all but 1 patient sample had single parasite clones by Msp-1 and Msp-2 analysis. Chloroquine susceptibility was determined for 36 samples using the DELI test. Of these, 11 samples demonstrated in vitro chloroquine resistance (IC50>100 nm) and 25 samples exhibited in vitro chloroquine sensitivity (IC50<70 nm) (Thomas et al. 2002).
Detection of pfcrt alleles
To test the hypothesis of sequential acquisition of polymorphism at codons A220S, Q271E, N326S and R371I and the potential association with in vitro chloroquine resistance, the identification of these downstream pfcrt polymorphisms was carried out using polymerase chain reaction (PCR) amplification of parasite DNA from each field isolate. Primers Pfcrt4571 F (5′-GCTCTTGTAGAAATGAAATTAATC-3′) and Pfcrt 6279 R (5′-ATAGAACCAAATAGGTAGC-3′) were designed to amplify a region that spanned all the corresponding nucleotides for pfcrt codons A220S, Q271E, N326S and R371I. Although the occurrence of mixed infection is rare, this strategy ensured that all polymorphisms detected came from a single clone. The cycling conditions were as follows: 95 °C for 1 min followed by 32 cycles of denaturation at 95 °C for 1 min, annealing at 40 °C for 30 sec, extension at 62 °C for 1.5 min and a final extension at 62 °C for 30 min. Negative controls without DNA were included in all sets of PCR reactions. A 1600-base pair (bp) fragment for each isolate was electrophoresed on a 1% agarose gel and the DNA fragment was gel isolated using the SNAP Gel Purification Kit (Invitrogen, Carlsbad, CA).
This purified 1600 bp fragment was cloned into the TOPO-TA vector for sequencing, transfected into E. coli and grown overnight in Terrific Broth (Invitrogen, Carlsbad, CA). Plasmid DNA was isolated from E. coli using the Wizard Plus SV miniprep kit (Promega, Madison, WI). Clones were screened for the presence of a complete fragment either by PCR amplification using the universal primers M13 F and M13 R or by restriction digest using EcoRI (New England Biolabs, Beverly, MA). Plasmids with a 1600 bp insert were then selected for sequence analysis.
Two to three clones containing pfcrt sequence were generated for each field isolate and each clone was sequenced on both forward and reverse strands (High-Throughput DNA Sequencing Facility, The Dana-Farber/Harvard Cancer Center, Boston, MA). Multiple sequencing primers were used to obtain complete sequence of the 1600-bp fragment with each clone having, on average, 5-fold sequence coverage. Sequences were aligned using the SeqMan™ and MegAlign™ programs (DNASTAR, Madison WI).
Statistical analysis
To determine whether there was a statistical association between in vitro chloroquine resistance as measured by IC50 values and polymorphisms at these loci, the Wilcoxon rank sum test (Mann–Whitney) was performed using STATA (Stata, College Station, TX).
Results
PCR amplification of the 1600-bp fragment was successful in 38 of the 43 parasite DNA samples from which the pfcrt codon 76 had been previously analysed (Fig. 1). Of the 7 isolates that contained the K76 allele, all had the wild-type alleles at codons A220S, Q271E, N326S and R371I. The absence of polymorphisms was completely linked in this subset. Of the 31 isolates with the T76 allele, 2 of them had polymorphisms at codons A220S, Q271E and R371I, while the remaining 29 isolates contained polymorphisms at A220S, Q271E, N326S and R371I.
Fig. 1.
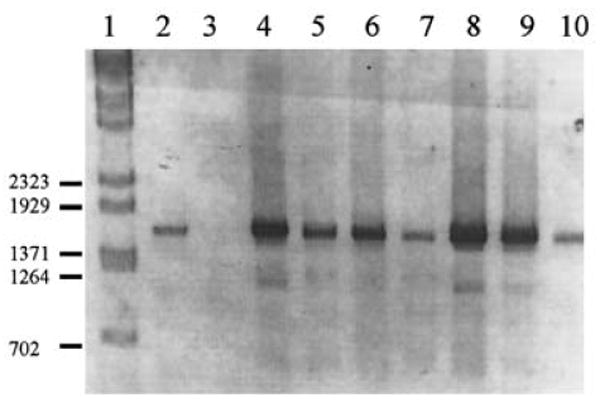
Representative agarose gel electrophoresis of 1600-bp PCR products. Lane 1, Lambda BstE II marker; lane 2, amplification from 3D7 genomic DNA; lane 3, no DNA control; lanes 4–10, amplification products generated from field isolates.
The 33 samples for which both sequence information and IC50 data was available was further analysed (Fig. 2). The median IC50 value of the 6 isolates with no mutations was 26 nm chloroquine. The IC50 of the 2 isolates with 4 mutations was 269 nm chloroquine, and 38 nm chloroquine in the 25 isolates with all 5 mutations. The Wilcoxon rank sum test was performed to determine if the number of polymorphisms had a significant association with IC50 value. Median chloroquine IC50 value was not significantly correlated between isolates that have wild-type sequence in all positions versus those with all 5 mutations (P=0.25). Similarly, there was no significant correlation of chloroquine IC50 value and the presence of zero versus 4 mutations (P=0.09). No additional mutations in coding regions were detected. A single patient was previously found to have more than 1 clone based on Msp-1 and Msp-2 typing and this isolate had both wild-type and mutant alleles at codon 76. Analysis of the downstream alleles showed that 1 isolate contained wild-type alleles at A220S, Q271E, N326S and R371I, while the second clone was wild-type in all 4 of these alleles. Finally, all 38 isolates encoded isoleucine at codon 356.
Fig. 2.
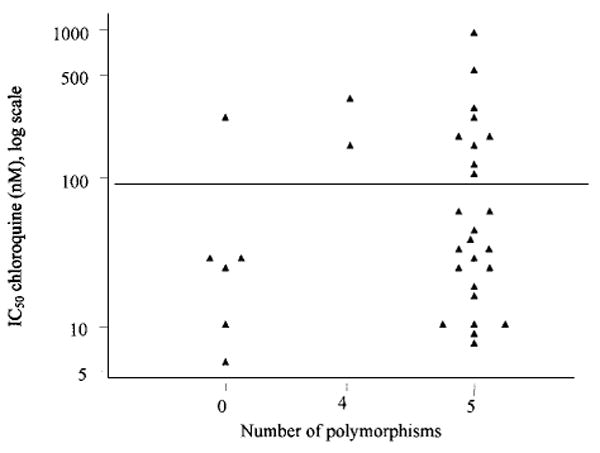
Dotplot of chloroquine IC50 versus of number of polymorphism in pfcrt codons K76T, A220S, Q271E, N326S and R371I from field isolates in Senegal.
Discussion
Mutations in pfcrt have been correlated with in vitro chloroquine resistance in different parts of the world. Yet field studies report isolates containing the pfcrt K76T mutation that are in vivo or in vitro sensitive. To test if additional polymorphisms reported in pfcrt are required for in vitro chloroquine resistance, these alleles were analysed in isolates from Senegal. The presence or absence of polymorphisms at codons K76T, A220S, Q271E, N326S and R371I were almost completely linked. Therefore, no correlation of chloroquine IC50 and these additional alleles could be established. This is in apparent contrast to a recent report of an allelic exchange of pfcrt containing these mutations into a chloroquine-sensitive line which conferred in vitro resistance. (Sidhu, Verdier-Pinard & Fidock, 2002). The field data generally support the central role of pfcrt in conferring chloroquine resistance, though there are a subset of isolates containing the pfcrt mutations which are chloroquine sensitive. This may reflect a genetically diverse origin of chloroquine resistance. Alternatively, these isolates may simply have not adapted to the short-term culture needed for the in vitro test and appear chloroquine sensitive. It will be important to confirm this interesting subset of isolates, chloroquine IC50 and polymorphisms. We have demonstrated that the occurrence of sequentially acquired mutations within pfcrt leading chloroquine resistance does not occur in this geographical area. Similarly, a study of 21 isolates from distinct regions in Southeast Asia, including PNG and Thailand, showed no correlation between polymorphisms at these codons and modulation of in vitro chloroquine resistance (Chen et al. 2001). Nevertheless, occurrence of these multiple polymorphisms is very intriguing. Previous studies on the prevalence of single nucleotide polymorphisms in malaria have determined that this is an infrequent occurrence, and when found it is typically in genes that are under selective pressure (Rich et al. 1998).
In this study it is striking to observe that the number of polymorphisms across these codons in pfcrt in both in vitro chloroquine-sensitive and resistant isolates was not distributed randomly; they were either completely wild-type or contained 4–5 polymorphisms in the same codons. This finding suggests that certain combinations of polymorphisms result in a selective disadvantage for the parasite and are therefore eliminated. This is an interesting consideration since the function of pfcrt remains unknown. Interestingly, patterns of polymorphisms in pfcrt have been correlated with geographical location. A study comparing in vitro chloroquine-resistant isolates between PNG, Thailand and Brazil found distinct patterns of pfcrt polymorphisms.
M74/N75/Q271/D326/L356/R371 were found in all isolates from PNG as well as the 7G8 strain from Brazil, whereas I74/E75/E271/S326/T(I) 356/I371 were found in all but 1 isolate from Thailand. From this genotyping the authors concluded that the PNG and Brazilian strains may have been derived from a common origin, whereas the Thailand strain arose independently (Chen et al. 2001). While these polymorphisms were not found in isolates analysed in this study, the pattern of polymorphisms in resistant isolates matches the pattern of codon polymorphisms described in another well-characterized African strain from Mali (T76/S220/E271/S326/I371). This includes the high prevalence of a wild-type allele encoding isoleucine at codon 356 (Djimde et al. 2001a). This is consistent with a recent report of 87 P. falciparum worldwide isolates that were found to have linkage disequilibrium at these amino acids in chloroquine-resistant strains (Wootton et al. 2002).
Due to the high prevalence of complete linkage of codons A220S, Q271E, N326S, R371I with K76T, it is concluded that further analysis of these additional polymorphisms in the T76 background will be unlikely to uncover further modulators of in vitro chloroquine resistance. These data suggest that in vitro resistance to chloroquine occurs within the genetic background of pfcrt K76T mutation but that a subset of isolates may require additional mutations in loci outside the pfcrt gene.
Finally, these data are consistent with previous analysis from other field sites suggesting that the polymorphisms in pfcrt can be used as surrogate markers of geographical origin, particularly to track the pattern of polymorphisms associated with chloroquine resistance.
References
- Basco LK, Ringwald P. Analysis of the key pfcrt point mutation and in vitro and in vivo response to chloroquine in Yaounde, Cameroon. Journal of Infectious Disease. 2001;183:28–31. doi: 10.1086/320726. [DOI] [PubMed] [Google Scholar]
- Chen N, Russell B, Staley J, Kotecka B, Nasveld P, Cheng Q. Sequence polymorphisms in pfcrt are strongly associated with chloroquine resistance in Plasmodium falciparum. Journal of Infectious Disease. 2001;183:1543–1545. doi: 10.1086/320206. [DOI] [PubMed] [Google Scholar]
- Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Dicko A, SU XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV, Coulibaly D. A molecular marker for chloroquine-resistant falciparum malaria. New England Journal of Medicine. 2001a;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- Djimde A, Doumbo OK, Steketee RW, Plowe CV. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet. 2001b;358:890–891. doi: 10.1016/S0140-6736(01)06040-8. [DOI] [PubMed] [Google Scholar]
- Dorsey G, Kamya MR, Singh A, Rosenthal PJ. Polymorphisms in the Plasmodium falciparum pfcrt and pfmdr-1 genes and clinical response to chloroquine in Kampala, Uganda. Journal of Infectious Disease. 2001;183:1417–1420. doi: 10.1086/319865. [DOI] [PubMed] [Google Scholar]
- Gaye O, Soumare M, Sambou B, Faye O, Dieng Y, Diouf M, Bah IB, Dieng T, N'Dir O, Diallo S. Heterogeneity of chloroquine resistant malaria in Senegal. Bulletin de la Société de Pathologie Exotique. 1999;92:149–152. [PubMed] [Google Scholar]
- Kyosiimire-Lugemwa J, Nalunkuma-Kazibwe AJ, Mujuzi G, Mulindwa H, Talisuna A, Egwang TG. The Lys-76-Thr mutation in PfCRT and chloroquine resistance in Plasmodium falciparum isolates from Uganda. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2002;96:91–95. doi: 10.1016/s0035-9203(02)90252-x. [DOI] [PubMed] [Google Scholar]
- Maguire JD, Susanti AI, Krisin, Sismadi P, Fryauff DJ, Baird JK. The T76 mutation in the pfcrt gene of Plasmodium falciparum and clinical chloroquine resistance phenotypes in Papua, Indonesia. Annals of Tropical Medicine and Parasitology. 2001;95:559–572. doi: 10.1080/00034980120092516. [DOI] [PubMed] [Google Scholar]
- Mayor AG, Gomez-Olive X, Aponte JJ, Casimiro S, Mabunda S, Dgedge M, Barreto A, Alonso PL. Prevalence of the K76T mutation in the putative Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene and its relation to chloroquine resistance in Mozambique. Journal of Infectious Disease. 2001;183:1413–1416. doi: 10.1086/319856. [DOI] [PubMed] [Google Scholar]
- Moreno A, Brasseur P, Cuzin-Ouattara N, Blanc C, Druilhe P. Evaluation under field conditions of the colourimetric DELI-microtest for the assessment of Plasmodium falciparum drug resistance. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2001;95:100–103. doi: 10.1016/s0035-9203(01)90351-7. [DOI] [PubMed] [Google Scholar]
- Pillai DR, Labbe AC, Vanisaveth V, Hongvangthong B, Pomphid AS, Inkathone S, Zhong K, Kain KC. Plasmodium falciparum malaria in Laos: chloroquine treatment outcome and predictive value of molecular markers. Journal of Infectious Disease. 2001;183:789–795. doi: 10.1086/318836. [DOI] [PubMed] [Google Scholar]
- Rich SM, Licht MC, Hudson RR, Ayala FJ. Malaria's Eve: evidence of a recent population bottleneck throughout the world populations of Plasmodium falciparum. Proceedings of the National Academy of Sciences, USA. 1998;95:4425–4430. doi: 10.1073/pnas.95.8.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SM, Ndir O, Dieng T, Mboup S, Wypij D, Maguire JH, Wirth DF. In vitro chloroquine susceptibility and PCR analysis of pfcrt and pfmdr1 polymorphisms in Plasmodium falciparum isolates from Senegal. American Journal of Tropical Medicine and Hygiene. 2002;66:474–480. doi: 10.4269/ajtmh.2002.66.474. [DOI] [PubMed] [Google Scholar]
- Vieira PP, Das Gracas Alecrim M, DA Silva LH, Gonzalez-Jimenez I, Zalis MG. Analysis of the PfCRT K76T mutation in Plasmodium falciparum isolates from the Amazon region of Brazil. Journal of Infectious Disease. 2001;183:1832–1833. doi: 10.1086/320739. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Feng X, Ferdig MT, Cooper RA, MU J, Baruch DI, Magill AJ, SU XZ. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature, London. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]


